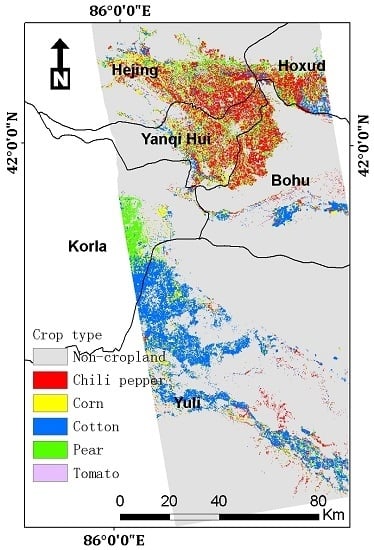
Figure 1.
The study area across Yuli, Korla, Bohu, Yanqi, Hejing, and Hoxud counties in Bayin Guoyu Mongolian Autonomous Prefecture, Xinjiang Uygur Autonomous Region, China. The in-situ collected sample points of major crop types are highlighted. The overlapped area of Sentinel-1 IW2 sub-swath (in the blue rectangle) and mosaicked coverage of Sentinel-2 scenes (in the black rectangle) is the area of interest in this study. The background image is a true color Google Earth high-resolution image. Map data: Google Earth, Image © 2020 Europa Technologies.
Figure 1.
The study area across Yuli, Korla, Bohu, Yanqi, Hejing, and Hoxud counties in Bayin Guoyu Mongolian Autonomous Prefecture, Xinjiang Uygur Autonomous Region, China. The in-situ collected sample points of major crop types are highlighted. The overlapped area of Sentinel-1 IW2 sub-swath (in the blue rectangle) and mosaicked coverage of Sentinel-2 scenes (in the black rectangle) is the area of interest in this study. The background image is a true color Google Earth high-resolution image. Map data: Google Earth, Image © 2020 Europa Technologies.
Figure 2.
The workflow of crop type mapping by the integration of time series Sentinel-1 and Sentinel-2 features.
Figure 2.
The workflow of crop type mapping by the integration of time series Sentinel-1 and Sentinel-2 features.
Figure 3.
The F1 score of cropland recorded in each iteration of the recursive feature increment (RFI) process, reaching the maximum at the 114th iteration. Therefore, the top 114 features in the importance ranking were chosen as the optimal feature set for land cover classification in step 1.
Figure 3.
The F1 score of cropland recorded in each iteration of the recursive feature increment (RFI) process, reaching the maximum at the 114th iteration. Therefore, the top 114 features in the importance ranking were chosen as the optimal feature set for land cover classification in step 1.
Figure 4.
The feature importance scores of the top six features selected by the RFI method for cropland extraction, in descending order. Each importance score was normalized and converted to percentage. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 4.
The feature importance scores of the top six features selected by the RFI method for cropland extraction, in descending order. Each importance score was normalized and converted to percentage. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 5.
Boxplots of the top six most important features of different land cover types. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 5.
Boxplots of the top six most important features of different land cover types. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 6.
(a) Land cover classification map; (b) cropland mask extracted from the land cover classification results.
Figure 6.
(a) Land cover classification map; (b) cropland mask extracted from the land cover classification results.
Figure 7.
The mean overall accuracy (OA) and mean kappa coefficient of fivefold cross-validation recorded for each iteration during the RFI feature selection process.
Figure 7.
The mean overall accuracy (OA) and mean kappa coefficient of fivefold cross-validation recorded for each iteration during the RFI feature selection process.
Figure 8.
The feature importance score of the top six features selected by the RFI method for crop type classification, in descending order. Each importance score was normalized and converted to a percentage. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 8.
The feature importance score of the top six features selected by the RFI method for crop type classification, in descending order. Each importance score was normalized and converted to a percentage. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 9.
Boxplots of different crop types of the top six important features selected by the RFI approach. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the band 11 values from the 3 June 2018 acquisition of Sentinel-2; (b) boxplots of the band 6 values from the 17 August 2018 acquisition of Sentinel-2; (c) boxplots of the NDVIre2n indices derived from the 1 September 2018 acquisition of Sentinel-2; (d) boxplots of the VH intensity values from the 26 March 2018 acquisition of Sentinel-1; (e) boxplots of the band 8 values from the 23 July 2018 acquisition of Sentinel-2; (f) boxplots of the band 7 values from the 17 August 2018 acquisition of Sentinel-2.
Figure 9.
