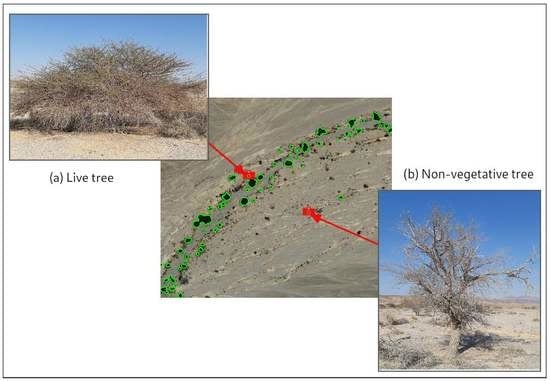
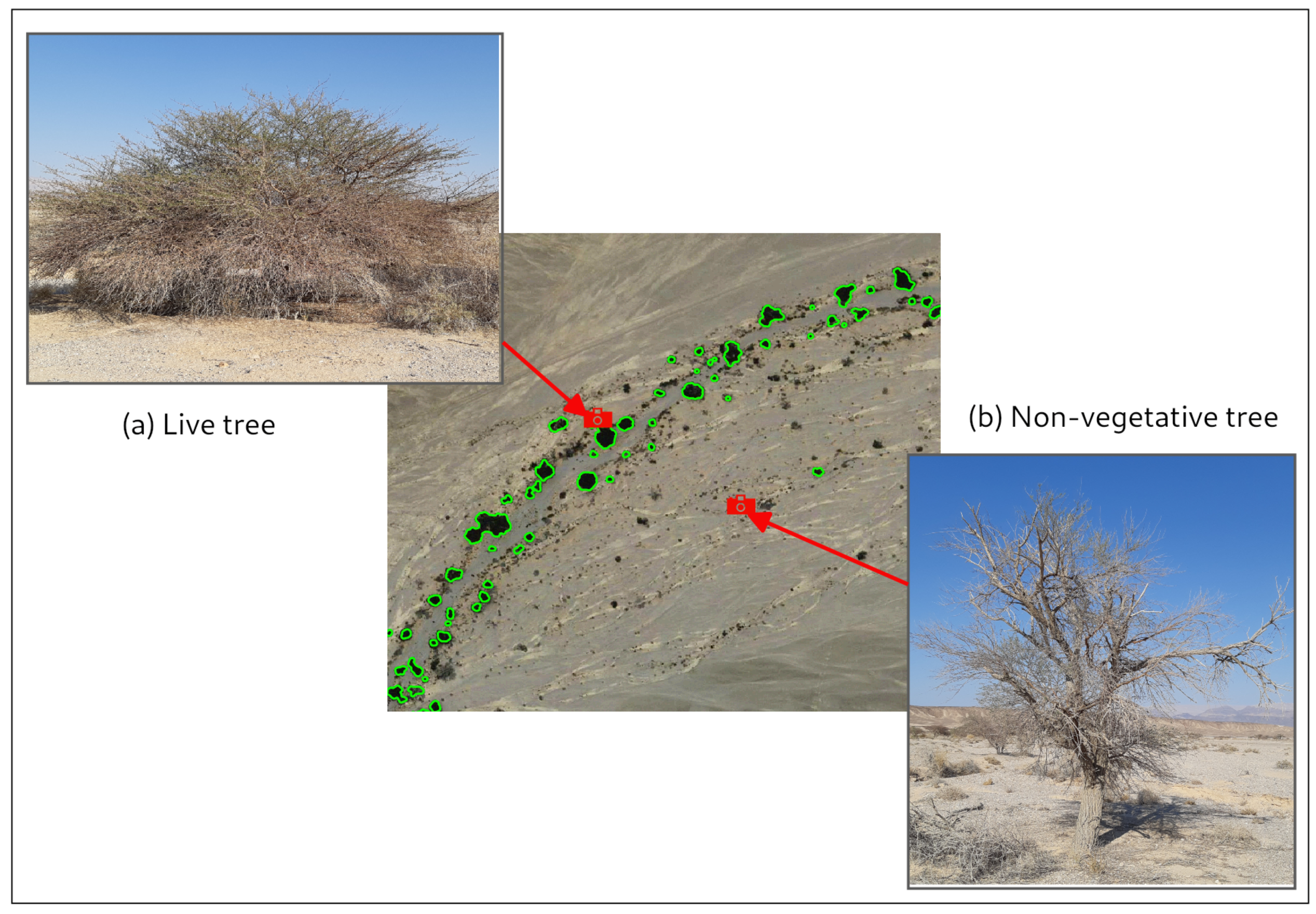
Identifying Vegetation in Arid Regions Using Object-Based Image Analysis with RGB-Only Aerial Imagery
Abstract
:1. Introduction
1.1. Vegetation Indices
1.2. Image Texture
1.3. OBIA Applied to Vegetation Classification
1.4. Segmentation and Classification
1.5. Objectives
2. Materials and Methods
2.1. Study Areas
Aerial Photographs
2.2. Preprocessing
2.2.1. Image Texture
2.2.2. Unsupervised Parameter Optimization
2.2.3. Superpixels
2.3. Segmentation and Classification
2.4. Post-Processing
2.5. Validation
2.6. Implementation
- a small, representative subset of the full study area for USPO;
- a layer of training points for supervised classification (Section 2.3);
- the true tree locations from monitoring campaigns;
- the validation zones as described above in Section 2.5.
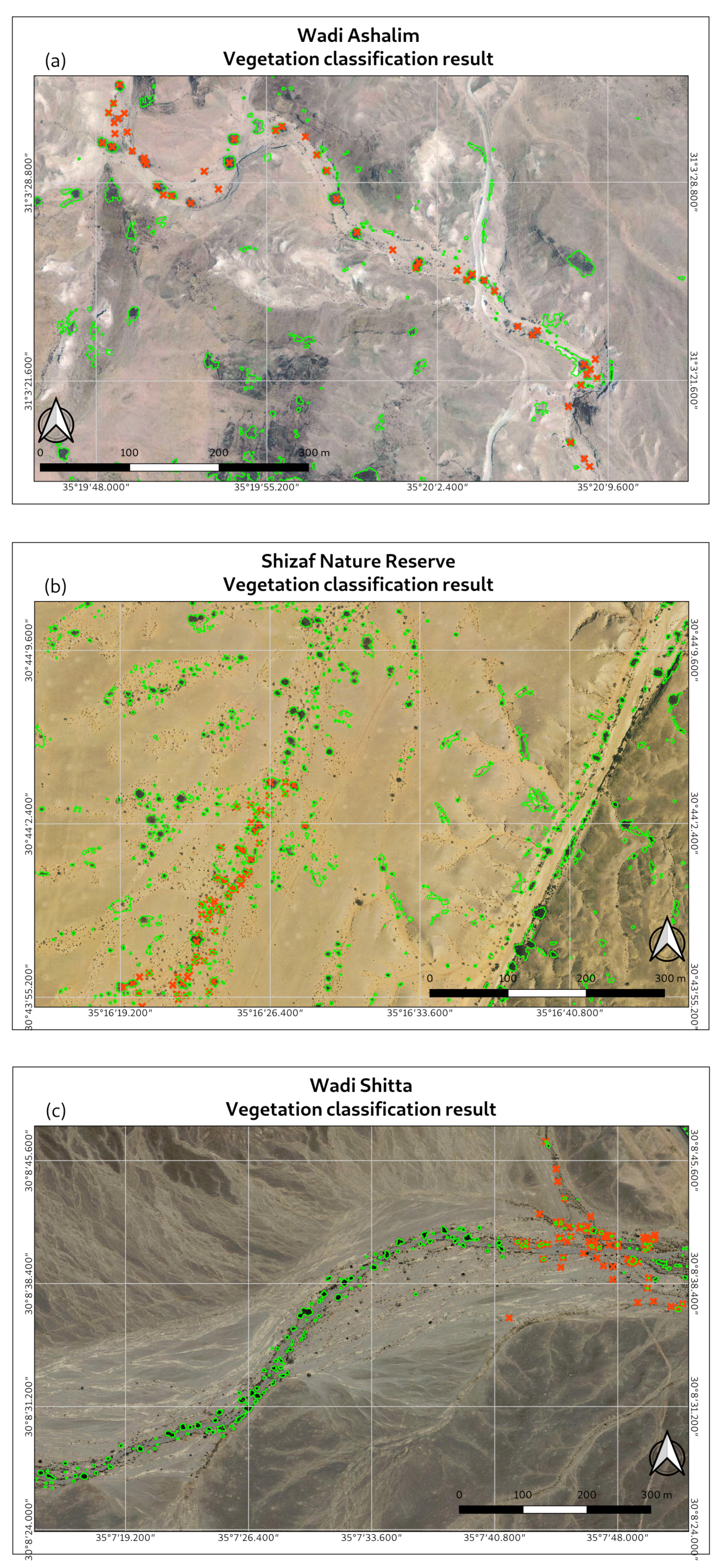
3. Results
- sections of aerial photographs with modeled vegetation and true tree locations;
- graphs showing receiver operating characteristic (ROC) curves;
- a table summarizing AUC values for all validation zones.
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| MDPI | Multidisciplinary Digital Publishing Institute |
| OBIA | object-based image analysis |
| GEOBIA | geographic object-based image analysis |
| NDVI | Normalized differential vegetation index |
| VI | Vegetation index |
| NIR | near infrared |
| LIDAR | light detection and ranging |
| GLCM | gray-level co-occurrence matrix |
| RF | Random forest |
| RGB | red, green, blue |
| SLIC | simple iterative linear clustering |
| TPR | true positive rate |
| FPR | false positive rate |
| ROC | receiver operating characteristic |
| AUC | area under the curve |
References
- Rouse, J.; Haas, R.; Schell, J.; Deering, D. Monitoring Vegetation Systems in the Great Plains with ERTS; Remote Sensing Center, Texas A&M University: College Station, TX, USA, 1974. [Google Scholar]
- Isaacson, S.; Rachmilevitch, S.; Ephrath, J.E.; Maman, S.; Blumberg, D.G. Monitoring tree population dynamics in arid zone through multiple temporal scales: Integration of spatial analysis change detection and field long term monitoring. ISPRS Int. Arch. Photogramm. Remote. Sens. Spat. Inf. Sci. 2016, XLI-B7, 513–515. [Google Scholar] [CrossRef]
- Wiegand, K.; Schmidt, H.; Jeltsch, F.; Ward, D. Linking a spatially-explicit model of acacias to GIS and remotely-sensed data. Folia Geobot. 2000, 35, 211–230. [Google Scholar] [CrossRef]
