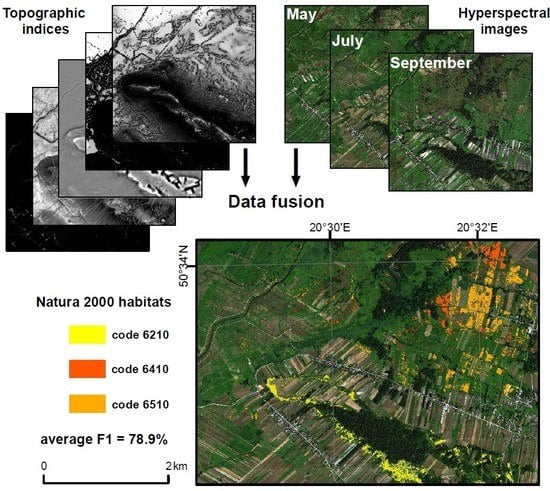
Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats
Abstract
:1. Introduction
2. Materials and Methods
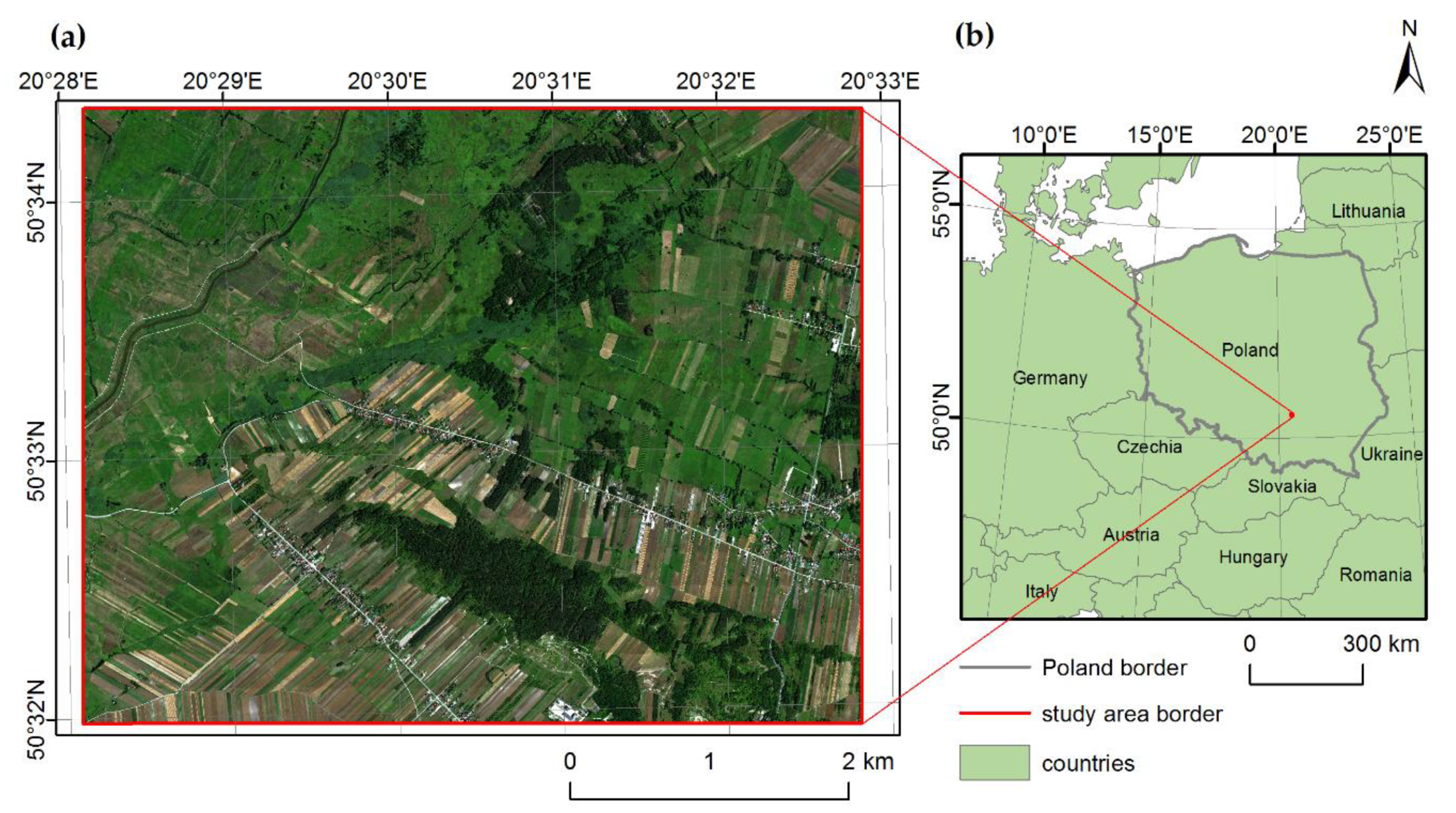
2.1. Study Area and Focus
2.2. Remote Sensing Data
2.2.1. Hyperspectral Data
2.2.2. Airborne Laser Scanning Data
2.2.3. Data Fusion
2.3. Reference Data
2.4. Classification and Iterative Accuracy Assessment
3. Results
3.1. Single Date Hyperspectral Data Classification
3.2. Hyperspectral and Topographic Data Fusion Classification
3.3. Multitemporal Hyperspectral Data Fusion Classification
3.4. Multitemporal Hyperspectral and Topographic Data Fusion Classification