Boxplots of different crop types of the top six important features selected by the RFI approach. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the band 11 values from the 3 June 2018 acquisition of Sentinel-2; (b) boxplots of the band 6 values from the 17 August 2018 acquisition of Sentinel-2; (c) boxplots of the NDVIre2n indices derived from the 1 September 2018 acquisition of Sentinel-2; (d) boxplots of the VH intensity values from the 26 March 2018 acquisition of Sentinel-1; (e) boxplots of the band 8 values from the 23 July 2018 acquisition of Sentinel-2; (f) boxplots of the band 7 values from the 17 August 2018 acquisition of Sentinel-2.
Figure 10.
Crop distribution map of the study area derived from random forest (RF) classification using the combined Sentinel-1 and Sentinel-2 feature set.
Figure 10.
Crop distribution map of the study area derived from random forest (RF) classification using the combined Sentinel-1 and Sentinel-2 feature set.
Figure 11.
A subset of VH intensity on 26 March 2018 extracted from (a) Original synthetic aperture radar (SAR) intensity; (b) SAR intensity filtered by the refined Lee method; (c) SAR intensity filtered by the SHP distributed scatterer interferometry (DSI) method.
Figure 11.
A subset of VH intensity on 26 March 2018 extracted from (a) Original synthetic aperture radar (SAR) intensity; (b) SAR intensity filtered by the refined Lee method; (c) SAR intensity filtered by the SHP distributed scatterer interferometry (DSI) method.
Figure 12.
A subset of VV coherence on 7 April 2019 extracted from (a) Coherence coefficient estimated by a 7 × 7 sliding window; (b) Coherence coefficient estimated by the SHP DSI method with bias mitigation.
Figure 12.
A subset of VV coherence on 7 April 2019 extracted from (a) Coherence coefficient estimated by a 7 × 7 sliding window; (b) Coherence coefficient estimated by the SHP DSI method with bias mitigation.
Figure 13.
Comparison of crop type mapping results using different feature combinations. Four groups of features were tested, the first group contains only SAR (Sentinel-1) features, the second group contains only optical (Sentinel-2) features without the red-edge contribution, the third group has all of the Sentinel-2 features, the fourth group includes both SAR and optical (Sentinel-1 and 2) features.
Figure 13.
Comparison of crop type mapping results using different feature combinations. Four groups of features were tested, the first group contains only SAR (Sentinel-1) features, the second group contains only optical (Sentinel-2) features without the red-edge contribution, the third group has all of the Sentinel-2 features, the fourth group includes both SAR and optical (Sentinel-1 and 2) features.
Figure 14.
Accumulated feature importance scores of features selected for (a) step 1: cropland extraction and (b) step 2: crop type discrimination, calculated for each subcategory regardless of acquisition time. Each importance score was normalized and converted to a percentage. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 14.
Accumulated feature importance scores of features selected for (a) step 1: cropland extraction and (b) step 2: crop type discrimination, calculated for each subcategory regardless of acquisition time. Each importance score was normalized and converted to a percentage. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 15.
The accumulated importance scores of features selected in step 1: cropland extraction in three groups. The first group contains only Sentinel-1 features; the second group comprises only Sentinel-2 features exclusive of the red-edge features; the third group includes only the red-edge features. (a) Feature scores in the three groups calculated regardless of the acquisition time; (b) Feature scores in the three groups calculated for each month. The VH amplitude dispersion, as a single-phase feature, is plotted on the rightmost bar. Each importance score was normalized and converted to a percentage.
Figure 15.
The accumulated importance scores of features selected in step 1: cropland extraction in three groups. The first group contains only Sentinel-1 features; the second group comprises only Sentinel-2 features exclusive of the red-edge features; the third group includes only the red-edge features. (a) Feature scores in the three groups calculated regardless of the acquisition time; (b) Feature scores in the three groups calculated for each month. The VH amplitude dispersion, as a single-phase feature, is plotted on the rightmost bar. Each importance score was normalized and converted to a percentage.
Figure 16.
The accumulated importance scores of features selected in step 2: crop type discrimination in three groups. The first group contains only Sentinel-1 features; the second group comprises only Sentinel-2 features exclusive of the red-edge features; the third group includes only the red-edge features. (a) Feature scores in the three groups calculated regardless of the acquisition time; (b) Feature scores in the three groups calculated for each month. The VH amplitude dispersion, as a single-phase feature, is plotted on the rightmost bar. Each importance score was normalized and converted to a percentage.
Figure 16.