- Pham, T.D.; Yokoya, N.; Bui, D.T.; Yoshino, K.; Friess, D.A. Remote Sensing Approaches for Monitoring Mangrove Species, Structure, and Biomass: Opportunities and Challenges. Remote Sens. 2019, 11, 230. [Google Scholar] [CrossRef]
- Paz-Kagan, T.; Silver, M.; Panov, N.; Karnieli, A. Multispectral Approach for Identifying Invasive Plant Species Based on Flowering Phenology Characteristics. Remote Sens. 2019, 11, 953. [Google Scholar] [CrossRef]
- Hong, D.; Yokoya, N.; Chanussot, J.; Zhu, X.X. CoSpace: Common Subspace Learning From Hyperspectral-Multispectral Correspondences. IEEE Trans. Geosci. Remote Sens. 2019, 57, 4349–4359. [Google Scholar] [CrossRef] [Green Version]
- Hong, D.; Yokoya, N.; Chanussot, J.; Zhu, X.X. An Augmented Linear Mixing Model to Address Spectral Variability for Hyperspectral Unmixing. IEEE Trans. Image Process. 2019, 28, 1923–1938. [Google Scholar] [CrossRef] [PubMed]
- Shoshany, M.; Karnibad, L. Remote Sensing of Shrubland Drying in the South-East Mediterranean, 1995–2010: Water-Use-Efficiency-Based Mapping of Biomass Change. Remote Sens. 2015, 7, 2283–2301. [Google Scholar] [CrossRef]
- Karnieli, A.; Agam, N.; Pinker, R.T.; Anderson, M.; Imhoff, M.L.; Gutman, G.G.; Panov, N.; Goldberg, A. Use of NDVI and Land Surface Temperature for Drought Assessment: Merits and Limitations. J. Clim. 2010, 23, 618–633. [Google Scholar] [CrossRef]
- Mbow, C.; Fensholt, R.; Rasmussen, K.; Diop, D. Can vegetation productivity be derived from greenness in a semi-arid environment? Evidence from ground-based measurements. J. Arid Environ. 2013, 97, 56–65. [Google Scholar] [CrossRef]
- Théau, J.; Sankey, T.T.; Weber, K.T. Multi-sensor analyses of vegetation indices in a semi-arid environment. GISci. Remote Sens. 2010, 47, 260–275. [Google Scholar] [CrossRef]
- Peng, D.; Wu, C.; Li, C.; Zhang, X.; Liu, Z.; Ye, H.; Luo, S.; Liu, X.; Hu, Y.; Fang, B. Spring green-up phenology products derived from MODIS NDVI and EVI: Intercomparison, interpretation and validation using National Phenology Network and AmeriFlux observations. Ecol. Indic. 2017, 77, 323–336. [Google Scholar] [CrossRef]
- Huete, A.R. A soil-adjusted vegetation index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Qi, J.; Chehbouni, A.; Huete, A.; Kerr, Y.; Sorooshian, S. A Modified Soil Adjusted Vegetation Index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Huete, A.R.; Liu, H.Q.; Batchily, K.; Van Leeuwen, W. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Motohka, T.; Nasahara, K.N.; Oguma, H.; Tsuchida, S. Applicability of Green-Red Vegetation Index for Remote Sensing of Vegetation Phenology. Remote Sens. 2010, 2, 2369–2387. [Google Scholar] [CrossRef] [Green Version]