3.5. Comparison of Results for All Datasets
4. Discussion
4.1. Optimal Term for Hyperspectral Data Acquisition
4.2. Hyperspectral and Topographic Data Fusion
4.3. Multitemporal Data Fusion
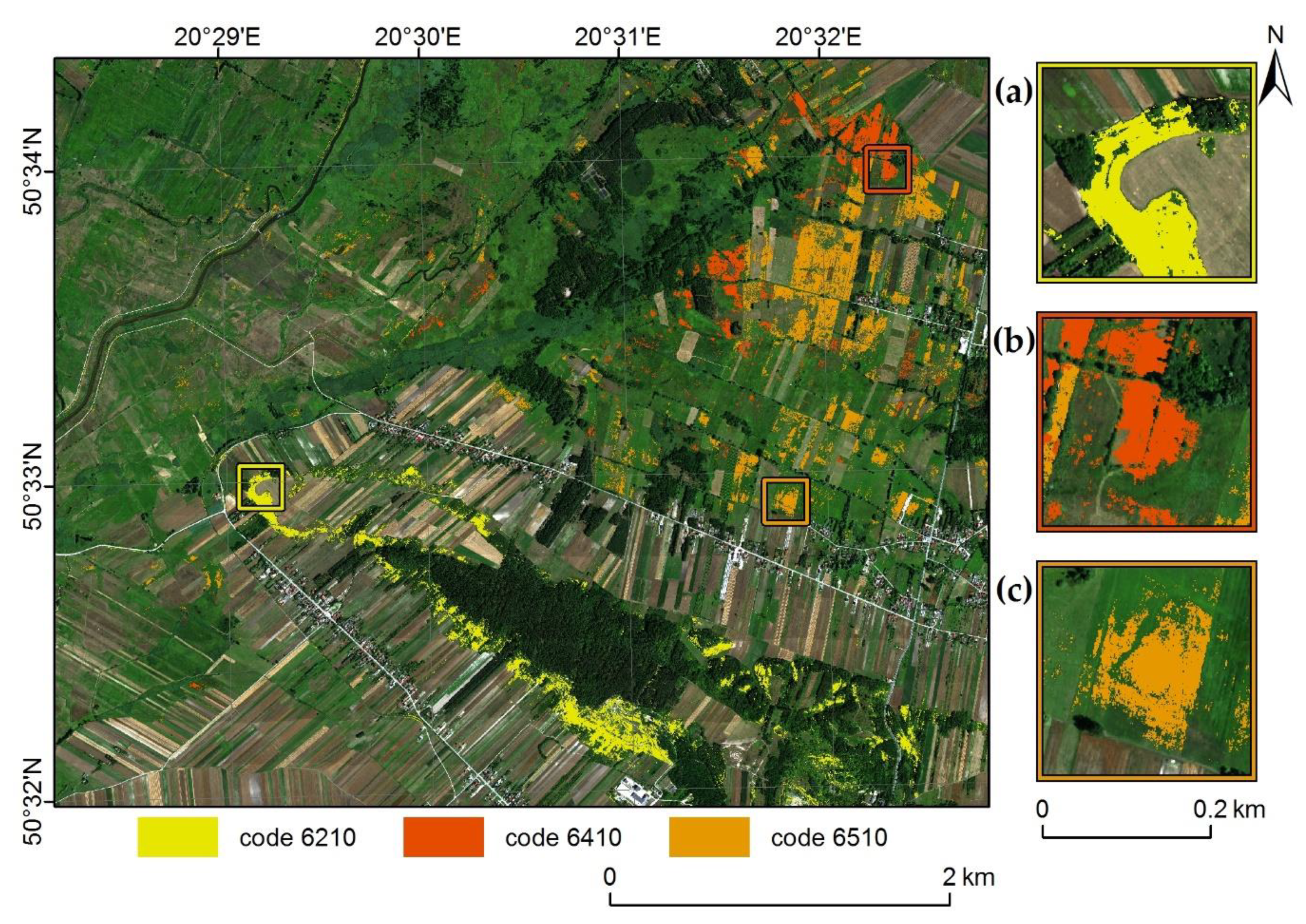
4.4. Habitat Classification Performance
5. Conclusions
- Summer and early autumn were indicated as the optimal times to obtain remote sensing data to map grassland Natura 2000 habitats. Autumn is the best time of year to identify habitat 6510, while for habitats 6210 and 6410, results were slightly better for summer than autumn. Spring was the worst period of the year to identify non-forest Natura 2000 habitats.
