Spectral-Spatial Hyperspectral Image Classification with Superpixel Pattern and Extreme Learning Machine
Abstract
1. Introduction
2. Related Work
2.1. Superpixel Segmentation Method
2.2. Principal Component Analysis
2.3. Extreme Learning Machine
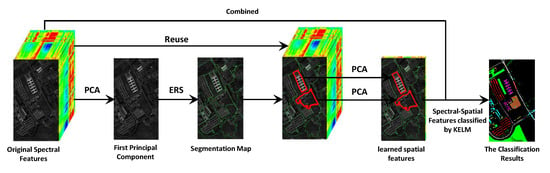
3. Proposed Spectral-Spatial Classification Model
| Algorithm 1 Pseudocode. for SP-KELM |
| Input: HSI cube ; Number of hidden nodes Q; Coefficient C; Number of superpixel segmentation ; Dimension of spatial features ; Output: The predicted labels for each tesing pixel in HSI cube;
|
3.1. Superpixel Based Spatial Features
3.2. Kernel Based Extreme Learning Machine
4. Experiments
4.1. Hyperspectral Datasets
4.2. Competing Methods and Experimental Setting
4.3. Experimental Comparison
4.4. Investigation on the Number of Superpixels
4.5. Investigation on the Dimension of Superpixel Patterns
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Zhang, L.; Zhang, L.; Du, B. Deep learning for remote sensing data: A technical tutorial on the state of the art. IEEE Geosci. Remote Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Ma, J.; Jiang, J.; Zhou, H.; Zhao, J.; Guo, X. Guided locality preserving feature matching for remote sensing image registration. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4435–4447. [Google Scholar] [CrossRef]
- Ma, J.; Zhao, J.; Jiang, J.; Zhou, H.; Guo, X. Locality preserving matching. Int. J. Comput. Vis. 2019, 127, 512–531. [Google Scholar] [CrossRef]
- Jiang, J.; Chen, C.; Yu, Y.; Jiang, X.; Ma, J. Spatial-aware collaborative representation for hyperspectral remote sensing image classification. IEEE Geosci. Remote Sens. Lett. 2017, 14, 404–408. [Google Scholar] [CrossRef]
- Jiang, X.; Song, X.; Zhang, Y.; Jiang, J.; Gao, J.; Cai, Z. Laplacian Regularized Spatial-Aware Collaborative Graph for Discriminant Analysis of Hyperspectral Imagery. Remote Sens. 2019, 11, 29. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef]
- Bazi, Y.; Alajlan, N.; Melgani, F.; Hichri, H.; Malek, S.; Yager, R.R. Differential evolution extreme learning machine for the classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1066–1070. [Google Scholar] [CrossRef]
- Fauvel, M.; Tarabalka, Y.; Benediktsson, J.A.; Chanussot, J.; Tilton, J.C. Advances in spectral-spatial classification of hyperspectral images. Proc. IEEE 2013, 101, 652–675. [Google Scholar] [CrossRef]
- Fan, H.; Chang, L.; Guo, Y.; Kuang, G.; Ma, J. Spatial-Spectral Total Variation Regularized Low-Rank Tensor Decomposition for Hyperspectral Image Denoising. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6196–6213. [Google Scholar] [CrossRef]
- Liu, T.; Gu, Y.; Jia, X.; Benediktsson, J.A.; Chanussot, J. Class-specific sparse multiple kernel learning for spectral–spatial hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2016, 54, 7351–7365. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Gomez-Chova, L.; Muñoz-Marí, J.; Vila-Francés, J.; Calpe-Maravilla, J. Composite kernels for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2006, 3, 93–97. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, J.; Chen, C.P. Extreme learning machine with composite kernels for hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2351–2360. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Su, H.; Du, Q. Local Binary Patterns and Extreme Learning Machine for Hyperspectral Imagery Classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 3681–3693. [Google Scholar] [CrossRef]