- McKinnon, T.; Huff, P. Comparing RGB-Based Vegetation Indices With NDVI For Drone Based Agricultural Sensing. Agribotix. Com. 2017, 1–8. [Google Scholar]
- Blaschke, T.; Hay, G.J.; Kelly, M.; Lang, S.; Hofmann, P.; Addink, E.; Queiroz Feitosa, R.; van der Meer, F.; van der Werff, H.; van Coillie, F.; et al. Geographic Object-Based Image Analysis—Towards a new paradigm. ISPRS J. Photogramm. Remote Sens. 2014, 87, 180–191. [Google Scholar] [CrossRef]
- Cheng, G.; Han, J. A survey on object detection in optical remote sensing images. ISPRS J. Photogramm. Remote Sens. 2016, 117, 11–28. [Google Scholar] [CrossRef] [Green Version]
- Myint, S.W.; Gober, P.; Brazel, A.; Grossman-Clarke, S.; Weng, Q. Per-pixel vs. object-based classification of urban land cover extraction using high spatial resolution imagery. Remote Sens. Environ. 2011, 115, 1145–1161. [Google Scholar] [CrossRef]
- Hussain, M.; Chen, D.; Cheng, A.; Wei, H.; Stanley, D. Change detection from remotely sensed images: From pixel-based to object-based approaches. ISPRS J. Photogramm. Remote Sens. 2013, 80, 91–106. [Google Scholar] [CrossRef]
- Feng, Q.; Liu, J.; Gong, J. UAV Remote Sensing for Urban Vegetation Mapping Using Random Forest and Texture Analysis. Remote Sens. 2015, 7, 1074–1094. [Google Scholar] [CrossRef] [Green Version]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Laws, K.I. Goal-Directed Textured-Image Segmentation. In Applications of Artificial Intelligence II; SPIE: Bellingham, WA, USA, 1985; Volume 548. [Google Scholar]
- Selvarajah, S.; Kodituwakku, S.R. Analysis and comparison of texture features for content based image retrieval. Int. J. Latest Trends Comput. 2011, 2, 108–113. [Google Scholar]
- Ruiz, L.A.; Fdez-Sarría, A.; Recio, J.A. Texture feature extraction for classification of remote sensing data using wavelet decomposition: A comparative study. In Proceedings of the 20th ISPRS Congress, Istanbul, Turkey, 12–23 July 2004; Volume 35, pp. 1109–1114. [Google Scholar]
- Marceau, D.J.; Howarth, P.J.; Dubois, J.M.M.; Gratton, D.J. Evaluation of the Grey-Level Co-Occurrence Matrix Method For Land-Cover Classification Using SPOT Imagery. IEEE Trans. Geosci. Remote Sens. 1990, 28, 513–519. [Google Scholar] [CrossRef]
