A Novel Hyperspectral Image Simulation Method Based on Nonnegative Matrix Factorization
Abstract
1. Introduction
2. Materials and Methods
2.1. Datasets
2.2. Methods
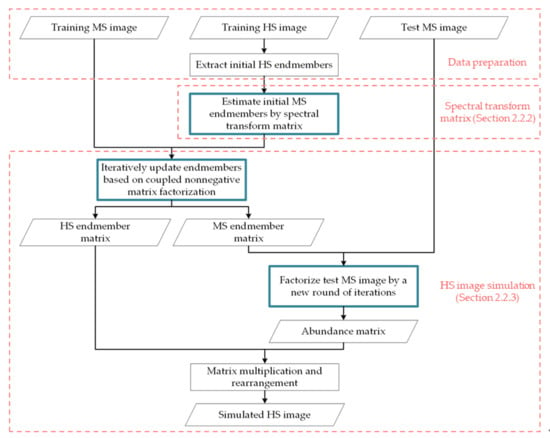
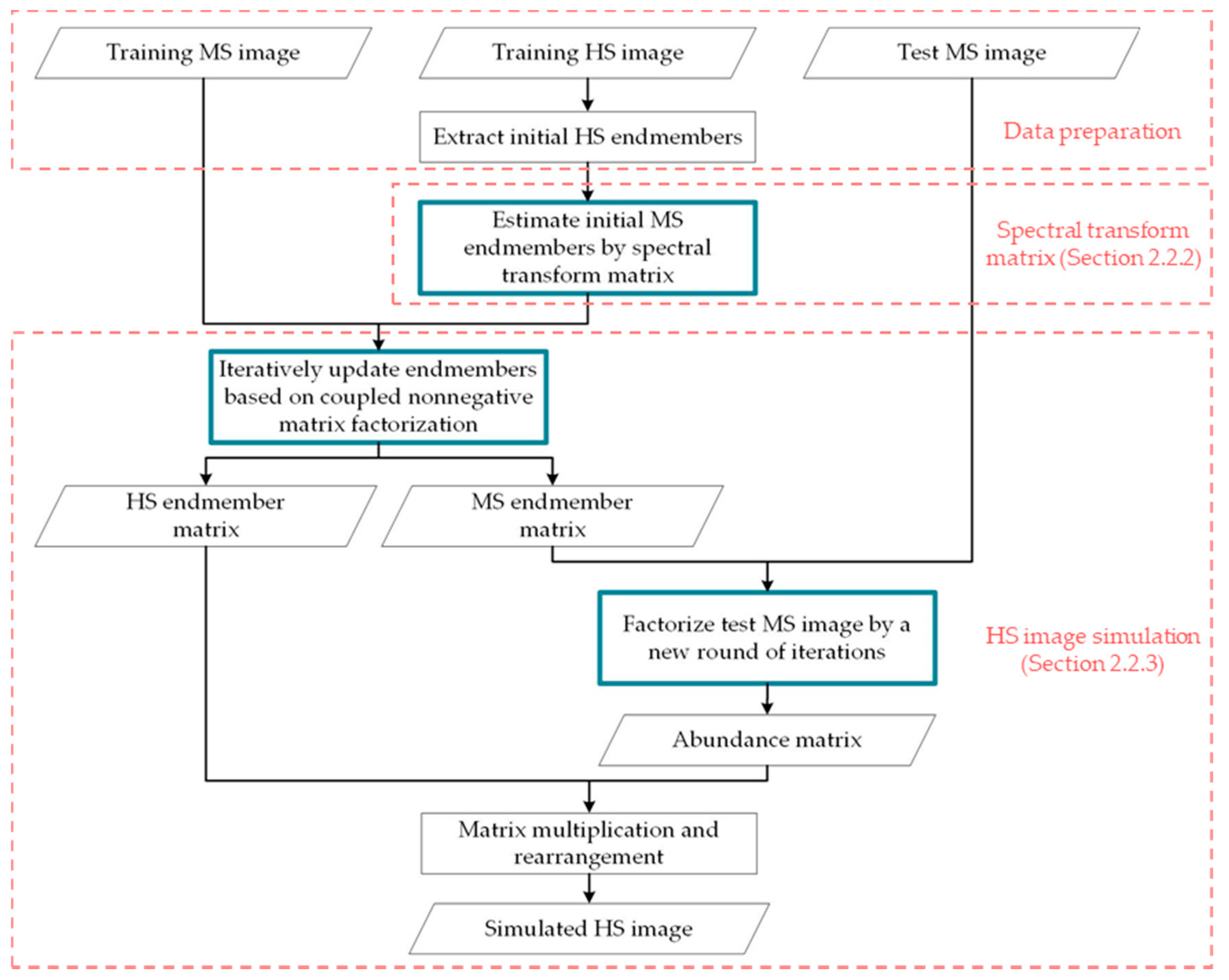
2.2.1. Nonnegative Matrix Factorization of Images
2.2.2. Estimation of Spectral Transformation Matrix
2.2.3. HS Image Simulation and Iterative Calculation Scheme
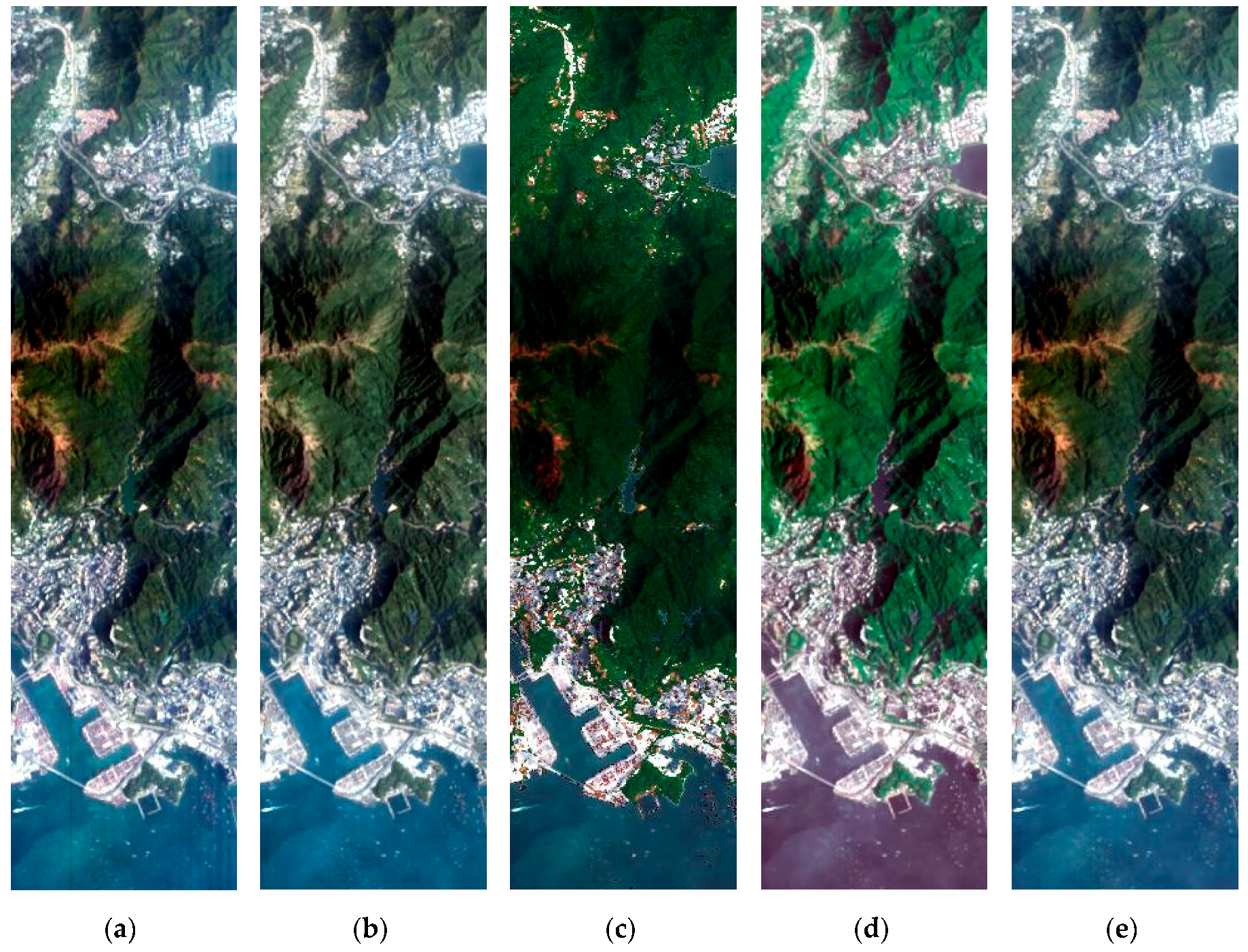