- Jain, A.K.; Ratha, N.K.; Lakshmanan, S. Object detection using Gabor filters. Pattern Recognit. 1997, 30, 295–309. [Google Scholar] [CrossRef]
- Chen, C.; Li, W.; Tramel, E.W.; Cui, M.; Prasad, S.; Fowler, J.E. Spectral-spatial preprocessing using multihypothesis prediction for noise-robust hyperspectral image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1047–1059. [Google Scholar] [CrossRef]
- Chen, C.; Li, W.; Su, H.; Liu, K. Spectral-spatial classification of hyperspectral image based on kernel extreme learning machine. Remote Sens. 2014, 6, 5795–5814. [Google Scholar] [CrossRef]
- Tarabalka, Y.; Fauvel, M.; Chanussot, J.; Benediktsson, J.A. SVM and MRF-based method for accurate classification of hyperspectral images. IEEE Geosci. Remote Sens. Lett. 2010, 7, 736–740. [Google Scholar] [CrossRef]
- Li, W.; Prasad, S.; Fowler, J.E. Hyperspectral image classification using Gaussian mixture models and Markov random fields. IEEE Geosci. Remote Sens. Lett. 2014, 11, 153–157. [Google Scholar] [CrossRef]
- Li, J.; Bioucas-Dias, J.M.; Plaza, A. Spectral-spatial hyperspectral image segmentation using subspace multinomial logistic regression and Markov random fields. IEEE Trans. Geosci. Remote Sens. 2012, 50, 809–823. [Google Scholar] [CrossRef]
- Li, S.; Lu, T.; Fang, L.; Jia, X.; Benediktsson, J.A. Probabilistic fusion of pixel-level and superpixel-level hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2016, 54, 7416–7430. [Google Scholar] [CrossRef]
- Priya, T.; Prasad, S.; Wu, H. Superpixels for Spatially Reinforced Bayesian Classification of Hyperspectral Images. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1071–1075. [Google Scholar] [CrossRef]
- Roscher, R.; Waske, B. Superpixel-based classification of hyperspectral data using sparse representation and conditional random fields. In Proceedings of the 2014 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Quebec City, QC, Canada, 13–18 July 2014; pp. 3674–3677. [Google Scholar]
- Zhan, T.; Sun, L.; Xu, Y.; Yang, G.; Zhang, Y.; Wu, Z. Hyperspectral Classification via Superpixel Kernel Learning-Based Low Rank Representation. Remote Sens. 2018, 10, 1639. [Google Scholar] [CrossRef]
- Sun, H.; Ren, J.; Zhao, H.; Zabalza, J.; Marshall, S. Superpixel based Feature Specific Sparse Representation for Spectral-Spatial Classification of Hyperspectral Images. Remote Sens. 2019, 11, 536. [Google Scholar] [CrossRef]
- Fang, L.; Li, S.; Duan, W.; Ren, J.; Benediktsson, J.A. Classification of hyperspectral images by exploiting spectral-spatial information of superpixel via multiple kernels. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6663–6674. [Google Scholar] [CrossRef]