- Maillard, P. Comparing Texture Analysis Methods through Classification. Photogramm. Eng. Remote Sens. 2003, 69, 357–367. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Gong, P.; Clinton, N.; Biging, G.; Kelly, M.; Schirokauer, D. Object-based detailed vegetation classification with airborne high spatial resolution remote sensing imagery. Photogramm. Eng. Remote Sens. 2006, 72, 799–811. [Google Scholar] [CrossRef]
- Lucas, R.; Rowlands, A.; Brown, A.; Keyworth, S.; Bunting, P. Rule-based classification of multi-temporal satellite imagery for habitat and agricultural land cover mapping. ISPRS J. Photogramm. Remote Sens. 2007, 62, 165–185. [Google Scholar] [CrossRef]
- Blaschke, T.; Lang, S.; Hay, G.J. (Eds.) Pixels to Objects to Information: Spatial Context to Aid in Forest Characterization with Remote Sensing. In Object-Based Image Analysis: Spatial Concepts for Knowledge-Driven Remote Sensing Applications; Springer: Berlin/Heidelberg, Germany, 2008; pp. 345–363. [Google Scholar] [CrossRef]
- Cleve, C.; Kelly, M.; Kearns, F.R.; Moritz, M. Classification of the wildland–urban interface: A comparison of pixel- and object-based classifications using high-resolution aerial photography. Comput. Environ. Urban Syst. 2008, 32, 317–326. [Google Scholar] [CrossRef]
- Moffett, K.B.; Gorelick, S.M. Distinguishing wetland vegetation and channel features with object-based image segmentation. Int. J. Remote Sens. 2013, 34, 1332–1354. [Google Scholar] [CrossRef]
- Karlson, M.; Reese, H.; Ostwald, M. Tree Crown Mapping in Managed Woodlands (Parklands) of Semi-Arid West Africa Using WorldView-2 Imagery and Geographic Object Based Image Analysis. Sensors 2014, 14, 22643–22669. [Google Scholar] [CrossRef]
- Juel, A.; Groom, G.B.; Svenning, J.C.; Ejrnæs, R. Spatial application of Random Forest models for fine-scale coastal vegetation classification using object based analysis of aerial orthophoto and DEM data. Int. J. Appl. Earth Obs. Geoinf. 2015, 42, 106–114. [Google Scholar] [CrossRef]
- Alsharrah, S.A.; Bruce, D.A.; Bouabid, R.; Somenahalli, S.; Corcoran, P.A. High-Spatial Resolution Multispectral and Panchromatic Satellite Imagery for Mapping Perennial Desert Plants. In Proceedings of the SPIE; SPIE: Bellingham, WA, USA, 2015; p. 96440Z. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Strager, M.P.; Warner, T.A.; Ramezan, C.A.; Morgan, A.N.; Pauley, C.E. Large-Area, High Spatial Resolution Land Cover Mapping Using Random Forests, GEOBIA, and NAIP Orthophotography: Findings and Recommendations. Remote Sens. 2019, 11, 1409. [Google Scholar] [CrossRef]