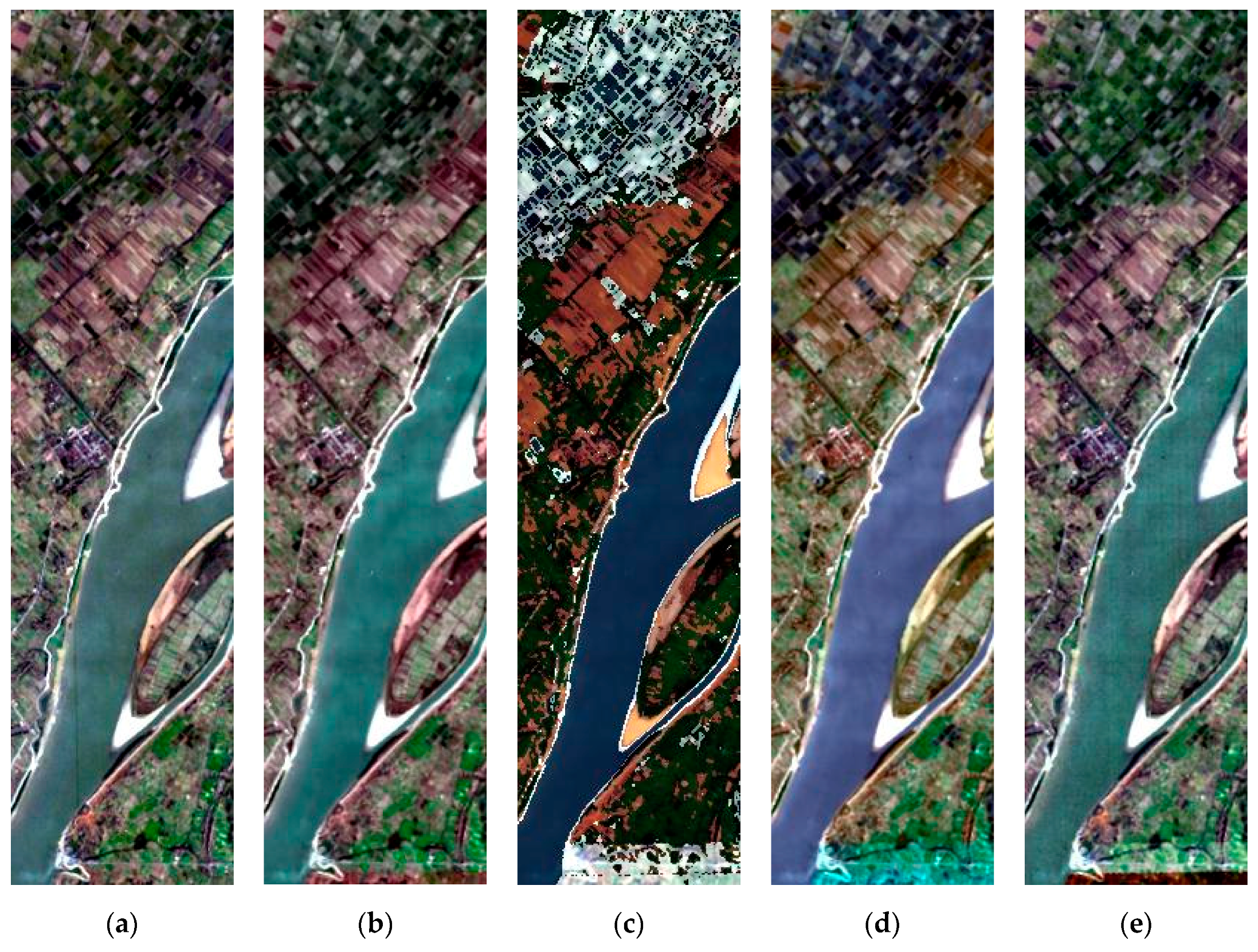
3. Results
3.1. Evaluation and Comparative Methods
3.2. Experimental Results
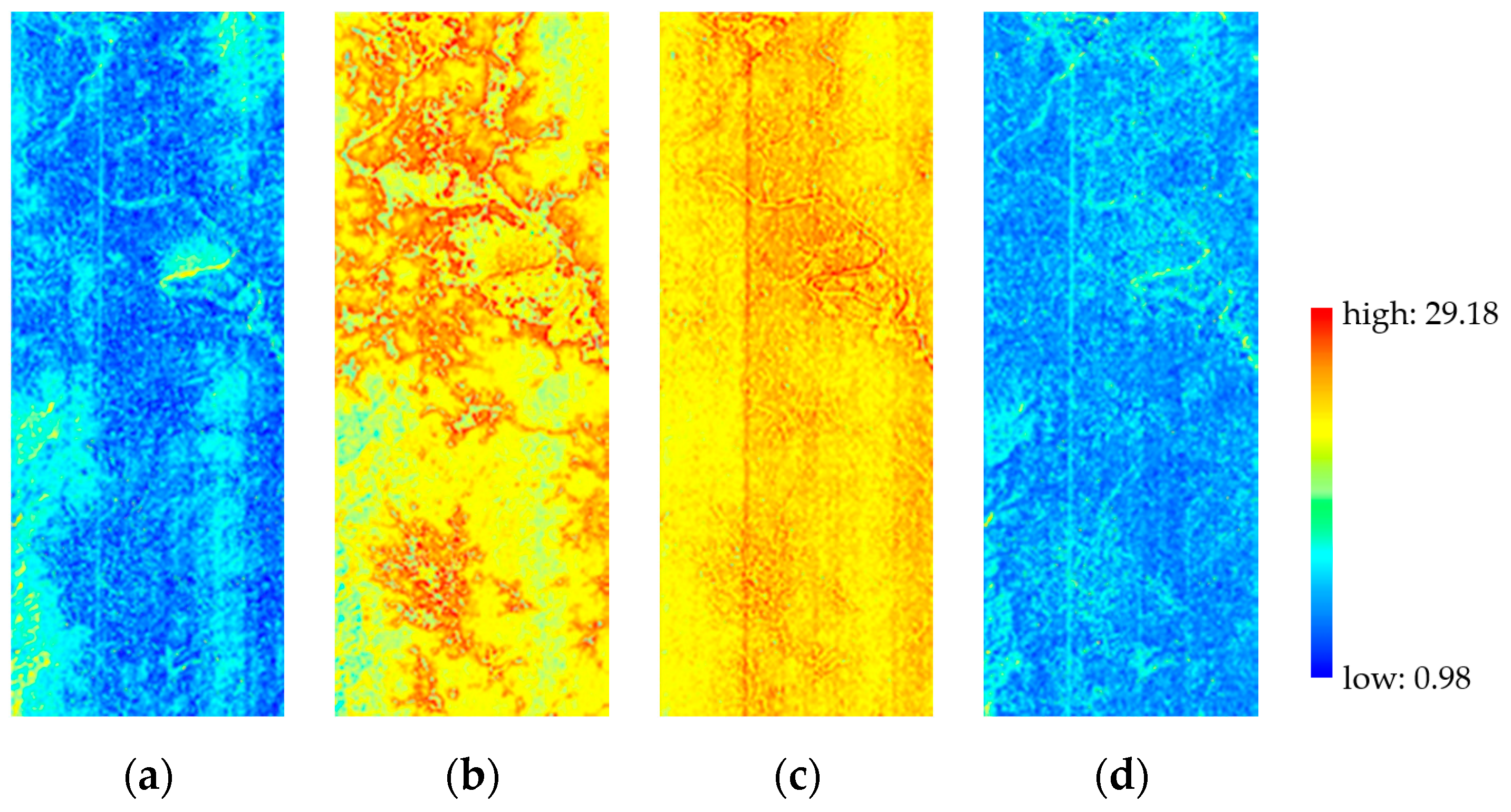
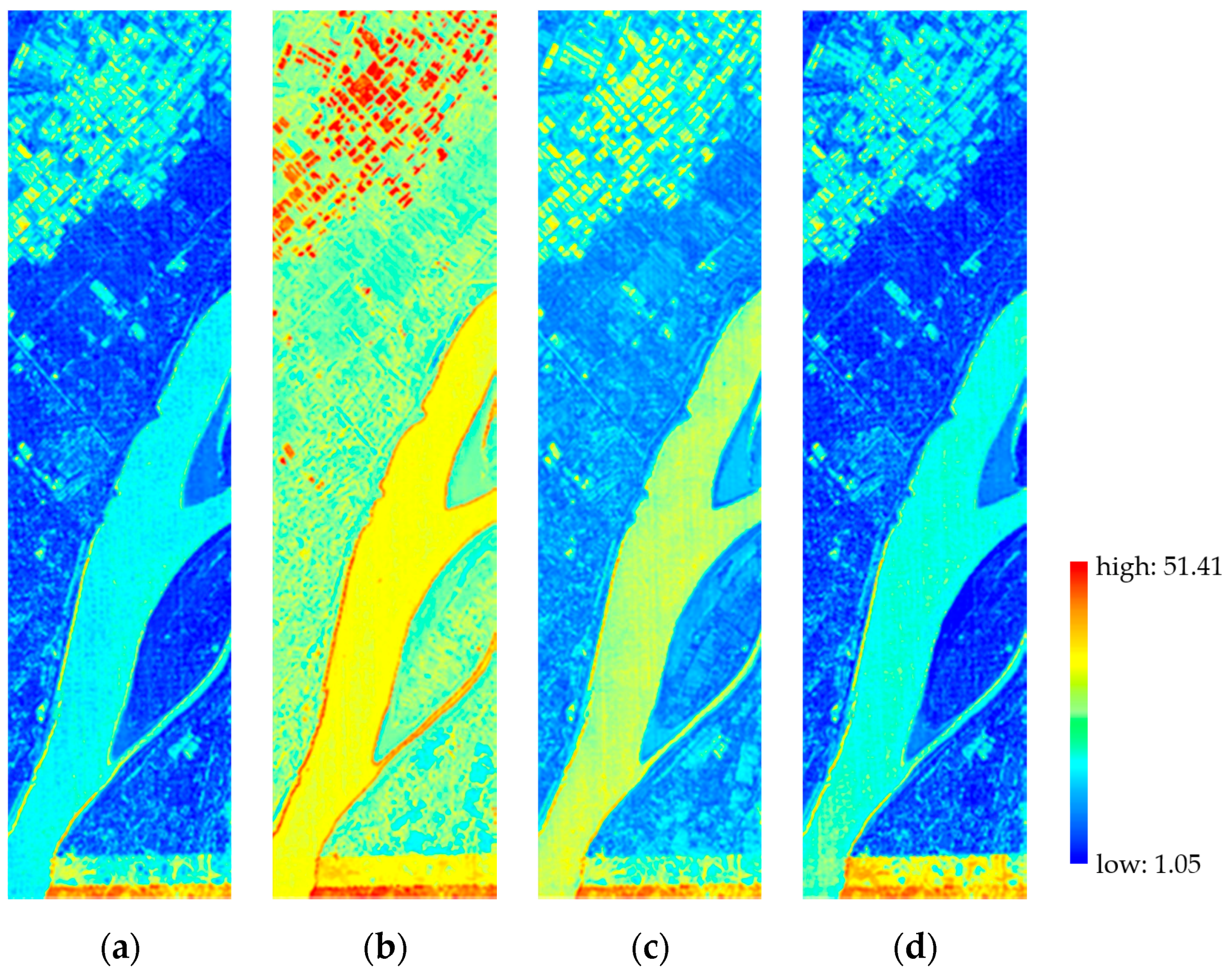
3.2.1. Spatial Performance
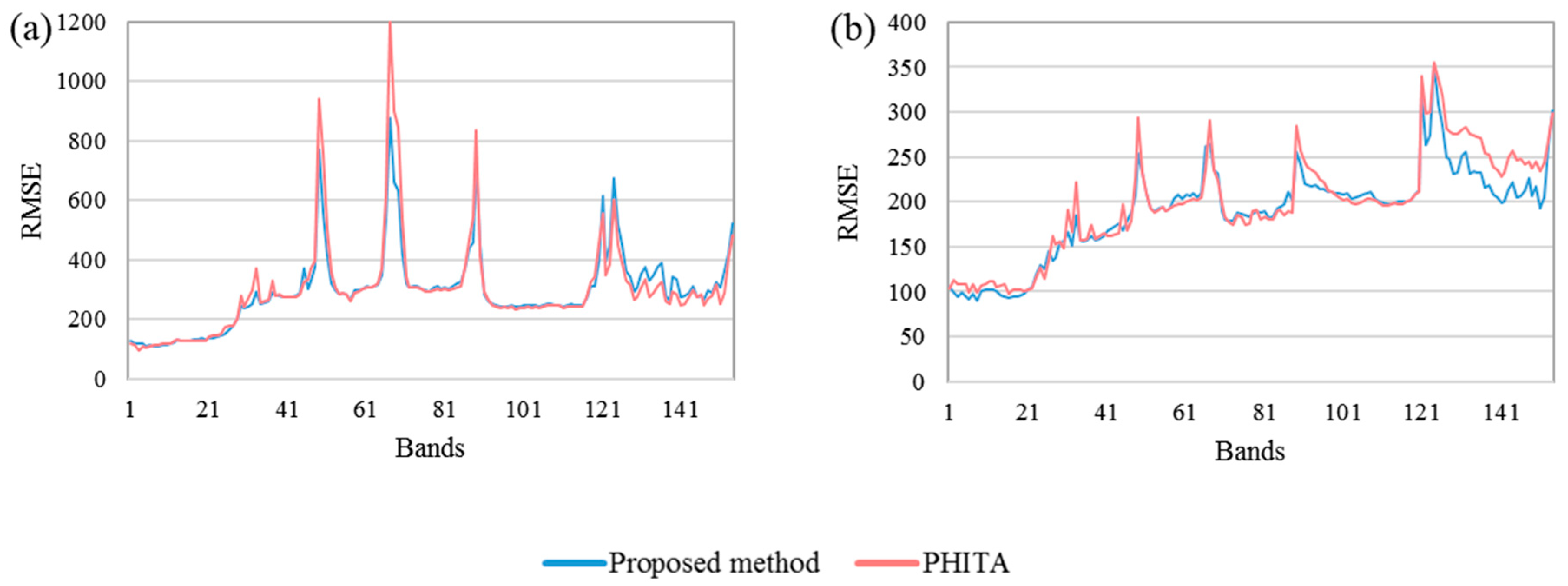
3.2.2. Spectral Performance
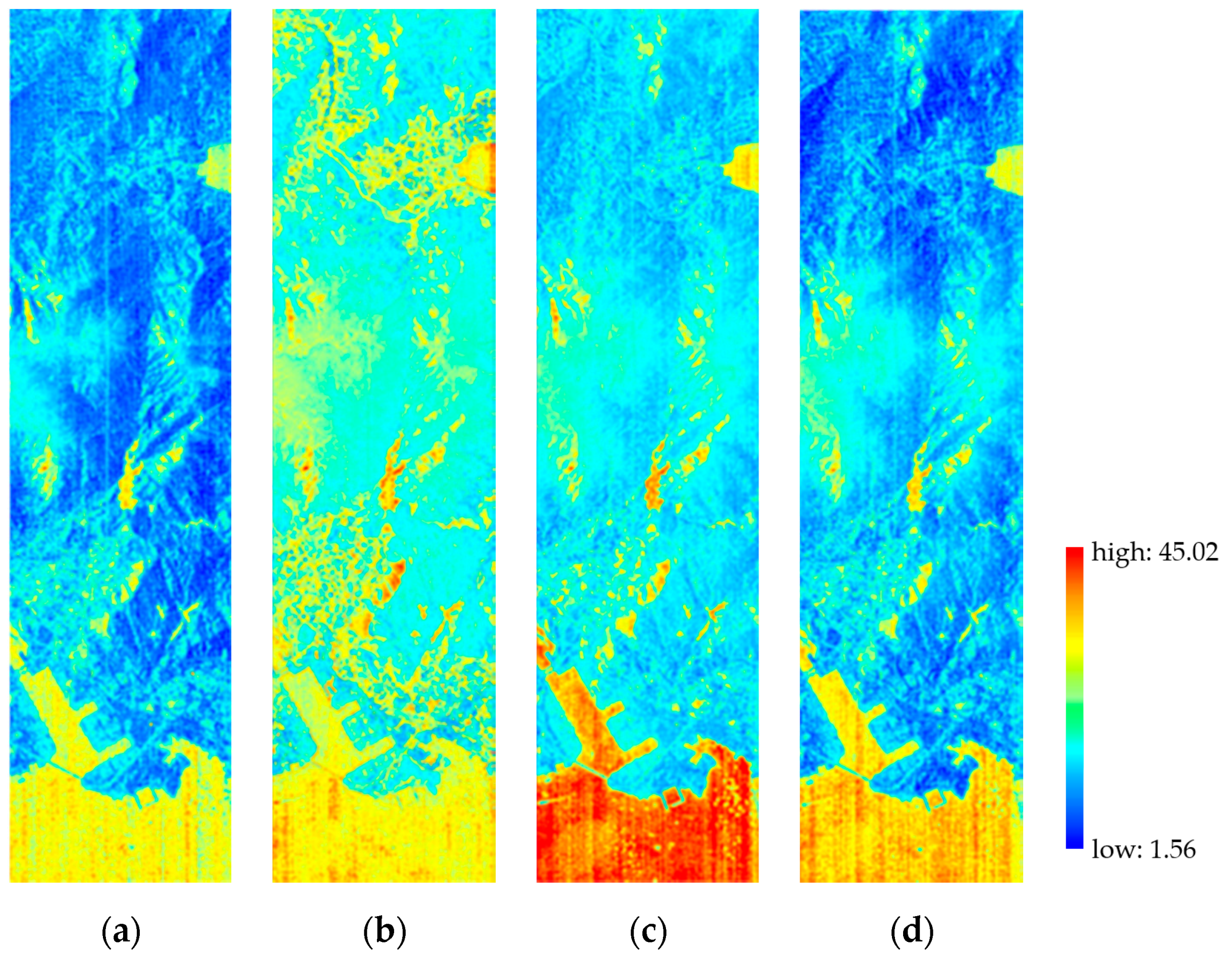
3.2.3. Overall Quality
- The proposed method builds the spectral relation between the MS and HS bands in the same wavelength ranges. The simulated images can be generated by using prior HS endmembers extracted from training HS images. In this manner, the relations between the bands of spectral endmembers are preserved, and fine spectral features are achieved in the simulated images.
- The proposed method reconstructs the simulated images by combining the spectrums of related materials, pixel by pixel. Following this strategy, the simulation of each pixel is independent, which can improve the spectral quality of the areas with complex materials and objects.
- We utilize iterative schemes to optimize the endmembers and abundance matrixes of the images. This method reduces the residual error in unmixing and reconstruction, thereby further improving the global quality of the results. Our method achieves the best performance in Table 3.