- The use of fused multitemporal hyperspectral data allowed higher classification accuracy to be achieved than the use of data from a single collection. The fusion of hyperspectral data obtained from spring, summer, and autumn always provided the best results, regardless of habitat type. However, the fusion of data from summer and autumn provided comparable results to the fusion of data from all terms, and from a practical point of view it could be used instead. The difference in F1 accuracy between the classification results achieved by the merger of two (summer and autumn) versus three terms, regardless of the Natura 2000 habitat, is no more than 2%.
- For semi-natural dry grasslands and scrubland facies on calcareous substrates (code 6210), whose presence is most strongly associated with appropriate terrain conditions, the classification of hyperspectral data fused with topographic indices was much more effective than the classification of single date hyperspectral data, and comparable to data fusion performed with all available data.
- For Molinia meadows (code 6410), the best classification accuracy was obtained using a fusion of three-term multitemporal hyperspectral data and topographic indices; however, multitemporal fusion of only hyperspectral data from three terms can be assessed as comparably suitable.
- For lowland hay meadows (code 6510), whose presence is not related to particular topographical conditions, it was confirmed that fusion of hyperspectral data and topographic indices did not influence classification results, both single-date and multitemporal. The highest accuracy was obtained using fused multitemporal hyperspectral data from all terms; however, fused summer and autumn data provided also satisfactory results.
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Turner, B.; Ii, B.T. Land Change Science. Int. Encycl. Hum. Geogr. 2009, 107–111. [Google Scholar] [CrossRef]
- European Comission Council. Directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora (OJ L 206 22.07.1992 p. 7). Doc. Eur. Community Environ. Law 2010, 568–583. [Google Scholar] [CrossRef]
- Dimopoulous, P.; Chytry, M.; Loidi Arregui, J.J.; Etlicher, B.; Mazagol, P.-O.; Sacca, C.; Just, A.; Debarros, G.; Millet, J.; Savia, L.; et al. The survey of mapping projects in European countries: A focus on mapping methodology. Terr. Habitat Mapp. Eur. Overv. 2014, 55–72. [Google Scholar] [CrossRef]
- Zlinszky, A.; Schroiff, A.; Kania, A.; Deák, B.; Mücke, W.; Vári, Á.; Székely, B.; Pfeifer, N. Categorizing grassland vegetation with full-waveform airborne laser scanning: A feasibility study for detecting natura 2000 habitat types. Remote Sens. 2014, 6, 8056–8087. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Raczko, E.; Ochtyra, A.; Jarocińska, A. Classification of High-Mountain Vegetation Communities within a Diverse Giant Mountains Ecosystem Using Airborne APEX Hyperspectral Imagery. Remote Sens. 2018, 10, 570. [Google Scholar] [CrossRef]
- Wald, L. Some terms of reference in data fusion. IEEE Trans. Geosci. Remote Sens. 1999, 37, 1190–1193. [Google Scholar] [CrossRef] [Green Version]
- Serpico, S.; Bruzzone, L. A new search algorithm for feature selection in hyperspectral remote sensing images. IEEE Trans. Geosci. Remote Sens. 2001, 39, 1360–1367. [Google Scholar] [CrossRef]