- Kim, M.; Madden, M.; Xu, B. GEOBIA Vegetation Mapping in Great Smoky Mountains National Park with Spectral and Non-spectral Ancillary Information. Photogramm. Eng. Remote Sens. 2010, 76, 137–149. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Bohlman, S.; Zare, A.; White, E. Individual Tree-Crown Detection in RGB Imagery Using Semi-Supervised Deep Learning Neural Networks. Remote Sens. 2019, 11, 1309. [Google Scholar] [CrossRef]
- Ozdemir, I.; Karnieli, A. Predicting forest structural parameters using the image texture derived from WorldView-2 multispectral imagery in a dryland forest, Israel. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 701–710. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; López-Granados, F.; Peña, J. An automatic object-based method for optimal thresholding in UAV images: Application for vegetation detection in herbaceous crops. Comput. Electron. Agric. 2015, 114, 43–52. [Google Scholar] [CrossRef]
- Espindola, G.M.; Camara, G.; Reis, I.A.; Bins, L.S.; Monteiro, A.M. Parameter selection for region-growing image segmentation algorithms using spatial autocorrelation. Int. J. Remote Sens. 2006, 27, 3035–3040. [Google Scholar] [CrossRef]
- Cánovas-García, F.; Alonso-Sarría, F. Optimal Combination of Classification Algorithms and Feature Ranking Methods for Object-Based Classification of Submeter Resolution Z/I-Imaging DMC Imagery. Remote Sens. 2015, 7, 4651–4677. [Google Scholar] [CrossRef] [Green Version]
- Malatesta, L.; Attorre, F.; Altobelli, A.; Adeeb, A.; De Sanctis, M.; Taleb, N.M.; Scholte, P.T.; Vitale, M. Vegetation mapping from high-resolution satellite images in the heterogeneous arid environments of Socotra Island (Yemen). J. Appl. Remote Sens. 2013, 7. [Google Scholar] [CrossRef]
- Rapinel, S.; Clément, B.; Magnanon, S.; Sellin, V.; Hubert-Moy, L. Identification and mapping of natural vegetation on a coastal site using a Worldview-2 satellite image. J. Environ. Manag. 2014, 144, 236–246. [Google Scholar] [CrossRef] [Green Version]
- Mboga, N.; Georganos, S.; Grippa, T.; Lennert, M.; Vanhuysse, S.; Wolff, E. Fully Convolutional Networks and Geographic Object-Based Image Analysis for the Classification of VHR Imagery. Remote Sens. 2019, 11, 597. [Google Scholar] [CrossRef]
- Li, M.; Ma, L.; Blaschke, T.; Cheng, L.; Tiede, D. A systematic comparison of different object-based classification techniques using high spatial resolution imagery in agricultural environments. Int. J. Appl. Earth Obs. Geoinf. 2016, 49, 87–98. [Google Scholar] [CrossRef]