- Fang, L.; Li, S.; Kang, X.; Benediktsson, J.A. Spectral-spatial classification of hyperspectral images with a superpixel-based discriminative sparse model. IEEE Trans. Geosci. Remote Sens. 2015, 53, 4186–4201. [Google Scholar] [CrossRef]
- Li, S.; Jia, X.; Zhang, B. Superpixel-based Markov random field for classification of hyperspectral images. In Proceedings of the 2013 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Melbourne, VIC, Australia, 21–26 July 2013; pp. 3491–3494. [Google Scholar]
- Zhang, S.; Li, S.; Fu, W.; Fang, L. Multiscale superpixel-based sparse representation for hyperspectral image classification. Remote Sens. 2017, 9, 139. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, J.; Chen, C.; Wang, Z.; Cai, Z.; Wang, L. SuperPCA: A superpixelwise PCA approach for unsupervised feature extraction of hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2018, 56, 4581–4593. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, J.; Wang, Z.; Chen, C.; Liu, X. Hyperspectral Image Classification in the Presence of Noisy Labels. IEEE Trans. Geosci. Remote Sens. 2019, 57, 851–865. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhou, H.; Ding, X.; Zhang, R. Extreme learning machine for regression and multiclass classification. IEEE Trans. Syst. Man Cybern. Part B (Cybern.) 2012, 42, 513–529. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Cai, Z.; Zhang, P.; Chen, L. Memetic extreme learning machine. Pattern Recognit. 2016, 58, 135–148. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, J.; Zhou, C.; Cai, Z. Instance cloned extreme learning machine. Pattern Recognit. 2017, 68, 52–65. [Google Scholar] [CrossRef]
- Jia, L.; Li, M.; Zhang, P.; Wu, Y. SAR image change detection based on correlation kernel and multistage extreme learning machine. IEEE Trans. Geosci. Remote Sens. 2016, 54, 5993–6006. [Google Scholar] [CrossRef]
- Tang, J.; Deng, C.; Huang, G.B.; Zhao, B. Compressed-domain ship detection on spaceborne optical image using deep neural network and extreme learning machine. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1174–1185. [Google Scholar] [CrossRef]
- Yang, Z.; Jie, L.; Min, H. Remote Sensing Image Transfer Classification Based on Weighted Extreme Learning Machine. IEEE Geosci. Remote Sens. Lett. 2016, 13, 1405–1409. [Google Scholar]
- Chen, C.; Zhang, B.; Su, H.; Li, W.; Wang, L. Land-use scene classification using multi-scale completed local binary patterns. Signal Image Video Process. 2016, 10, 745–752. [Google Scholar] [CrossRef]
- Leo, G. Random walks for image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2006, 28, 1768–1783. [Google Scholar]
- Shen, J.; Du, Y.; Wang, W.; Li, X. Lazy random walks for superpixel segmentation. IEEE Trans. Image Process. 2014, 23, 1451–1462. [Google Scholar] [CrossRef]
- Kaut, H.; Singh, R. A review on image segmentation techniques. Pattern Recognit. 1993, 26, 1277–1294. [Google Scholar]
- Felzenszwalb, P.F.; Huttenlocher, D.P. Efficient Graph-Based Image Segmentation. Int. J. Comput. Vis. 2004, 59, 167–181. [Google Scholar] [CrossRef]