- Mücher, C.A.; Kooistra, L.; Vermeulen, M.; Haest, B.; Spanhove, T.; Delalieux, S.; Vanden Borre, J.; Schmidt, A.M. Object identification and characterization with hyperspectral imagery to identify structure and function of Natura 2000 habitats. In Proceedings of the GEOBIA 2010—The Geographic Object-Based Image Analysis Conference, Ghent, Belgium, 29 June–2 July 2010; Volume XXXVIII-4/C7, pp. 1–6. [Google Scholar]
- Onojeghuo, A.O.; Onojeghuo, A.R. Object-based habitat mapping using very high spatial resolution multispectral and hyperspectral imagery with LiDAR data. Int. J. Appl. Earth Obs. Geoinf. 2017, 59, 79–91. [Google Scholar] [CrossRef]
- Delalieux, S.; Somers, B.; Haest, B.; Spanhove, T.; Vanden Borre, J.; Mücher, C.A. Heathland conservation status mapping through integration of hyperspectral mixture analysis and decision tree classifiers. Remote Sens. Environ. 2012, 126, 222–231. [Google Scholar] [CrossRef]
- Hufkens, K.; Thoonen, G.; Borre, J.V.; Scheunders, P.; Ceulemans, R. Habitat reporting of a heathland site: Classification probabilities as additional information, a case study. Ecol. Inform. 2010, 5, 248–255. [Google Scholar] [CrossRef]
- Burai, P.; Deák, B.; Valkó, O.; Tomor, T. Classification of Herbaceous Vegetation Using Airborne Hyperspectral Imagery. Remote Sens. 2015, 7, 2046–2066. [Google Scholar] [CrossRef] [Green Version]
- Marcinkowska-Ochtyra, A.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Wojtuń, B.; Rogass, C.; Mielke, C.; Lavender, S. Subalpine and alpine vegetation classification based on hyperspectral APEX and simulated EnMAP images. Int. J. Remote Sens. 2017, 38, 1839–1864. [Google Scholar] [CrossRef] [Green Version]
- Jędrych, M.; Zagajewski, B.; Marcinkowska-Ochtyra, A. Application of Sentinel-2 and EnMAP new satellite data to the mapping of alpine vegetation of the Karkonosze Mountains. Pol. Cartogr. Rev. 2017, 49, 107–119. [Google Scholar] [CrossRef] [Green Version]
- Kupková, L.; Červená, L.; Suchá, R.; Jakešová, L.; Zagajewski, B.; Březina, S.; Albrechtová, J. Classification of tundra vegetation in the Krkonoše Mts. National park using APEX, AISA dual and sentinel-2A data. Eur. J. Remote Sens. 2017, 50, 29–46. [Google Scholar] [CrossRef]
- Feilhauer, H.; Dahlke, C.; Doktor, D.; Lausch, A.; Schmidtlein, S.; Schulz, G.; Stenzel, S. Mapping the local variability of Natura 2000 habitats with remote sensing. Appl. Veg. Sci. 2014, 17, 765–779. [Google Scholar] [CrossRef]
- Sławik, Ł.; Niedzielko, J.; Kania, A.; Piórkowski, H.; Kopeć, D. Multiple Flights or Single Flight Instrument Fusion of Hyperspectral and ALS Data? A Comparison of their Performance for Vegetation Mapping. Remote Sens. 2019, 11, 970. [Google Scholar] [CrossRef]
- Landmann, T.; Piiroinen, R.; Makori, D.M.; Abdel-Rahman, E.M.; Makau, S.; Pellikka, P.; Raina, S.K. Application of hyperspectral remote sensing for flower mapping in African savannas. Remote Sens. Environ. 2015, 166, 50–60. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Calpe-Maravilla, J.; Olivas, E.S.; Alonso-Chorda, L.; Gómez-Chova, L.; Martín-Guerrero, J.D.; Moreno, J. Robust support vector method for hyperspectral data classification and knowledge discovery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1530–1542. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Xie, Z. Object-based Vegetation Mapping in the Kissimmee River Watershed Using HyMap Data and Machine Learning Techniques. Wetlands 2013, 33, 233–244. [Google Scholar] [CrossRef]
- Chan, J.C.-W.; Beckers, P.; Spanhove, T.; Borre, J.V. An evaluation of ensemble classifiers for mapping Natura 2000 heathland in Belgium using spaceborne angular hyperspectral (CHRIS/Proba) imagery. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 13–22. [Google Scholar] [CrossRef]