- Grippa, T.; Lennert, M.; Beaumont, B.; Vanhuysse, S.; Stephenne, N.; Wolff, E. An Open-Source Semi-Automated Processing Chain for Urban Object-Based Classification. Remote Sens. 2017, 9, 358. [Google Scholar] [CrossRef]
- Biau, G. Analysis of a Random Forests Model. J. Mach. Learn. Res. 2012, 13, 1063–1095. [Google Scholar]
- Nicolas, G.; Robinson, T.P.; Wint, G.R.W.; Conchedda, G.; Cinardi, G.; Gilbert, M. Using Random Forest to Improve the Downscaling of Global Livestock Census Data. PLoS ONE 2016, 11, e0150424. [Google Scholar] [CrossRef]
- Cherlet, M.; Hutchinson, C.; Reynolds, J.; Hill, J.; Sommer, S.; von Maltitz, G. World Atlas of Desertification; Publications Office of the European Union: Luxembourg, 2018. [Google Scholar] [CrossRef]
- Lahav-Ginott, S.; Kadmon, R.; Gersani, M. Evaluating the viability of Acacia populations in the Negev Desert: A remote sensing approach. Biol. Conserv. 2001, 98, 127–137. [Google Scholar] [CrossRef]
- Ward, D.; Rohner, C. Anthropogenic Causes of high mortality and low recruitment in three Acacia tree taxa in the Negev desert, Israel. Biodivers. Conserv. 1997, 6, 877–893. [Google Scholar] [CrossRef]
- Johnson, B.A.; Bragais, M.; Endo, I.; Magcale-Macandog, D.B.; Macandog, P.B.M. Image Segmentation Parameter Optimization Considering Within- and Between-Segment Heterogeneity at Multiple Scale Levels: Test Case for Mapping Residential Areas Using Landsat Imagery. ISPRS Int. J. Geoinf. 2015, 4, 2292–2305. [Google Scholar] [CrossRef] [Green Version]
- Georganos, S.; Lennert, M.; Grippa, T.; Vanhuysse, S.; Johnson, B.; Wolff, E. Normalization in Unsupervised Segmentation Parameter Optimization: A Solution Based on Local Regression Trend Analysis. Remote Sens. 2018, 10, 222. [Google Scholar] [CrossRef]
- Ren, X.; Malik, J. Learning a classification model for segmentation. In Proceedings of the Ninth IEEE International Conference on Computer Vision, Nice, France, 13–16 October 2003; IEEE: Piscataway, NJ, USA, 2003; Volume 1, pp. 10–17. [Google Scholar] [CrossRef]
- Stutz, D.; Hermans, A.; Leibe, B. Superpixels: An evaluation of the state-of-the-art. Comput. Vis. Image Underst. 2018, 166, 1–27. [Google Scholar] [CrossRef] [Green Version]
- Achanta, R.; Shaji, A.; Smith, K.; Lucchi, A.; Fua, P.; Süsstrunk, S. SLIC Superpixels Compared to State-of-the-Art Superpixel Methods. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 34, 2274–2282. [Google Scholar] [CrossRef] [PubMed]