- Verdoja, F.; Grangetto, M. Fast Superpixel-Based Hierarchical Approach to Image Segmentation. In Proceedings of the International Conference on Image Analysis and Processing (ICIAP), Genoa, Italy, 7–11 September 2015; pp. 364–374. [Google Scholar]
- Yan, Q.; Li, X.; Shi, J.; Jia, J. Hierarchical Saliency Detection. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Portland, OR, USA, 23–28 June 2013; pp. 1155–1162. [Google Scholar]
- Shi, J.; Jitendra, M. Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 888–905. [Google Scholar]
- Alex, L.; Adrian, S.; Kutulakos, K.N.; Fleet, D.J.; Dickinson, S.J.; Kaleem, S. TurboPixels: Fast superpixels using geometric flows. IEEE Trans. Pattern Anal. Mach. Intell. 2009, 31, 2290–2297. [Google Scholar]
- Liu, M.Y.; Tuzel, O.; Ramalingam, S.; Chellappa, R. Entropy rate superpixel segmentation. In Proceedings of the 2011 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Colorado Springs, CO, USA, 20–25 June 2011; pp. 2097–2104. [Google Scholar]
- Lunga, D.; Prasad, S.; Crawford, M.M.; Ersoy, O. Manifold-learning-based feature extraction for classification of hyperspectral data: A review of advances in manifold learning. IEEE Signal Process. Mag. 2014, 31, 55–66. [Google Scholar] [CrossRef]
- Zhou, Y.; Peng, J.; Chen, C.P. Dimension reduction using spatial and spectral regularized local discriminant embedding for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 1082–1095. [Google Scholar] [CrossRef]
- Bachmann, C.M.; Ainsworth, T.L.; Fusina, R.A. Exploiting manifold geometry in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2005, 43, 441–454. [Google Scholar] [CrossRef]
- He, X.; Cai, D.; Yan, S.; Zhang, H.J. Neighborhood preserving embedding. In Proceedings of the 10th IEEE International Conference on Computer Vision, Beijing, China, 17–21 October 2005; Volume 2, pp. 1208–1213. [Google Scholar]
- He, X.; Niyogi, P. Locality preserving projections. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2004; pp. 153–160. [Google Scholar]
- Schölkopf, B.; Smola, A.; Müller, K.R. Nonlinear component analysis as a kernel eigenvalue problem. Neural Comput. 1998, 10, 1299–1319. [Google Scholar] [CrossRef]
- Hossain, M.A.; Pickering, M.; Jia, X. Unsupervised feature extraction based on a mutual information measure for hyperspectral image classification. In Proceedings of the 2011 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Vancouver, BC, Canada, 24–29 July 2011; pp. 1720–1723. [Google Scholar]
- Liao, W.; Pizurica, A.; Philips, W.; Pi, Y. A fast iterative kernel PCA feature extraction for hyperspectral images. In Proceedings of the 17th IEEE International Conference on Image Processing (ICIP), Hong Kong, China, 26–29 September 2010; pp. 1317–1320. [Google Scholar]
- Laparra, V.; Malo, J.; Camps-Valls, G. Dimensionality reduction via regression in hyperspectral imagery. IEEE J. Sel. Top. Signal Process. 2015, 9, 1026–1036. [Google Scholar] [CrossRef]
- Prasad, S.; Bruce, L.M. Limitations of principal components analysis for hyperspectral target recognition. IEEE Geosci. Remote Sens. Lett. 2008, 5, 625–629. [Google Scholar] [CrossRef]