- Corbane, C.; Alleaume, S.; Deshayes, M. Mapping natural habitats using remote sensing and sparse partial least square discriminant analysis. Int. J. Remote Sens. 2013, 34, 7625–7647. [Google Scholar] [CrossRef] [Green Version]
- Stenzel, S.; Feilhauer, H.; Mack, B.; Metz, A.; Schmidtlein, S. Remote sensing of scattered Natura 2000 habitats using a one-class classifier. Int. J. Appl. Earth Obs. Geoinf. 2014, 33, 211–217. [Google Scholar] [CrossRef]
- Marcinkowska, A.; Zagajewski, B.; Ochtyra, A.; Jarocińska, A.; Raczko, E.; Kupková, L.; Stych, P.; Meuleman, K. Mapping vegetation communities of the Karkonosze National Park using APEX hyperspectral data and Support Vector Machines. Misc. Geogr. 2014, 18, 23–29. [Google Scholar] [CrossRef] [Green Version]
- Kopeć, D.; Michalska-Hejduk, D.; Sławik, Ł.; Berezowski, T.; Borowski, M.; Rosadziński, S.; Chormański, J. Application of multisensoral remote sensing data in the mapping of alkaline fens Natura 2000 habitat. Ecol. Indic. 2016, 70, 196–208. [Google Scholar] [CrossRef]
- Zhang, C. Combining Hyperspectral and Lidar Data for Vegetation Mapping in the Florida Everglades. Photogramm. Eng. Remote Sens. 2014, 80, 733–743. [Google Scholar] [CrossRef] [Green Version]
- Zagajewski, B.; Tømmervik, H.; Bjerke, J.W.; Raczko, E.; Bochenek, Z.; Kłos, A.; Jarocińska, A.; Lavender, S.; Ziółkowski, D. Intraspecific Differences in Spectral Reflectance Curves as Indicators of Reduced Vitality in High-Arctic Plants. Remote Sens. 2017, 9, 1289. [Google Scholar] [CrossRef]
- Buck, O.; Millán, V.E.G.; Klink, A.; Pakzad, K. Using information layers for mapping grassland habitat distribution at local to regional scales. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 83–89. [Google Scholar] [CrossRef]
- Marcinkowska-Ochtyra, A.; Jarocińska, A.; Bzdęga, K.; Tokarska-Guzik, B. Classification of Expansive Grassland Species in Different Growth Stages Based on Hyperspectral and LiDAR Data. Remote Sens. 2018, 10, 2019. [Google Scholar] [CrossRef]
- Alcantara, C.; Kuemmerle, T.; Prishchepov, A.V.; Radeloff, V.C. Mapping abandoned agriculture with multi-temporal MODIS satellite data. Remote Sens. Environ. 2012, 124, 334–347. [Google Scholar] [CrossRef]
- Prishchepov, A.V.; Radeloff, V.C.; Dubinin, M.; Alcantara, C. The effect of Landsat ETM/ETM+ image acquisition dates on the detection of agricultural land abandonment in Eastern Europe. Remote Sens. Environ. 2012, 126, 195–209. [Google Scholar] [CrossRef]
- Rapinel, S.; Mony, C.; Lecoq, L.; Clément, B.; Thomas, A.; Hubert-Moy, L. Evaluation of Sentinel-2 time-series for mapping floodplain grassland plant communities. Remote Sens. Environ. 2019, 223, 115–129. [Google Scholar] [CrossRef]
- Schuster, C.; Schmidt, T.; Conrad, C.; Kleinschmit, B.; Forster, M. Grassland habitat mapping by intra-annual time series analysis—Comparison of RapidEye and TerraSAR-X satellite data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 25–34. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (randomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Govender, M.; Chetty, K.; Bulcock, H. A review of hyperspectral remote sensing and its application in vegetation and water resource studies. Water SA 2007, 33. [Google Scholar] [CrossRef]
- Pohl, C.; Van Genderen, J.L. Review article Multisensor image fusion in remote sensing: Concepts, methods and applications. Int. J. Remote Sens. 1998, 19, 823–854. [Google Scholar] [CrossRef] [Green Version]