The accumulated importance scores of features selected in step 2: crop type discrimination in three groups. The first group contains only Sentinel-1 features; the second group comprises only Sentinel-2 features exclusive of the red-edge features; the third group includes only the red-edge features. (a) Feature scores in the three groups calculated regardless of the acquisition time; (b) Feature scores in the three groups calculated for each month. The VH amplitude dispersion, as a single-phase feature, is plotted on the rightmost bar. Each importance score was normalized and converted to a percentage.
Figure 17.
Heat maps showing correlation between selected features. (a) Correlation heat map of selected features in step 1; (b) correlation heat map of selected features in step 2. In both (a,b), the feature indexes follow the feature rankings obtained through the RF feature importance score. Correlation close to 1 or −1 indicates high positive or negative correlation between features. The diagonal elements in both correlation matrices are self-correlation coefficients, so they constantly equal one. (c) The histogram of correlation between selected features in step 1; (d) the histogram of correlation between selected features in step 2. In both (c,d), the histograms were calculated after removing the diagonal elements of the correlation matrices and converting a negative correlation coefficient to corresponding positive values. Thus, the ‘0’ in the histogram indicates low correlation, while ‘1’ indicates high correlation.
Figure 17.
Heat maps showing correlation between selected features. (a) Correlation heat map of selected features in step 1; (b) correlation heat map of selected features in step 2. In both (a,b), the feature indexes follow the feature rankings obtained through the RF feature importance score. Correlation close to 1 or −1 indicates high positive or negative correlation between features. The diagonal elements in both correlation matrices are self-correlation coefficients, so they constantly equal one. (c) The histogram of correlation between selected features in step 1; (d) the histogram of correlation between selected features in step 2. In both (c,d), the histograms were calculated after removing the diagonal elements of the correlation matrices and converting a negative correlation coefficient to corresponding positive values. Thus, the ‘0’ in the histogram indicates low correlation, while ‘1’ indicates high correlation.
Figure 18.
Boxplots of selected VH intensity features of different crop types, spanning the time from 26 March 2018 to 13 May 2018, as well as a feature on 16 October 2018. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. In (a–e), pear values show an apparent distinction from other crops, while in (f), chili can be easier recognized.
Figure 18.
Boxplots of selected VH intensity features of different crop types, spanning the time from 26 March 2018 to 13 May 2018, as well as a feature on 16 October 2018. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. In (a–e), pear values show an apparent distinction from other crops, while in (f), chili can be easier recognized.
Figure 19.
Boxplots of the top three scoring master versus slave VH intensity ratio of different crop types. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the master vs. slave VH intensity ratio derived from the 29 August 2018 acquisition of Sentinel-1; (b) boxplots of the master vs. slave VH intensity ratio derived from 10 September 2018 acquisition of Sentinel-1; (c) boxplots of the master vs. slave VH intensity ratio derived from 4 October 2018 acquisition of Sentinel-1.
Figure 19.
Boxplots of the top three scoring master versus slave VH intensity ratio of different crop types. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the master vs. slave VH intensity ratio derived from the 29 August 2018 acquisition of Sentinel-1; (b) boxplots of the master vs. slave VH intensity ratio derived from 10 September 2018 acquisition of Sentinel-1; (c) boxplots of the master vs. slave VH intensity ratio derived from 4 October 2018 acquisition of Sentinel-1.
Figure 20.
Boxplots of selected SAR features of different crop types. (a) Coherence coefficient in VV polarization mode; (b) amplitude dispersion in VH polarization mode. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 20.
Boxplots of selected SAR features of different crop types. (a) Coherence coefficient in VV polarization mode; (b) amplitude dispersion in VH polarization mode. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”.
Figure 21.
Boxplots of selected red-edge indices showing a good capability to distinguish corn from other crops. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the NDVIre2n indices derived from the 1 September 2018 acquisition of Sentinel-2; (b) boxplots of the NDVIre2n indices derived from the 17 August 2018 acquisition of Sentinel-2; (c) boxplots of the CIre indices derived from the 17 August 2018 acquisition of Sentinel-2; (d) boxplots of the NDVIre1n indices derived from the 17 August 2018 acquisition of Sentinel-2; (e) boxplots of the MSRre indices derived from the 17 August 2018 acquisition of Sentinel-2; (f) boxplots of the NDre2 indices derived from the 17 August 2018 acquisition of Sentinel-2; (g) boxplots of the NDVIre2 indices derived from the 17 August 2018 acquisition of Sentinel-2; (h) boxplots of the MSRren indices derived from the 17 August 2018 acquisition of Sentinel-2; (i) boxplots of the NDVIre2 indices derived from the 01 September 2018 acquisition of Sentinel-2.