4. Discussion
4.1. Result Analysis
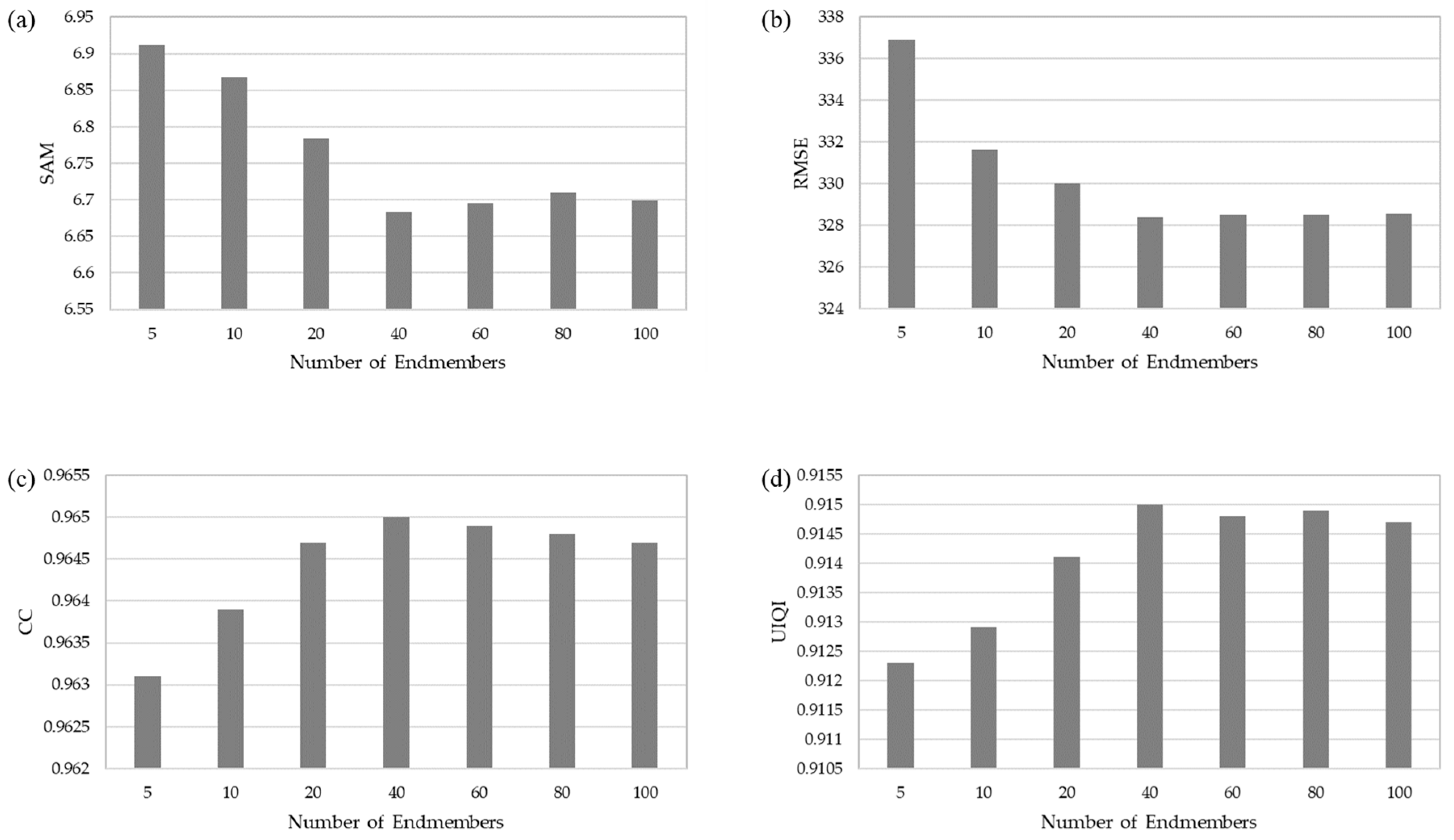
4.2. Sensitivity Analysis
4.2.1. Iteration Times
4.2.2. Number of Endmembers
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Pelta, R.; Ben-Dor, E. Assessing the detection limit of petroleum hydrocarbon in soils using hyperspectral remote-sensing. Remote Sens. Environ. 2019, 224, 145–153. [Google Scholar] [CrossRef]
- Li, N.; Huang, X.; Zhao, H.; Qiu, X.; Deng, K.; Jia, G.; Li, Z.; Fairbairn, D.; Gong, X. A Combined Quantitative Evaluation Model for the Capability of Hyperspectral Imagery for Mineral Mapping. Sensors 2019, 19, 328. [Google Scholar] [CrossRef] [PubMed]
- Veraverbeke, S.; Dennison, P.; Gitas, I.; Hulley, G.; Kalashnikova, O.; Katagis, T.; Kuai, L.; Meng, R.; Roberts, D.; Stavros, N. Remote Sensing of Environment Hyperspectral remote sensing of fire: State-of-the-art and future perspectives. Remote Sens. Environ. 2018, 216, 105–121. [Google Scholar] [CrossRef]
- Tan, Y.; Sun, J.; Zhang, B.; Chen, M.; Liu, Y.; Liu, X. Sensitivity of a Ratio Vegetation Index Derived from Hyperspectral Remote Sensing to the Brown Planthopper Stress on Rice Plants. Sensors 2019, 19, 375. [Google Scholar] [CrossRef]
- Hoang, N.T.; Koike, K. Transformation of Landsat imagery into pseudo-hyperspectral imagery by a multiple regression-based model with application to metal deposit-related minerals mapping. ISPRS J. Photogramm. Remote Sens. 2017, 133, 157–173. [Google Scholar] [CrossRef]
- Miura, T.; Huete, A.; Yoshioka, H. An empirical investigation of cross-sensor relationships of NDVI and red/near-infrared reflectance using EO-1 Hyperion data. Remote Sens. Environ. 2006, 100, 223–236. [Google Scholar] [CrossRef]
- USGS Earth Observing 1 (EO-1). Available online: https://archive.usgs.gov/archive/sites/eo1.usgs.gov/index.html (accessed on 21 May 2019).
- Zhu, X.; Chen, J.; Gao, F.; Chen, X.; Masek, J.G. An enhanced spatial and temporal adaptive reflectance fusion model for complex heterogeneous regions. Remote Sens. Environ. 2010, 114, 2610–2623. [Google Scholar] [CrossRef]
- Ju, J.; Roy, D.P. The availability of cloud-free Landsat ETM+ data over the conterminous United States and globally. Remote Sens. Environ. 2008, 112, 1196–1211. [Google Scholar] [CrossRef]
- Sun, X.; Zhang, L.; Yang, H.; Wu, T.; Cen, Y.; Guo, Y. Enhancement of Spectral Resolution for Remotely Sensed Multispectral Image. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2198–2211. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First experience with Sentinel-2 data for crop and tree species classifications in central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Dierckx, W.; Sterckx, S.; Benhadj, I.; Livens, S.; Duhoux, G.; Van Achteren, T.; Francois, M.; Mellab, K.; Saint, G. PROBA-V mission for global vegetation monitoring: standard products and image quality. Int. J. Remote Sens. 2014, 35, 2589–2614. [Google Scholar] [CrossRef]
- Hoan, N.T.; Tateishi, R. Cloud Removal of Optical Image Using SAR Data for ALOS Applications. Experimenting on Simulated ALOS Data. J. Remote Sens. Soc. Japan 2009, 29, 410–417. [Google Scholar]
- Wulder, M.A.; White, J.C.; Loveland, T.R.; Woodcock, C.E.; Belward, A.S.; Cohen, W.B.; Fosnight, E.A.; Shaw, J.; Masek, J.G.; Roy, D.P. The global Landsat archive: Status, consolidation, and direction. Remote Sens. Environ. 2016, 185, 271–283. [Google Scholar] [CrossRef]
- Huo, H.; Guo, J.; Li, Z. Hyperspectral Image Classification for Land Cover Based on an Improved Interval Type-II Fuzzy C-Means Approach. Sensors 2018, 18, 363. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.-Q.; Zhao, X.; Su, H.-Y.; Li, B.; Hu, Y.-M.; Cui, X.-S. Predicting Spatial Variations in Soil Nutrients with Hyperspectral Remote Sensing at Regional Scale. Sensors 2018, 18, 3086. [Google Scholar] [CrossRef] [PubMed]
- Fan, L.; Zhao, J.; Xu, X.; Liang, D.; Yang, G.; Feng, H.; Yang, H.; Wang, Y.; Chen, G.; Wei, P. Hyperspectral-based Estimation of Leaf Nitrogen Content in Corn Using Optimal Selection of Multiple Spectral Variables. Sensors 2019, 19, 2898. [Google Scholar] [CrossRef]
- Chen, F.; Niu, Z.; Sun, G.Y.; Wang, C.Y.; Teng, J. Using low-spectral-resolution images to acquire simulated hyperspectral images. Int. J. Remote Sens. 2008, 29, 2963–2980. [Google Scholar] [CrossRef]
- Liu, B.; Zhang, L.; Zhang, X.; Zhang, B.; Tong, Q. Simulation of EO-1 Hyperion Data from ALI Multispectral Data Based on the Spectral Reconstruction Approach. Sensors 2009, 9, 3090–3108. [Google Scholar] [CrossRef]
- Winter, M.E.; Winter, E.M.; Beaven, S.G.; Ratkowski, A.J. Hyperspectral image sharpening using multispectral data. In Proceedings of the IEEE Aerospace Conference, Big Sky, MT, USA, 3–10 March 2007; pp. 1–9. [Google Scholar]