- Kycko, M.; Zagajewski, B.; Lavender, S.; Romanowska, E.; Zwijacz-Kozica, M. The Impact of Tourist Traffic on the Condition and Cell Structures of Alpine Swards. Remote Sens. 2018, 10, 220. [Google Scholar] [CrossRef]
- Ghamisi, P.; Rasti, B.; Yokoya, N.; Wang, Q.; Hofle, B.; Bruzzone, L.; Bovolo, F.; Chi, M.; Anders, K.; Gloaguen, R.; et al. Multisource and Multitemporal Data Fusion in Remote Sensing: A Comprehensive Review of the State of the Art. IEEE Geosci. Remote Sens. Mag. 2019, 7, 6–39. [Google Scholar] [CrossRef] [Green Version]
- Yeadon, M.; King, M. A method for synchronising digitised video data. J. Biomech. 1999, 32, 983–986. [Google Scholar] [CrossRef]
- Ka̧cki, Z.; Michalska-Hejduk, D. Assessment of biodiversity in Molinia meadows in Kampinoski National Park based on biocenotic indicators. Pol. J. Environ. Stud. 2010, 19, 351–362. [Google Scholar]
- Nimbalkar, P.; Jarocinska, A.; Zagajewski, B. Optimal Band Configuration for the Roof Surface Characterization Using Hyperspectral and LiDAR Imaging. J. Spectrosc. 2018, 2018, 1–15. [Google Scholar] [CrossRef]
- Sabat-Tomala, A.; Jarocińska, A.M.; Zagajewski, B.; Magnuszewski, A.S.; Sławik, Ł.M.; Ochtyra, A.; Raczko, E.; Lechnio, J.R. Application of HySpex hyperspectral images for verification of a two-dimensional hydrodynamic model. Eur. J. Remote Sens. 2018, 51, 637–649. [Google Scholar] [CrossRef] [Green Version]
- Schläpfer, D.; Richter, R. Geo-atmospheric processing of airborne imaging spectrometry data. Part 1: Parametric orthorectification. Int. J. Remote Sens. 2002, 23, 2609–2630. [Google Scholar] [CrossRef]
- Richter, R.; Schläpfer, D. ATCOR-4 User Guide; German Aerospace Center: Cologne, Germany, 2016. [Google Scholar]
- Green, A.; Berman, M.; Switzer, P.; Craig, M. A transformation for ordering multispectral data in terms of image quality with implications for noise removal. IEEE Trans. Geosci. Remote Sens. 1988, 26, 65–74. [Google Scholar] [CrossRef] [Green Version]
- Conrad, O.; Bechtel, B.; Bock, M.; Dietrich, H.; Fischer, E.; Gerlitz, L.; Wehberg, J.; Wichmann, V.; Böhner, J. System for Automated Geoscientific Analyses (SAGA) v. 2.1.4. Geosci. Model Dev. 2015, 8, 1991–2007. [Google Scholar] [CrossRef] [Green Version]
- Dowling, T.I.; Gallant, J.C. A multiresolution index of valley bottom flatness for mapping depositional areas. Water Resour. Res. 2003, 39. [Google Scholar] [CrossRef]
- Guisan, A.; Weiss, S.B. GLM versus CCA spatial modeling of plant species distribution. Plant Ecol. 1999, 143, 107–122. [Google Scholar] [CrossRef]
- Weiss, A. Topographic position and landforms analysis. Poster Present. ESRI User Conf. San Diego, CA (2001). Available online: http://www.jennessent.com/downloads/TPI-poster-TNC_18x22.pdf (accessed on 28 September 2019).
- Beven, K.J.; Kirkby, M.J. A physically based, variable contributing area model of basin hydrology. Hydrol. Sci. Bull. 1979, 24, 43–69. [Google Scholar] [CrossRef]
- Moore, I.D.; Grayson, R.B.; Ladson, A.R. Digital terrain modelling: A review of hydrological, geomorphological, and biological applications. Hydrol. Process. 1991, 5, 3–30. [Google Scholar] [CrossRef]
- Shaw, R.L.; Booth, A.; Sutton, A.J.; Miller, T.; Smith, J.A.; Young, B.; Jones, D.R.; Dixon-Woods, M. Finding qualitative research: An evaluation of search strategies. BMC Med. Res. Methodol. 2004, 4, 5. [Google Scholar] [CrossRef]
- Böhner, J.; Köthe, R.; Conrad, O.; Gross, J.; Ringeler, A.; Selige, T. Soil regionalisation by means of terrain analysis and process parameterisation. Eur. Soil Bur. 2001, 7, 213–222. [Google Scholar]