- Huang, G.B.; Zhu, Q.Y.; Siew, C.K. Extreme learning machine: Theory and applications. Neurocomputing 2006, 70, 489–501. [Google Scholar] [CrossRef]
- Zhang, R.; Lan, Y.; Huang, G.B.; Xu, Z.B. Universal approximation of extreme learning machine with adaptive growth of hidden nodes. IEEE Trans. Neural Netw. Learn. Syst. 2012, 23, 365. [Google Scholar] [CrossRef]
- Huang, G.; Huang, G.B.; Song, S.; You, K. Trends in extreme learning machines: A review. Neural Netw. 2015, 61, 32–48. [Google Scholar] [CrossRef]
- Xun, L.; Deng, C.; Wang, S.; Huang, G.B.; Zhao, B.; Lauren, P. Fast and Accurate Spatiotemporal Fusion Based Upon Extreme Learning Machine. IEEE Geosci. Remote Sens. Lett. 2017, 13, 2039–2043. [Google Scholar]
- Samat, A.; Du, P.; Liu, S.; Li, J.; Liang, C. E2LMs: Ensemble Extreme Learning Machines for Hyperspectral Image Classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1060–1069. [Google Scholar] [CrossRef]
- Samat, A.; Gamba, P.; Du, P.; Luo, J. Active extreme learning machines for quad-polarimetric SAR imagery classification. Int. J. Appl. Earth Obs. Geoinf. 2015, 35, 305–319. [Google Scholar] [CrossRef]
- Agarwal, A.; El-Ghazawi, T.; El-Askary, H.; Le-Moigne, J. Efficient hierarchical-PCA dimension reduction for hyperspectral imagery. In Proceedings of the 2007 IEEE International Symposium on Signal Processing and Information Technology, Giza, Egypt, 15–18 December 2007; pp. 353–356. [Google Scholar]








| Indian Pines | University of Pavia | Salinas Scene | |||
|---|---|---|---|---|---|
| Class Names | Numbers | Class Names | Numbers | Class Names | Numbers |
| 1. Alfalfa | 46 | 1. Asphalt | 6631 | 1. Broccoli green weeds 1 | 2009 |
| 2. Corn-notill | 1428 | 2. Bare soil | 18,649 | 2. Broccoli green weeds 2 | 3726 |
| 3. Corn-mintill | 830 | 3. Bitumen | 2099 | 3. Fallow | 1976 |
| 4. Corn | 237 | 4. Bricks | 3064 | 4. Fallow rough plow | 1394 |
| 5. Grass-pasture | 483 | 5. Gravel | 1345 | 5. Fallow smooth | 2678 |
| 6. Grass-trees | 730 | 6. Meadows | 5029 | 6. Stubble | 3959 |
| 7. Grass-pasture-mowed | 28 | 7. Metal sheets | 1330 | 7. Celery | 3579 |
| 8. Hay-windrowed | 478 | 8. Shadows | 3682 | 8. Grapes untrained | 11,271 |
| 9. Oats | 20 | 9. Trees | 947 | 9. Soil vineyard develop | 6203 |
| 10. Soybean-notill | 972 | 10. Corn senesced green weeds | 3278 | ||
| 11. Soybean-mintill | 2455 | 11. Lettuce romaine 4 wk | 1068 | ||
| 12. Soybean-clean | 593 | 12. Lettuce romaine 5 wk | 1927 | ||
| 13. Wheat | 205 | 13. Lettuce romaine 6 wk | 916 | ||
| 14. Woods | 1265 | 14. Lettuce romaine 7 wk | 1070 | ||
| 15. Buildings-Grass-Trees-Drives | 286 | 15. Vineyard untrained | 7268 | ||
| 16. Stone-Steel-Towers | 93 | 16. Vineyard vertical trellis | 1807 | ||
| Total Number | 10,249 | Total Number | 42,776 | Total Number | 54,129 |
| Class | #Samples | Spectral Approaches | Spectral-Spatial Approaches | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | SVM | ELM | KELM | PCA-KELM | CK-KELM | LBP-KELM | SP-KELM | |
| 1 | 23 | 23 | 93.04 | 86.52 | 92.61 | 93.04 | 99.57 | 100.00 | 100.00 |
| 2 | 30 | 1398 | 52.63 | 45.89 | 56.09 | 54.59 | 85.94 | 83.63 | 89.11 |
| 3 | 30 | 800 | 62.99 | 39.05 | 60.40 | 60.16 | 89.11 | 93.51 | 92.42 |