- Winter, M.E.; Winter, E.M.; Beaven, S.G.; Ratkowski, A.J. High-performance fusion of multispectral and hyperspectral data. In Proceedings of the Defense and Security Symposium, Orlando, FL, USA, 17–21 April 2006; Volume 6233. [Google Scholar]
- Zhang, Z.; Shi, Z. Nonnegative matrix factorization-based hyperspectral and panchromatic image fusion. Neural Comput. Appl. 2013, 23, 895–905. [Google Scholar] [CrossRef]
- Lin, C.; Ma, F.; Chi, C.; Hsieh, C. A Convex Optimization-Based Coupled Nonnegative Matrix Factorization Algorithm for Hyperspectral and Multispectral Data Fusion. IEEE Trans. Geosci. Remote Sens. 2018, 56, 1652–1667. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, M.; Yang, S.; Xing, Y.; Qu, R. Fusion of Panchromatic and Multispectral Images via Coupled Sparse Non-Negative Matrix Factorization. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 5740–5747. [Google Scholar] [CrossRef]
- Karoui, M.S.; Deville, Y.; Benhalouche, F.Z.; Boukerch, I. Hypersharpening by joint-criterion nonnegative matrix factorization. IEEE Trans. Geosci. Remote Sens. 2016, 55, 1660–1670. [Google Scholar] [CrossRef]
- Dong, W.; Fu, F.; Shi, G.; Cao, X.; Wu, J.; Li, G.; Li, X. Hyperspectral image super-resolution via non-negative structured sparse representation. IEEE Trans. Image Process. 2016, 25, 2337–2352. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Wright, J.; Huang, T.S.; Ma, Y. Image super-resolution via sparse representation. IEEE Trans. Image Process. 2010, 19, 2861–2873. [Google Scholar] [CrossRef] [PubMed]
- Nieves, J.L.; Valero, E.M.; Romero, J.; Henández-Andrés, J. Spectral recovery of artificial illuminants using a CCD colour camera with Non-negative Matrix Factorization and Independent Component Analysis. In Proceedings of the Conference on Colour in Graphics, Imaging, and Vision; Society for Imaging Science and Technology, Leeds, UK, 19–22 June 2006; pp. 237–240. [Google Scholar]
- Liu, X.; Xia, W.; Wang, B.; Zhang, L. An approach based on constrained nonnegative matrix factorization to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2011, 49, 757–772. [Google Scholar] [CrossRef]
- Huck, A.; Guillaume, M.; Blanc-Talon, J. Minimum dispersion constrained nonnegative matrix factorization to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2010, 48, 2590–2602. [Google Scholar] [CrossRef]
- Jia, S.; Qian, Y. Constrained nonnegative matrix factorization for hyperspectral unmixing. IEEE Trans. Geosci. Remote Sens. 2008, 47, 161–173. [Google Scholar] [CrossRef]
- Yokoya, N.; Yairi, T.; Iwasaki, A. Coupled nonnegative matrix factorization unmixing for hyperspectral and multispectral data fusion. IEEE Trans. Geosci. Remote Sens. 2012, 50, 528–537. [Google Scholar] [CrossRef]
- Poovalinga Ganesh, B.; Aravindan, S.; Raja, S.; Thirunavukkarasu, A. Hyperspectral satellite data (Hyperion) preprocessing—a case study on banded magnetite quartzite in Godumalai Hill, Salem, Tamil Nadu, India. Arab. J. Geosci. 2012, 6, 3249–3256. [Google Scholar] [CrossRef]
- Datt, B.; Mcvicar, T.; Van Niel, T.; L B Jupp, D.; Pearlman, J. Preprocessing EO-1 Hyperion hyperspectral data to support the application of agricultural indexes. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1246–1259. [Google Scholar] [CrossRef]
- Shi, C.; Wang, L. Incorporating spatial information in spectral unmixing: A review. Remote Sens. Environ. 2014, 149, 70–87. [Google Scholar] [CrossRef]
- Heylen, R.; Parente, M.; Member, S.; Gader, P. review of nonlinear HS-unmixing methods. IEEE Trans. Geosci. Remote Sens. 2014, 7, 1844–1868. [Google Scholar]
- Heylen, R.; Scheunders, P. A multilinear mixing model for nonlinear spectral unmixing. IEEE Trans. Geosci. Remote Sens. 2016, 54, 240–251. [Google Scholar] [CrossRef]
- Hoang, N.T.; Koike, K. Comparison of hyperspectral transformation accuracies of multispectral Landsat TM, ETM+, OLI and EO-1 ALI images for detecting minerals in a geothermal prospect area. ISPRS J. Photogramm. Remote Sens. 2018, 137, 15–28. [Google Scholar] [CrossRef]
- China Centre for Resource Satellite Data and Application. Available online: http://www.cresda.com/n16/ (accessed on 2 November 2018).
- Lee, D.D.; Seung, H.S. Learning the parts of objects by non-negative matrix factorization. Nature 1999, 401, 788–791. [Google Scholar] [CrossRef]
- Nascimento, J.M.P.; Dias, J.M.B. Vertex component analysis: A fast algorithm to unmix hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 898–910. [Google Scholar] [CrossRef]
- Styan, G.P.H. Hadamard products and multivariate statistical analysis. Linear Algebra Appl. 1973, 6, 217–240. [Google Scholar] [CrossRef]
- Wetzstein, G.; Lanman, D.; Hirsch, M.; Raskar, R. Tensor Displays: Compressive Light Field Synthesis Using Multilayer Displays with Directional Backlighting. 2012. Available online: http://hdl.handle.net/1721.1/92408 (accessed on 17 October 2019).