- Liaw, A.; Wiener, M. Classification and Regression by randomForest. R News 2002, 2–3, 18–22. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2018. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Breiman, L. Prediction Games and Arcing Algorithms. Neural Comput. 1999, 11, 1493–1517. [Google Scholar] [CrossRef] [PubMed]
- Cutler, D.R.; Edwards, T.C.; Beard, K.H.; Cutler, A.; Hess, K.T.; Gibson, J.; Lawler, J.J. Random forests for classification in ecology. Ecology 2007, 88, 2783–2792. [Google Scholar] [CrossRef]
- Kim, J.-H. Estimating classification error rate: Repeated cross-validation, repeated hold-out and bootstrap. Comput. Stat. Data Anal. 2009, 53, 3735–3745. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Champagne, C.; McNairn, H.; Daneshfar, B.; Shang, J. A bootstrap method for assessing classification accuracy and confidence for agricultural land use mapping in Canada. Int. J. Appl. Earth Obs. Geoinf. 2014, 29, 44–52. [Google Scholar] [CrossRef] [Green Version]
- Stehman, S.V.; Czaplewski, R.L. Design and analysis for thematic map accuracy assessment: Fundamental principles. Remote Sens. Environ. 1998. [Google Scholar] [CrossRef]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Van Rijsbergen, C.J. Information Retrieval, 2nd ed.; Butterworths: London, UK, 1979. [Google Scholar]
- Potvin, C.; Roff, D.A. Distribution-Free and Robust Statistical Methods: Viable Alternatives to Parametric Statistics. Ecology 1993, 74, 1617–1628. [Google Scholar] [CrossRef]
- Maślanka, M. Factors influencing ground point density from Airborne Laser Scanning—A case study with ISOK Project data. Ann. Geomat. 2016, 14, 511–519. [Google Scholar]
- Szostak, M.; Wężyk, P.; Pająk, M.; Haryło, P.; Lisańczuk, M. Determination of the spatial structure of vegetation on the repository of the mine “Fryderyk” in Tarnowskie Góry, based on airborne laser scanning from the ISOK project and digital orthophotomaps. Geodesy Cartogr. 2015, 64, 87–99. [Google Scholar] [CrossRef]
- Raab, C.; Stroh, H.G.; Tonn, B.; Meißner, M.; Rohwer, N.; Balkenhol, N.; Isselstein, J. Mapping semi-natural grassland communities using multi-temporal RapidEye remote sensing data. Int. J. Remote Sens. 2018, 39, 5638–5659. [Google Scholar] [CrossRef]
- Haest, B.; Borre, J.V.; Spanhove, T.; Thoonen, G.; Delalieux, S.; Kooistra, L.; Mücher, C.A.; Paelinckx, D.; Scheunders, P.; Kempeneers, P. Habitat Mapping and Quality Assessment of NATURA 2000 Heathland Using Airborne Imaging Spectroscopy. Remote Sens. 2017, 9, 266. [Google Scholar] [CrossRef]
- Woodcock, C.E.; Strahler, A.H. The factor of scale in remote sensing. Remote Sens. Environ. 1987, 21, 311–332. [Google Scholar] [CrossRef]
- Mack, B.; Roscher, R.; Stenzel, S.; Feilhauer, H.; Schmidtlein, S.; Waske, B. Mapping raised bogs with an iterative one-class classification approach. ISPRS J. Photogramm. Remote Sens. 2016, 120, 53–64. [Google Scholar] [CrossRef]
- Kopeć, D.; Zakrzewska, A.; Halladin-Dąbrowska, A.; Wylazłowska, J.; Kania, A.; Niedzielko, J. Using Airborne Hyperspectral Imaging Spectroscopy to Accurately Monitor Invasive and Expansive Herb Plants: Limitations and Requirements of the Method. Sensors 2019, 19, 2871. [Google Scholar] [CrossRef]



| Application | Product | Acronym | Reference |
|---|---|---|---|
| Morphometric analysis | Multiresolution Index of Valley Bottom Flatness | MRVBF | [48] |
| Multiresolution Index of the Ridge Top Flatness | MRRTF | ||