Figure 21.
Boxplots of selected red-edge indices showing a good capability to distinguish corn from other crops. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the NDVIre2n indices derived from the 1 September 2018 acquisition of Sentinel-2; (b) boxplots of the NDVIre2n indices derived from the 17 August 2018 acquisition of Sentinel-2; (c) boxplots of the CIre indices derived from the 17 August 2018 acquisition of Sentinel-2; (d) boxplots of the NDVIre1n indices derived from the 17 August 2018 acquisition of Sentinel-2; (e) boxplots of the MSRre indices derived from the 17 August 2018 acquisition of Sentinel-2; (f) boxplots of the NDre2 indices derived from the 17 August 2018 acquisition of Sentinel-2; (g) boxplots of the NDVIre2 indices derived from the 17 August 2018 acquisition of Sentinel-2; (h) boxplots of the MSRren indices derived from the 17 August 2018 acquisition of Sentinel-2; (i) boxplots of the NDVIre2 indices derived from the 01 September 2018 acquisition of Sentinel-2.
![Remotesensing 12 00158 g021]()
Figure 22.
Boxplots of selected red-edge indices revealing a clear distinction between the pear and other crops. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the NDVIre1 indices derived from the 9 May 2018 acquisition of Sentinel-2; (b) boxplots of the NDVIre1n indices derived from the 6 October 2018 acquisition of Sentinel-2; (c) boxplots of the NDVIre2 indices derived from the 9 May 2018 acquisition of Sentinel-2; (d) boxplots of the CIre indices derived from the 06 October 2018 acquisition of Sentinel-2.
Figure 22.
Boxplots of selected red-edge indices revealing a clear distinction between the pear and other crops. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the NDVIre1 indices derived from the 9 May 2018 acquisition of Sentinel-2; (b) boxplots of the NDVIre1n indices derived from the 6 October 2018 acquisition of Sentinel-2; (c) boxplots of the NDVIre2 indices derived from the 9 May 2018 acquisition of Sentinel-2; (d) boxplots of the CIre indices derived from the 06 October 2018 acquisition of Sentinel-2.
Figure 23.
Boxplots of selected red-edge features showing good separability of chili pepper. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the band 6 values from the 17 August 2018 acquisition of Sentinel-2; (b) boxplots of the band 6 values from the 1 September 2018 acquisition of Sentinel-2; (c) boxplots of the band 7 values from the 17 August 2018 acquisition of Sentinel-2.
Figure 23.
Boxplots of selected red-edge features showing good separability of chili pepper. Each feature is named by its subcategory and acquisition time in the format of “yyyymmdd”. (a) Boxplots of the band 6 values from the 17 August 2018 acquisition of Sentinel-2; (b) boxplots of the band 6 values from the 1 September 2018 acquisition of Sentinel-2; (c) boxplots of the band 7 values from the 17 August 2018 acquisition of Sentinel-2.
Table 1.
Phenological calendars of the major crop types in the study site.
Table 1.
Phenological calendars of the major crop types in the study site.
| | Jan | Feb | Mar | Apr | May | Jun | Jul | Aug | Sep | Oct | Nov | Dec |
|---|
| Cotton | | | | △ | △☆ | ☆ | ☆ | ☆ | ☆√ | | | |
| Corn (Spring) | | | | △ | △ | △☆ | ☆ | ☆√ | √ | | | |
| Corn (Summer) | | | | | | △ | △☆ | ☆ | √ | √ | | |
| Pear | | | ♤ | ☆ | ☆ | ☆ | ☆ | ☆ | √ | | | |
| Chili pepper | | ♠ | ♠ | ★ | ★☆ | ☆ | ☆ | ☆ | ☆√ | √ | | |
| Tomato | | ♠ | ♠ | ★ | ★☆ | ☆ | ☆ | √ | | | | |
Table 2.
Ground samples of the six land cover types in the study area.
Table 2.
Ground samples of the six land cover types in the study area.
| Land Cover Type | Sample Points |
|---|
| Cropland | 3817 |
| Forest | 1468 |
| Desert | 1468 |
| Urban area | 2349 |
| Waterbody | 3257 |
| Wetlands | 3125 |
Table 3.
Ground samples collected for major crop types in the study area.
Table 3.