- Loncan, L.; De Almeida, L.B.; Bioucas-Dias, J.M.; Briottet, X.; Chanussot, J.; Dobigeon, N.; Fabre, S.; Liao, W.; Licciardi, G.A.; Simoes, M.; et al. Hyperspectral Pansharpening: A Review. IEEE Geosci. Remote Sens. Mag. 2015, 3, 27–46. [Google Scholar] [CrossRef]
- Kruse, F.A.; Lefkoff, A.B.; Boardman, J.W.; Heidebrecht, K.B.; Shapiro, A.T.; Barloon, P.J.; Goetz, A.F.H. The spectral image processing system (SIPS)—interactive visualization and analysis of imaging spectrometer data. Remote Sens. Environ. 1993, 44, 145–163. [Google Scholar] [CrossRef]
- Aiazzi, B.; Alparone, L.; Baronti, S.; Garzelli, A.; Selva, M. Twenty-five years of pansharpening: A critical review and new developments. In Signal and Image Processing for Remote Sensing; CRC Press: Boca Raton, FL, USA, 2012; pp. 552–599. [Google Scholar]
- Wald, L. Quality of high resolution synthesised images: Is there a simple criterion? In Proceedings of the SEE/URISCA Third conference “Fusion of Earth data: merging point measurements, raster maps and remotely sensed images”, Sophia Antipolis, France, 26–28 January 2000; pp. 99–103. [Google Scholar]
- Wang, Z.; Bovik, A.C. A universal image quality index. IEEE Signal Process. Lett. 2002, 9, 81–84. [Google Scholar] [CrossRef]
- Sidike, P.; Asari, V.K.; Alam, M.S. Multiclass object detection with single query in hyperspectral imagery using class-associative spectral fringe-adjusted joint transform correlation. IEEE Trans. Geosci. Remote Sens. 2015, 54, 1196–1208. [Google Scholar] [CrossRef]
- Broadwater, J.; Chellappa, R. Hybrid detectors for subpixel targets. IEEE Trans. Pattern Anal. Mach. Intell. 2007, 29, 1891–1903. [Google Scholar] [CrossRef] [PubMed]
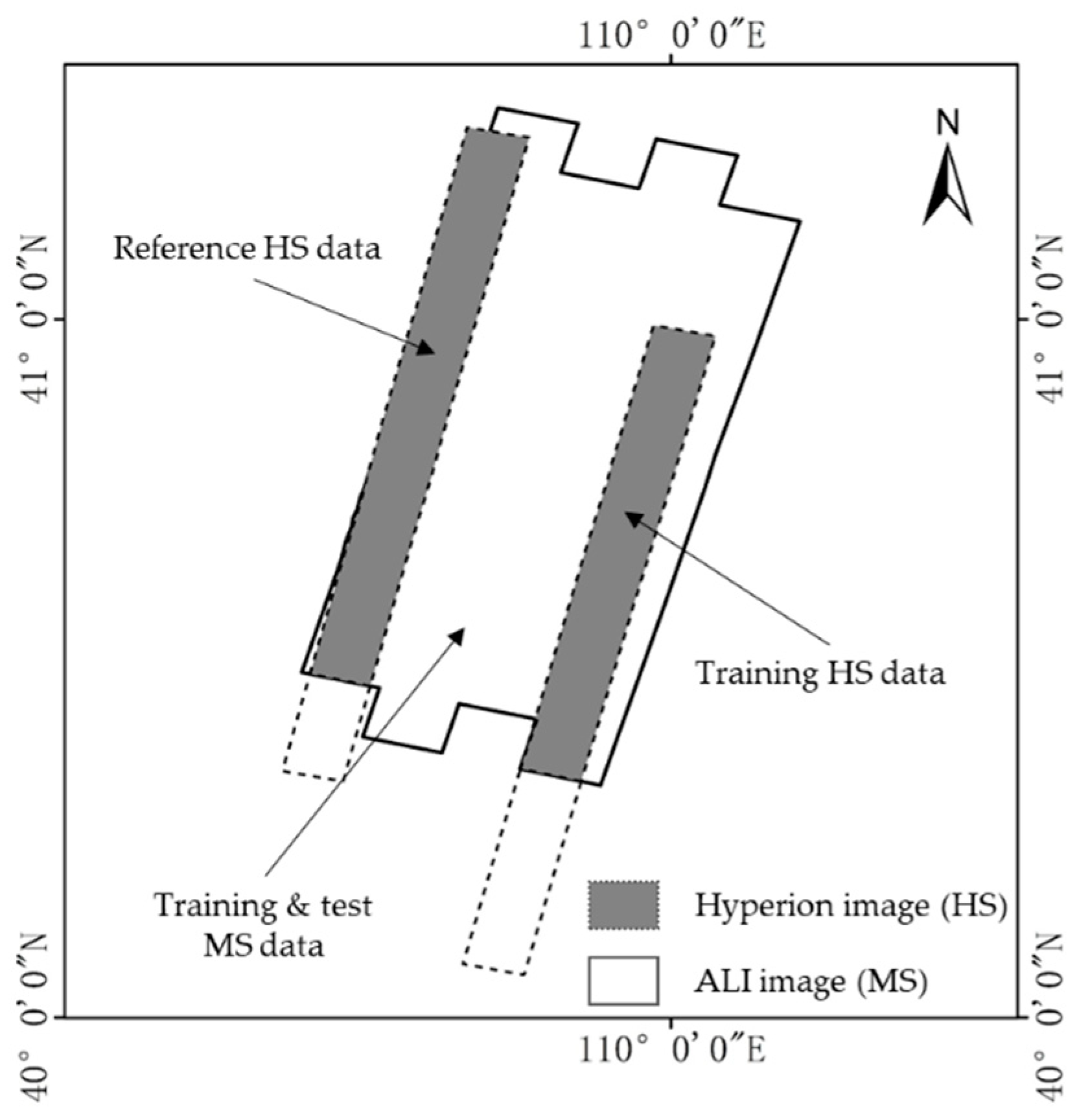
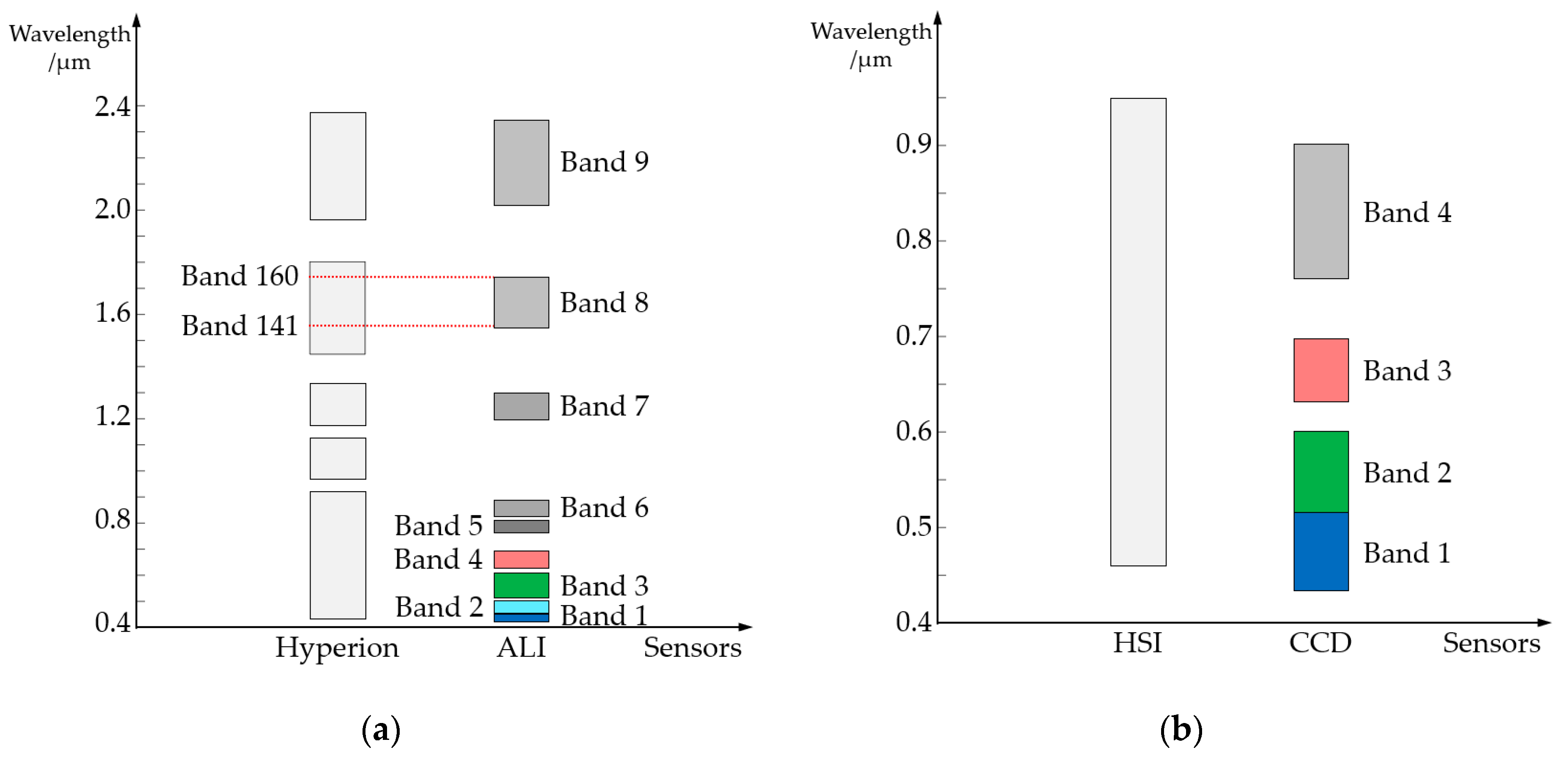






| Dataset | Sizes | Spatial Resolution | Training Area | Bands | Satellite & Sensors | Longitude/°E | Latitude/°N |
|---|---|---|---|---|---|---|---|
| 1 | MS 180 × 2000 | 30 m | 180 × 400 | 9 | EO-1 (ALI) | 114.22–114.28 | 22.33–25.87 |
| HS 180 × 2000 | 30 m | 154 | EO-1 (Hyperion) | ||||
| 2 | MS 318 × 480 | 30 m | 318 × 100 | 4 | HJ-1A (CCD) | 116.25–116.57 | 24.89–25.32 |
| HS 95 × 144 | 100 m | 92 | HJ-1A (HSI) | ||||
| 3 | MS 150 × 1850 | 30 m | 150 × 370 | 9 | EO-1 (ALI) | 113.96–114.00 | 29.95–30.45 |
| HS 150 × 1850 | 30 m | 154 | EO-1 (Hyperion) | ||||
| 4 | MS 180 × 600 | 30 m | 200 × 500 | 9 | EO-1 (ALI) | 109.45–109.74 | 40.44–41.08 |
| HS 180 × 600 | 30 m | 154 | EO-1 (Hyperion) |
| Datasets | Proposed Method | Chen | UPDM(Liu) | PHITA |
|---|---|---|---|---|
| 1 | 1.19 | 6.50 | 5.02 | 1.29 |
| 2 | 1.22 | 15.77 | 4.28 | 1.31 |
| 3 | 0.91 | 7.45 | 2.24 | 0.89 |
| 4 | 1.51 | 6.79 | 7.71 | 1.85 |
| Mean | 1.21 | 9.12 | 4.81 | 1.34 |