| Topographic Position Index | TPI | [49,50] | |
| Hydrologic analysis | Topographic Wetness Index | TWI | [51,52,53] |
| Modified Catchment Area | MCA | [53,54] |
| Dataset Name | MNF | TOPO | ||
|---|---|---|---|---|
| Spring (C1) | Summer (C2) | Autumn (C3) | ||
| C1_MNF | X | |||
| C2_MNF | X | |||
| C3_MNF | X | |||
| C1_MNF+TOPO | X | X | ||
| C2_MNF+TOPO | X | X | ||
| C3_MNF+TOPO | X | X | ||
| C1_C2_MNF7 | X | X | ||
| C1_C3_MNF | X | X | ||
| C2_C3_MNF | X | X | ||
| C1_C2_C3_MNF | X | X | X | |
| C1_C2_MNF+TOPO | X | X | X | |
| C1_C3+TOPO | X | X | X | |
| C2_C3+TOPO | X | X | X | |
| C1_C2_C3+TOPO | X | X | X | X |
| Class | No. of Polygons | |||
|---|---|---|---|---|
| C1 | C2 | C3 | Combined into Multitemporal Set | |
| 6210 | 323 | 339 | 321 | 347 |
| 6410 | 262 | 229 | 232 | 292 |
| 6510 | 250 | 236 | 250 | 267 |
| background | 892 | 868 | 896 | 954 |
| Dataset | OA [%] | Median F1 [%] for Natura 2000 Habitat | Mean F1 for 3 Natura 2000 Habitats [%] | ||
|---|---|---|---|---|---|
| 6210 | 6410 | 6510 | |||
| C1_MNF | 70.6 | 74.0 | 68.9 | 51.7 | 64.9 |
| C2_MNF | 75.1 | 79.5 | 75.2 | 59.5 | 71.4 |
| C3_MNF | 75.5 | 78.3 | 75.0 | 61.1 | 71.5 |
| C1_MNF+TOPO | 74.1 | 79.5 | 73.4 | 55.8 | 69.6 |
| C2_MNF+TOPO | 76.9 | 82.3 | 77.7 | 60.5 | 73.5 |
| C3_MNF+TOPO | 77.6 | 83.1 | 79.6 | 61.6 | 74.8 |
| C1_C2_MNF | 78.6 | 81.6 | 80.1 | 65.5 | 75.7 |
| C2_C3_MNF | 80.0 | 82.9 | 80.8 | 68.0 | 77.2 |
| C1_C3_MNF | 78.5 | 81.4 | 80.1 | 64.3 | 75.3 |
| C1_C2_C3_MNF | 81.0 | 83.7 | 82.4 | 69.9 | 78.7 |
| C1_C2_MNF+TOPO | 79.5 | 83.7 | 80.9 | 65.8 | 76.8 |
| C2_C3_MNF+TOPO | 79.8 | 83.1 | 81.2 | 67.9 | 77.4 |
| C1_C3_MNF+TOPO | 79.4 | 83.6 | 82.0 | 64.8 | 76.8 |
| C1_C2_C3_MNF+TOPO | 81.1 | 84.5 | 83.2 | 68.9 | 78.9 |
| C1_MNF | abc | |||||||||||||
| C2_MNF | abc | b | a | |||||||||||
| C3_MNF | b | abc | c | c | ||||||||||
| C1_MNF+TOPO | a | abc | ||||||||||||
| C2_MNF+TOPO | c | abc | ||||||||||||
| C3_MNF+TOPO | c | abc | a | b | a | |||||||||
| C1_C2_MNF | abc | ab | c | |||||||||||
| C2_C3_MNF | a | abc | b | abc | ||||||||||
| C1_C3_MNF | b | ab | abc | c | ||||||||||
| C1_C2_C3_MNF | abc | a | a | c | ||||||||||
| C1_C2_MNF+TOPO | c | b | a | abc | b | a | ||||||||
| C2_C3_MNF+TOPO | a | abc | b | abc | ||||||||||
| C1_C3_MNF+TOPO | c | a | a | abc | ||||||||||
| C1_C2_C3_MNF+TOPO | c | abc | ||||||||||||
| C1_MNF | C2_MNF | C3_MNF | C1_MNF+TOPO | C2_MNF+TOPO | C3_MNF+TOPO | C1_C2_MNF | C2_C3_MNF | C1_C3_MNF | C1_C2_C3_MNF | C1_C2_MNF+TOPO | C2_C3_MNF+TOPO | C1_C3_MNF+TOPO | C1_C2_C3_MNF+TOPO |
| Class | Errors [%] | Accuracy [%] | |
|---|---|---|---|
| Commission | Omission | F1 | |
| 6210 | 18.9 | 13.3 | 83.8 |
| 6410 | 17.0 | 14.6 | 84.2 |
| 6510 | 23.9 | 23.9 | 76.1 |
| background | 15.6 | 18.2 | 83.1 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Marcinkowska-Ochtyra, A.; Gryguc, K.; Ochtyra, A.; Kopeć, D.; Jarocińska, A.; Sławik, Ł. Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats. Remote Sens. 2019, 11, 2264. https://doi.org/10.3390/rs11192264
Marcinkowska-Ochtyra A, Gryguc K, Ochtyra A, Kopeć D, Jarocińska A, Sławik Ł. Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats. Remote Sensing. 2019; 11(19):2264. https://doi.org/10.3390/rs11192264
Chicago/Turabian StyleMarcinkowska-Ochtyra, Adriana, Krzysztof Gryguc, Adrian Ochtyra, Dominik Kopeć, Anna Jarocińska, and Łukasz Sławik. 2019. "Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats" Remote Sensing 11, no. 19: 2264. https://doi.org/10.3390/rs11192264