| 4 | 30 | 207 | 77.05 | 63.62 | 78.84 | 80.19 | 99.37 | 99.52 | 95.56 |
| 5 | 30 | 453 | 87.99 | 83.91 | 87.81 | 85.87 | 90.49 | 97.68 | 97.09 |
| 6 | 30 | 700 | 91.19 | 92.27 | 91.76 | 91.97 | 99.34 | 99.40 | 98.30 |
| 7 | 14 | 14 | 88.57 | 90.71 | 91.43 | 90.71 | 100.00 | 100.00 | 97.14 |
| 8 | 30 | 448 | 93.24 | 83.39 | 94.33 | 92.28 | 99.78 | 99.98 | 99.64 |
| 9 | 10 | 10 | 82.00 | 66.00 | 95.00 | 87.00 | 100.00 | 100.00 | 100.00 |
| 10 | 30 | 942 | 63.54 | 57.01 | 64.10 | 63.66 | 87.98 | 84.58 | 89.82 |
| 11 | 30 | 2425 | 52.88 | 46.29 | 54.41 | 53.30 | 82.39 | 82.29 | 90.18 |
| 12 | 30 | 563 | 61.51 | 60.02 | 73.11 | 69.57 | 93.02 | 84.30 | 92.50 |
| 13 | 30 | 175 | 97.43 | 99.26 | 98.40 | 98.46 | 99.89 | 100.00 | 99.43 |
| 14 | 30 | 1235 | 85.96 | 80.08 | 85.21 | 80.13 | 93.45 | 99.70 | 98.95 |
| 15 | 30 | 356 | 57.39 | 57.78 | 66.07 | 62.53 | 99.47 | 98.79 | 98.82 |
| 16 | 30 | 63 | 95.87 | 91.90 | 91.90 | 92.70 | 99.84 | 100.00 | 99.05 |
| OA (%) | 67.46 | 60.62 | 69.19 | 67.53 | 89.84 | 90.14 | 93.43 | ||
| AA (%) | 77.70 | 71.48 | 80.09 | 78.51 | 94.98 | 95.21 | 96.13 | ||
| Kappa | 0.6335 | 0.5576 | 0.6531 | 0.6353 | 0.8844 | 0.8878 | 0.9250 | ||
| Class | #Samples | Spectral Approaches | Spectral-Spatial Approaches | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | SVM | ELM | KELM | PCA-KELM | CK-KELM | LBP-KELM | SP-KELM | |
| 1 | 30 | 6601 | 69.47 | 37.37 | 62.56 | 68.94 | 83.83 | 81.39 | 83.29 |
| 2 | 30 | 18,619 | 72.76 | 71.24 | 71.67 | 76.07 | 93.31 | 86.43 | 90.12 |
| 3 | 30 | 2069 | 69.68 | 91.00 | 79.42 | 78.62 | 85.05 | 91.47 | 98.66 |
| 4 | 30 | 3034 | 93.19 | 94.11 | 92.99 | 93.88 | 95.85 | 97.25 | 92.27 |
| 5 | 30 | 1315 | 99.04 | 99.95 | 99.26 | 99.48 | 99.97 | 99.79 | 99.58 |
| 6 | 30 | 4999 | 69.21 | 64.50 | 71.59 | 76.02 | 93.35 | 96.03 | 94.11 |
| 7 | 30 | 1300 | 88.94 | 91.35 | 91.12 | 93.68 | 97.66 | 99.96 | 98.58 |
| 8 | 30 | 3652 | 78.00 | 26.69 | 67.90 | 77.95 | 86.69 | 98.41 | 99.00 |
| 9 | 30 | 917 | 98.66 | 90.84 | 96.04 | 99.81 | 90.46 | 99.95 | 93.82 |
| OA (%) | 75.46 | 65.88 | 73.79 | 78.29 | 91.33 | 89.94 | 91.49 | ||
| AA (%) | 82.11 | 74.12 | 81.40 | 84.94 | 91.80 | 94.52 | 94.38 | ||
| Kappa | 0.6879 | 0.5719 | 0.6685 | 0.7234 | 0.8865 | 0.8704 | 0.8892 | ||
| Class | #Samples | Spectral Approaches | Spectral-Spatial Approaches | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Train | Test | SVM | ELM | KELM | PCA-KELM | CK-KELM | LBP-KELM | SP-KELM | |
| 1 | 30 | 1979 | 98.71 | 99.68 | 99.52 | 99.69 | 99.45 | 100.00 | 100.00 |
| 2 | 30 | 3696 | 98.70 | 99.31 | 99.46 | 99.72 | 99.77 | 99.82 | 99.98 |
| 3 | 30 | 1946 | 94.29 | 91.96 | 92.02 | 99.18 | 98.75 | 99.92 | 99.67 |
| 4 | 30 | 1364 | 99.52 | 98.93 | 99.00 | 99.13 | 98.64 | 97.55 | 99.59 |
| 5 | 30 | 2648 | 96.36 | 98.65 | 97.67 | 97.16 | 99.23 | 98.49 | 99.24 |
| 6 | 30 | 3929 | 99.47 | 99.87 | 99.39 | 99.34 | 99.53 | 99.47 | 98.47 |
| 7 | 30 | 3549 | 99.33 | 99.43 | 99.49 | 99.39 | 98.14 | 99.83 | 98.11 |
| 8 | 30 | 11,241 | 67.03 | 75.70 | 75.68 | 73.27 | 84.93 | 89.25 | 94.70 |