Ground samples collected for major crop types in the study area.
| Crop Type | Fold | Training Samples | Testing Samples |
|---|
| No. of Fields | No. of Points Points | No. of Fields | No. of Points |
|---|
| Chili pepper | 1st | 57 | 148 | 18 | 37 |
| 2nd | 50 | 147 | 25 | 38 |
| 3rd | 68 | 147 | 7 | 38 |
| 4th | 63 | 151 | 12 | 34 |
| 5th | 71 | 147 | 4 | 38 |
| Corn | 1st | 23 | 89 | 9 | 22 |
| 2nd | 25 | 88 | 7 | 23 |
| 3rd | 30 | 90 | 2 | 21 |
| 4th | 29 | 87 | 3 | 24 |
| 5th | 21 | 90 | 11 | 21 |
| Cotton | 1st | 37 | 150 | 13 | 38 |
| 2nd | 43 | 151 | 7 | 37 |
| 3rd | 43 | 152 | 7 | 36 |
| 4th | 34 | 151 | 16 | 37 |
| 5th | 43 | 152 | 7 | 36 |
| Pear | 1st | 22 | 76 | 2 | 19 |
| 2nd | 20 | 78 | 4 | 17 |
| 3rd | 15 | 76 | 9 | 19 |
| 4th | 18 | 77 | 6 | 18 |
| 5th | 21 | 73 | 3 | 22 |
| Tomato | 1st | 19 | 104 | 2 | 31 |
| 2nd | 16 | 108 | 5 | 27 |
| 3rd | 19 | 108 | 2 | 27 |
| 4th | 15 | 110 | 6 | 25 |
| 5th | 15 | 110 | 6 | 25 |
Table 4.
The employed Sentinel-1 and Sentinel-2 acquisitions of each month.
Table 4.
The employed Sentinel-1 and Sentinel-2 acquisitions of each month.
| Acquisition Time | Mar | Apr | May | Jun | Jul | Aug | Sep | Oct |
|---|
| Sentinel-1A | 1 | 2 | 3 | 3 | 3 | 3 | 2 | 2 |
| Sentinel-2A | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 1 |
| Sentinel-2B | 0 | 0 | 0 | 1 | 1 | 0 | 1 | 1 |
Table 5.
Metadata of the data stack of Sentinel-1 (S1) interferometric wide swath (IW) single look complex (SLC) data using the parameters from the first image. These values remain very close for all subsequent acquisitions.
Table 5.
Metadata of the data stack of Sentinel-1 (S1) interferometric wide swath (IW) single look complex (SLC) data using the parameters from the first image. These values remain very close for all subsequent acquisitions.
| Sentinel-1 IW SLC Data |
|---|
| First acquisition | 26 March 2018 |
| Last acquisition | 16 October 2018 |
| Pass direction | Ascending |
| Polarization mode | VV + VH |
| Incidence angle (°) | 36.12–41.84 |
| Wavelength (cm) | 5.5 (C-band) |
| Range spacing (m) | 2.33 |
| Azimuth spacing (m) | 13.92 |
Table 6.
Central wavelength and bandwidth of different spectral bands of Sentinel-2 data used in this research.
Table 6.
Central wavelength and bandwidth of different spectral bands of Sentinel-2 data used in this research.
| Spatial Resolution (m) | Spectral Bands | S2A | S2B |
|---|
| Central Wavelength (nm) | Central Wavelength (nm) |
|---|
| 10 | B2 | Blue | 496.6 | 492.1 |
| B3 | Green | 560 | 559 |
| B4 | Red | 664.5 | 665 |
| B8 | Near-infrared (NIR) | 835.1 | 833 |
| 20 | B5 | Red-edge 1 | 703.9 | 703.8 |
| B6 | Red-edge 2 | 740.2 | 739.1 |
| B7 | Red-edge 3 | 782.5 | 779.7 |
| B8A | NIR narrow | 864.8 | 864 |
| B11 | Short-wave infrared (SWIR) 1 | 1613.7 | 1610.4 |
| B12 | SWIR 2 | 2202.4 | 2185.7 |
Table 7.
Spectral indices calculated from Sentinel-2 data.
Table 7.