| Datasets | Method | SAM | ERGAS | RMSE | CC | UIQI | ACE |
|---|---|---|---|---|---|---|---|
| 1 | Proposed | 6.683 | 16.542 | 328.4 | 0.965 | 0.915 | 0.21 |
| Chen | 12.911 | 48.459 | 1069.1 | 0.858 | 0.708 | 0.08 | |
| UPDM | 8.962 | 68.803 | 993.4 | 0.957 | 0.742 | 0.07 | |
| PHITA | 7.082 | 17.439 | 338.1 | 0.963 | 0.913 | 0.19 | |
| 2 | Proposed | 3.450 | 13.348 | 142.1 | 0.891 | 0.841 | 0.14 |
| Chen | 13.013 | 45.608 | 1641.7 | 0.498 | 0.350 | 0.11 | |
| UPDM | 13.802 | 78.619 | 440.9 | 0.875 | 0.709 | 0.11 | |
| PHITA | 3.196 | 13.310 | 142.8 | 0.886 | 0.834 | 0.13 | |
| 3 | Proposed | 4.791 | 12.167 | 191.3 | 0.972 | 0.926 | 0.22 |
| Chen | 15.132 | 60.631 | 1609.4 | 0.736 | 0.425 | 0.07 | |
| UPDM | 7.083 | 15.412 | 266.8 | 0.968 | 0.912 | 0.08 | |
| PHITA | 4.818 | 12.881 | 196.8 | 0.969 | 0.923 | 0.21 | |
| 4 | Proposed | 9.02 | 25.21 | 476.9 | 0.795 | 0.688 | 0.09 |
| Chen | 17.466 | 42.564 | 1787.5 | 0.642 | 0.455 | 0.07 | |
| UPDM | 13.833 | 65.187 | 1239.5 | 0.783 | 0.607 | 0.06 | |
| PHITA | 10.347 | 26.521 | 495.7 | 0.788 | 0.695 | 0.08 | |
| Mean | Proposed | 5.986 | 16.817 | 284.6 | 0.905 | 0.842 | 0.165 |
| Chen | 14.63 | 49.315 | 1526.9 | 0.683 | 0.484 | 0.082 | |
| UPDM | 10.92 | 57.005 | 735.1 | 0.896 | 0.742 | 0.08 | |
| PHITA | 6.360 | 17.538 | 293.4 | 0.901 | 0.841 | 0.152 | |
| Std | Proposed | 2.094 | 5.103 | 130.3 | 0.071 | 0.095 | 0.053 |
| Chen | 1.861 | 6.857 | 272.7 | 0.131 | 0.134 | 0.016 | |
| UPDM | 2.972 | 24.511 | 395.9 | 0.074 | 0.110 | 0.019 | |
| PHITA | 2.683 | 5.483 | 136.9 | 0.073 | 0.091 | 0.051 |
| Dataset | Proposed Method | Chen | UPDM(Liu) | PHITA |
|---|---|---|---|---|
| 1 | 151.54 | 44.49 | 141.75 | 30.16 |
| 2 | 47.77 | 10.24 | 12.34 | 8.26 |
| 3 | 121.42 | 34.18 | 49.69 | 28.91 |
| 4 | 57.91 | 10.47 | 6.4 | 5.6 |
| Mean | 94.66 | 24.84 | 52.55 | 18.23 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, Z.; Chen, Q.; Chen, Q.; Liu, X.; He, H. A Novel Hyperspectral Image Simulation Method Based on Nonnegative Matrix Factorization. Remote Sens. 2019, 11, 2416. https://doi.org/10.3390/rs11202416
Huang Z, Chen Q, Chen Q, Liu X, He H. A Novel Hyperspectral Image Simulation Method Based on Nonnegative Matrix Factorization. Remote Sensing. 2019; 11(20):2416. https://doi.org/10.3390/rs11202416
Chicago/Turabian StyleHuang, Zehua, Qi Chen, Qihao Chen, Xiuguo Liu, and Hao He. 2019. "A Novel Hyperspectral Image Simulation Method Based on Nonnegative Matrix Factorization" Remote Sensing 11, no. 20: 2416. https://doi.org/10.3390/rs11202416
APA StyleHuang, Z., Chen, Q., Chen, Q., Liu, X., & He, H. (2019). A Novel Hyperspectral Image Simulation Method Based on Nonnegative Matrix Factorization. Remote Sensing, 11(20), 2416. https://doi.org/10.3390/rs11202416