- Csillik, O. Fast Segmentation and Classification of Very High Resolution Remote Sensing Data Using SLIC Superpixels. Remote Sens. 2017, 9, 243. [Google Scholar] [CrossRef]
- Park, J.S.; Oh, S.J. A New Concave Hull Algorithm and Concaveness Measure for n-dimensional Datasets. J. Inf. Sci. Eng. 2012, 28, 14. [Google Scholar]
- R Development Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2008; ISBN 3-900051-07-0. [Google Scholar]
- Gombin, J.; Vaidyanathan, R.; Agafonkin, V. Concaveman: A Very Fast 2D Concave Hull Algorithm; R Package Version 1.0.0.; R Foundation for Statistical Computing: Vienna, Austria, 2017. [Google Scholar]
- GRASS Development Team. Geographic Resources Analysis Support System (GRASS GIS) Software, Version 7.2; Open Source Geospatial Foundation: Chicago, IL, USA, 2017. [Google Scholar]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Staben, G.W.; Lucieer, A.; Evans, K.G.; Scarth, P.; Cook, G.D. Obtaining biophysical measurements of woody vegetation from high resolution digital aerial photography in tropical and arid environments: Northern Territory, Australia. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 204–220. [Google Scholar] [CrossRef]
- Peters, A.J.; Eve, M.D.; Holt, E.H.; Whitford, W.G. Analysis of Desert Plant Community Growth Patterns with High Temporal Resolution Satellite Spectra. J. Appl. Ecol. 1997, 34, 418–432. [Google Scholar] [CrossRef]
- Joshi, P.K.K.; Roy, P.S.; Singh, S.; Agrawal, S.; Yadav, D. Vegetation cover mapping in India using multi-temporal IRS Wide Field Sensor (WiFS) data. Remote Sens. Environ. 2006, 103, 190–202. [Google Scholar] [CrossRef]
- Dorman, M.; Svoray, T.; Perevolotsky, A.; Sarris, D. Forest performance during two consecutive drought periods: Diverging long-term trends and short-term responses along a climatic gradient. For. Ecol. Manag. 2013, 310, 1–9. [Google Scholar] [CrossRef]
- Bajocco, S.; De Angelis, A.; Salvati, L. A satellite-based green index as a proxy for vegetation cover quality in a Mediterranean region. Ecol. Indic. 2012, 23, 578–587. [Google Scholar] [CrossRef]
- Fensholt, R.; Rasmussen, K.; Kaspersen, P.; Huber, S.; Horion, S.; Swinnen, E. Assessing Land Degradation/ Recovery in the African Sahel from Long-Term Earth Observation Based Primary Productivity and Precipitation Relationships. Remote Sens. 2013, 5, 664–686. [Google Scholar] [CrossRef]