Spectral indices calculated from Sentinel-2 data.
| Reference Spectral Indices | Formula |
| NDVI | Normalized Difference Vegetation Index | |
| NDWI | Normalized Difference Water Index | |
| Red-edge spectral indices | Formula |
| NDVIre1 | Normalized Difference Vegetation Index red-edge 1 [32] | |
| NDVIre1n | Normalized Difference Vegetation Index red-edge 1 narrow [6] | |
| NDVIre2 | Normalized Difference Vegetation Index red-edge 2 [6] | |
| NDVIre2n | Normalized Difference Vegetation Index red-edge 2 narrow [6] | |
| NDVIre3 | Normalized Difference Vegetation Index red-edge 3 [32] | |
| NDVIre3n | Normalized Difference Vegetation Index red-edge 3 narrow [6] | |
| CIre | Chlorophyll Index red-edge [33] | |
| NDre1 | Normalized Difference red-edge 1 [32] | |
| NDre2 | Normalized Difference red-edge 2 [34] | |
| MSRre | Modified Simple Ratio red-edge [35] | |
| MSRren | Modified Simple Ratio red-edge narrow [6] | |
Table 8.
Accuracy of step 1 land cover classification, assessed by stratified 10-fold cross-validation using the ground samples. OA: overall accuracy.
Table 8.
Accuracy of step 1 land cover classification, assessed by stratified 10-fold cross-validation using the ground samples. OA: overall accuracy.
| Mean OA (%) | Kappa Coefficient | F1 Score |
|---|
| 94.05% | 0.927 | Cropland | 0.942 |
| Forest | 0.835 |
| Desert | 0.908 |
| Urban area | 0.932 |
| Waterbody | 0.995 |
| Wetlands | 0.945 |
Table 9.
Accuracy of step 2 crop type classification using the optimal combination of S1 and S2 features, assessed by stratified fivefold cross-validation using the ground samples.
Table 9.
Accuracy of step 2 crop type classification using the optimal combination of S1 and S2 features, assessed by stratified fivefold cross-validation using the ground samples.
| Mean OA (%) | Kappa Coefficient | F1 Score |
|---|
| 86.98% | 0.83 | Chili pepper | 0.84 |
| Corn | 0.71 |
| Cotton | 0.97 |
| Pear | 0.94 |
| Tomato | 0.79 |
Table 10.
Accuracy of step 2 crop type classification using the optimal combination of S1 and S2 features, assessed by stratified fivefold cross-validation using one sample from each validation field.
Table 10.
Accuracy of step 2 crop type classification using the optimal combination of S1 and S2 features, assessed by stratified fivefold cross-validation using one sample from each validation field.
| Mean OA (%) | Kappa Coefficient | F1 Score |
|---|
| 83.22% | 0.77 | Chili pepper | 0.87 |
| Corn | 0.69 |
| Cotton | 0.91 |
| Pear | 0.89 |
| Tomato | 0.71 |
Table 11.
Accuracy metrics of crop type classification using SAR features processed by different filters. SHP: statistically homogeneous pixel.
Table 11.
Accuracy metrics of crop type classification using SAR features processed by different filters. SHP: statistically homogeneous pixel.
| | Mean OA | Kappa Coefficient | F1-Score Chili | F1-Score Corn | F1-Score Cotton | F1-Score Pear | F1-Score Tomato |
|---|
| Original | 60.20% | 0.48 | 0.55 | 0.33 | 0.67 | 0.72 | 0.58 |
| Refined Lee | 73.21% | 0.65 | 0.66 | 0.52 | 0.80 | 0.83 | 0.74 |
| SHP DSI | 79.46% | 0.73 | 0.75 | 0.60 | 0.88 | 0.86 | 0.77 |
Table 12.
Accuracy assessment of crop type discrimination using different groups of features. The mean overall accuracy (OA) and kappa coefficient were averaged over fivefold cross-validation in both multi-sample per field validation and one sample per field validation.
Table 12.
Accuracy assessment of crop type discrimination using different groups of features. The mean overall accuracy (OA) and kappa coefficient were averaged over fivefold cross-validation in both multi-sample per field validation and one sample per field validation.
| | Optimal Number of Features | Mean OA | Kappa Coefficient |
|---|
| Multi Samples per Field | One Sample per Field | Multi Samples per Field | One Sample per Field |
|---|
| Sentinel-1 | 133 | 79.46% | 76.91% | 0.73 | 0.69 |
| Sentinel-2 without red-edge features | 58 | 82.37% | 79.80% | 0.77 | 0.72 |
| Sentinel-2 | 104 | 85.43% | 81.64% | 0.81 | 0.75 |
| Sentinel-1 and Sentinel-2 | 113 | 86.98% | 83.22% | 0.83 | 0.77 |