| 9 | 30 | 6173 | 95.59 | 99.70 | 98.67 | 99.07 | 99.73 | 99.36 | 97.28 |
| 10 | 30 | 3248 | 91.74 | 90.90 | 92.76 | 92.16 | 96.15 | 98.24 | 97.67 |
| 11 | 30 | 1038 | 97.62 | 95.75 | 96.52 | 95.33 | 99.93 | 99.11 | 98.06 |
| 12 | 30 | 1897 | 99.86 | 83.11 | 99.98 | 100.00 | 99.98 | 97.64 | 97.75 |
| 13 | 30 | 886 | 98.01 | 97.96 | 97.98 | 97.78 | 99.50 | 96.65 | 98.21 |
| 14 | 30 | 1040 | 95.61 | 94.79 | 97.13 | 95.88 | 98.22 | 98.90 | 97.94 |
| 15 | 30 | 7238 | 72.67 | 56.44 | 68.19 | 71.18 | 84.10 | 86.97 | 99.43 |
| 16 | 30 | 1777 | 97.02 | 97.84 | 97.60 | 96.81 | 97.41 | 99.82 | 99.33 |
| OA (%) | 87.51 | 87.07 | 89.22 | 89.30 | 93.99 | 95.42 | 97.85 | ||
| AA (%) | 93.84 | 92.50 | 94.44 | 94.69 | 97.09 | 97.56 | 98.46 | ||
| Kappa | 0.8613 | 0.8559 | 0.8801 | 0.8811 | 0.9331 | 0.9490 | 0.9761 | ||
| Dataset | T.P.s/L.C | Spectral Approaches | Spectral-Spatial Approaches | |||||
|---|---|---|---|---|---|---|---|---|
| SVM | ELM | KELM | PCA-KELM | CK-KELM | LBP-KELM | SP-KELM | ||
| Indian Pines | 10 | 54.38 | 46.71 | 55.24 | 54.73 | 77.62 | 74.85 | 78.84 |
| 15 | 59.76 | 52.13 | 62.32 | 60.20 | 84.56 | 81.73 | 87.18 | |
| 20 | 63.00 | 54.45 | 64.81 | 63.11 | 86.15 | 86.09 | 90.53 | |
| 25 | 65.13 | 58.81 | 66.87 | 65.93 | 88.16 | 88.41 | 91.97 | |
| 30 | 67.46 | 60.62 | 69.19 | 67.53 | 89.84 | 90.14 | 93.43 | |
| University of Pavia | 10 | 64.38 | 61.53 | 66.06 | 71.79 | 77.02 | 72.24 | 80.26 |
| 15 | 66.71 | 63.45 | 68.46 | 74.37 | 81.71 | 79.39 | 84.94 | |
| 20 | 70.34 | 63.27 | 70.34 | 76.98 | 88.16 | 87.54 | 88.21 | |
| 25 | 75.16 | 63.33 | 72.34 | 78.48 | 88.83 | 88.66 | 89.99 | |
| 30 | 75.46 | 65.88 | 73.79 | 78.29 | 91.33 | 89.94 | 91.49 | |
| Salinas Scene | 10 | 84.33 | 84.78 | 85.82 | 85.62 | 90.73 | 89.29 | 88.58 |
| 15 | 85.56 | 86.04 | 87.96 | 86.75 | 92.32 | 91.56 | 92.81 | |
| 20 | 86.34 | 85.85 | 87.35 | 87.14 | 92.79 | 93.67 | 95.90 | |
| 25 | 87.97 | 87.56 | 89.23 | 89.30 | 93.81 | 94.66 | 97.49 | |
| 30 | 87.51 | 87.07 | 89.22 | 89.30 | 93.99 | 95.42 | 97.61 | |
| Dataset | CK-KELM | LBP-KELM | SP-KELM |
|---|---|---|---|
| Indian Pines | 200 | 1770 | 30 |
| University of Pavia | 103 | 1770 | 30 |
| Salinas Scene | 204 | 1770 | 30 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.; Jiang, X.; Wang, X.; Cai, Z. Spectral-Spatial Hyperspectral Image Classification with Superpixel Pattern and Extreme Learning Machine. Remote Sens. 2019, 11, 1983. https://doi.org/10.3390/rs11171983
Zhang Y, Jiang X, Wang X, Cai Z. Spectral-Spatial Hyperspectral Image Classification with Superpixel Pattern and Extreme Learning Machine. Remote Sensing. 2019; 11(17):1983. https://doi.org/10.3390/rs11171983
Chicago/Turabian StyleZhang, Yongshan, Xinwei Jiang, Xinxin Wang, and Zhihua Cai. 2019. "Spectral-Spatial Hyperspectral Image Classification with Superpixel Pattern and Extreme Learning Machine" Remote Sensing 11, no. 17: 1983. https://doi.org/10.3390/rs11171983
APA StyleZhang, Y., Jiang, X., Wang, X., & Cai, Z. (2019). Spectral-Spatial Hyperspectral Image Classification with Superpixel Pattern and Extreme Learning Machine. Remote Sensing, 11(17), 1983. https://doi.org/10.3390/rs11171983