- Zhang, G.; Biradar, C.M.; Xiao, X.; Dong, J.; Zhou, Y.; Qin, Y.; Zhang, Y.; Liu, F.; Ding, M.; Thomas, R.J. Exacerbated grassland degradation and desertification in Central Asia during 2000–2014. Ecol. Appl. 2018, 28, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Escobar-Flores, J.G.; Lopez-Sanchez, C.A.; Sandoval, S.; Marquez-Linares, M.A.; Wehenkel, C. Predicting Pinus monophylla forest cover in the Baja California Desert by remote sensing. PeerJ 2018, 6, e4603. [Google Scholar] [CrossRef] [PubMed]
- Moleele, N.; Ringrose, S.; Arnberg, W.; Lunden, B.; Vanderpost, C. Assessment of vegetation indexes useful for browse (forage) prediction in semi-arid rangelands. Int. J. Remote Sens. 2001, 22, 741–756. [Google Scholar] [CrossRef]






| Year Initialized | Shizaf | Shitta | Ashalim |
|---|---|---|---|
| 2007 | 2017 | 2012 | |
| Species | x | x | x |
| Number of trunks | x | ||
| Trunk circumference | x | x | |
| Age (est.) | x | ||
| Canopy height (est.) | x | x | x |
| Canopy area (est.) | x | x | |
| Canopy E–W | x | ||
| Canopy N–S | x | ||
| Mistletoe parasite (T/F) | x | ||
| Status (live/dead) | x | x | x |
| Monitoring date | x | x | |
| Continuous Monitoring (T/F) | x | ||
| Flowering | x |
| Study Area | Optimized Threshold |
|---|---|
| Ashalim | 0.11 |
| Shizaf | 0.13 |
| Shitta | 0.12 |
| Study Area | Validation Zone | AUC | Number of Trees |
|---|---|---|---|
| Ashalim | Wadi Amiaz | 0.818 | 62 |
| Ashalim | Wadi Ashalim | 0.749 | 85 |
| Ashalim | south | 0.850 | 66 |
| Shizaf | north | 0.712 | 134 |
| Shizaf | south | 0.731 | 159 |
| Shitta | east | 0.830 | 72 |
| Shitta | west | 0.730 | 82 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Silver, M.; Tiwari, A.; Karnieli, A. Identifying Vegetation in Arid Regions Using Object-Based Image Analysis with RGB-Only Aerial Imagery. Remote Sens. 2019, 11, 2308. https://doi.org/10.3390/rs11192308
Silver M, Tiwari A, Karnieli A. Identifying Vegetation in Arid Regions Using Object-Based Image Analysis with RGB-Only Aerial Imagery. Remote Sensing. 2019; 11(19):2308. https://doi.org/10.3390/rs11192308
Chicago/Turabian StyleSilver, Micha, Arti Tiwari, and Arnon Karnieli. 2019. "Identifying Vegetation in Arid Regions Using Object-Based Image Analysis with RGB-Only Aerial Imagery" Remote Sensing 11, no. 19: 2308. https://doi.org/10.3390/rs11192308
APA StyleSilver, M., Tiwari, A., & Karnieli, A. (2019). Identifying Vegetation in Arid Regions Using Object-Based Image Analysis with RGB-Only Aerial Imagery. Remote Sensing, 11(19), 2308. https://doi.org/10.3390/rs11192308