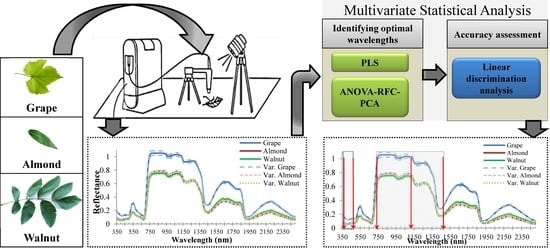
Optimal Spectral Wavelengths for Discriminating Orchard Species Using Multivariate Statistical Techniques
Abstract
:1. Introduction
2. Materials and Methods
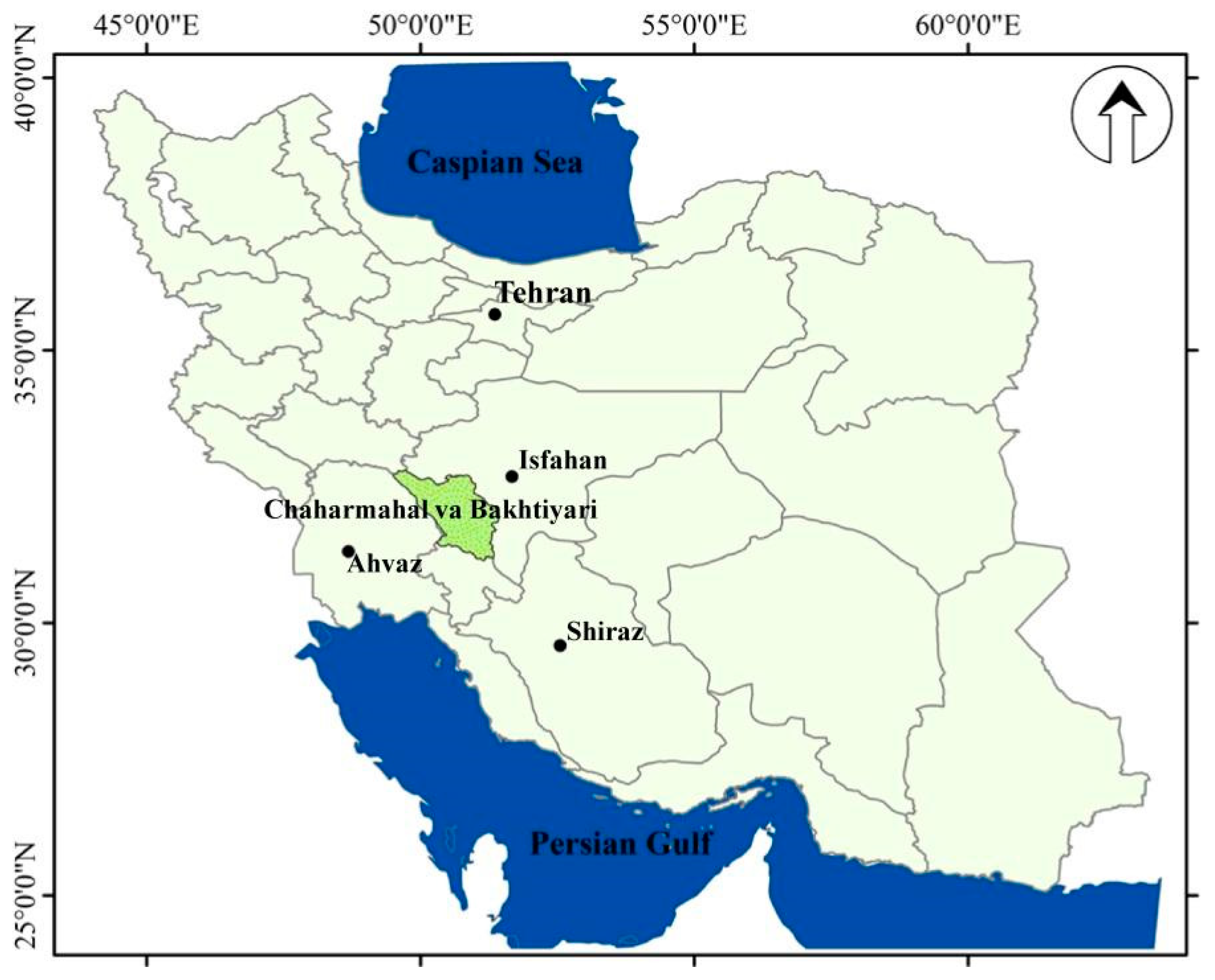
2.1. Study Area
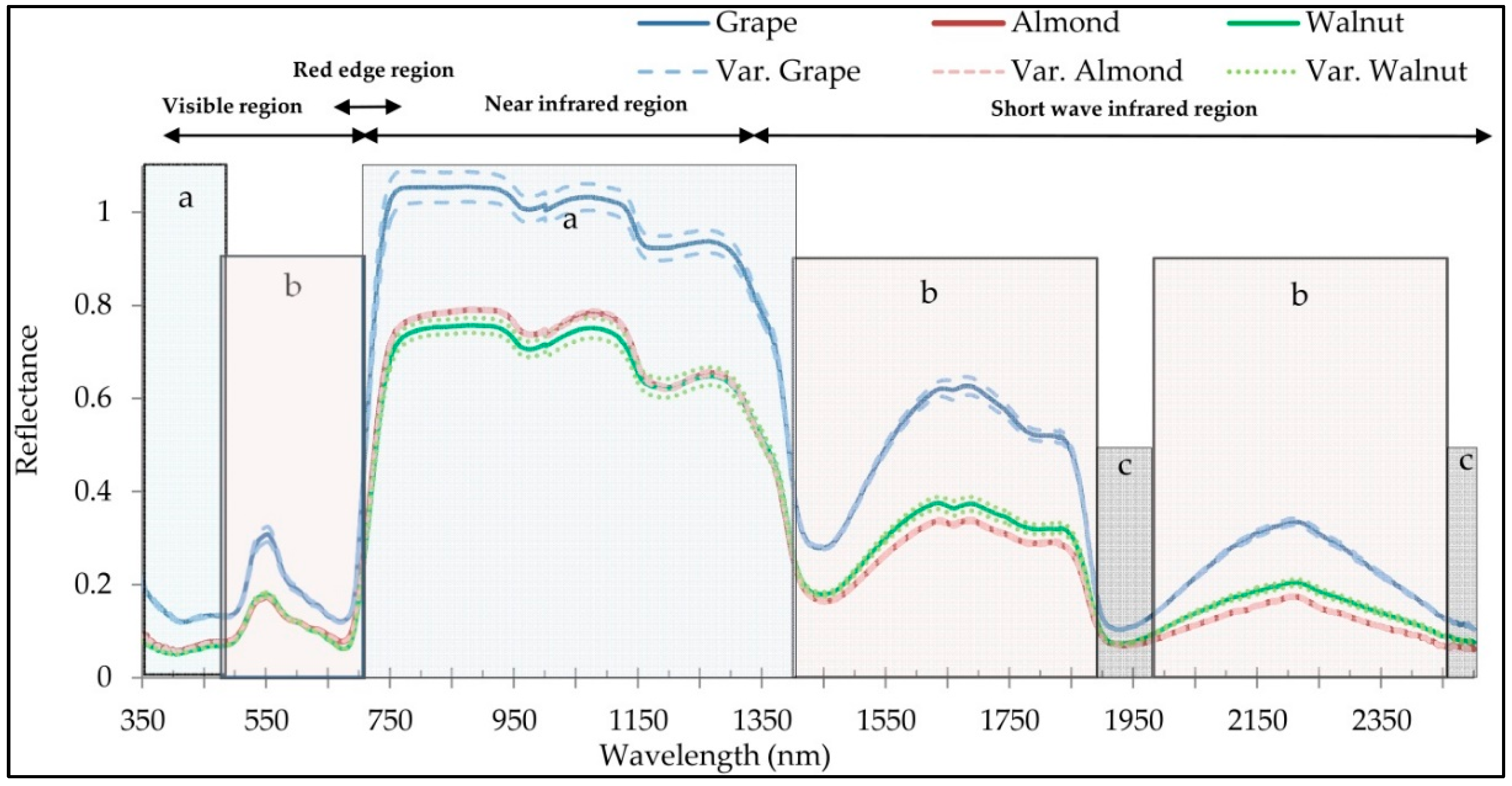
2.2. Field Spectral Acquisition
2.3. Identifying Optimal Wavelengths
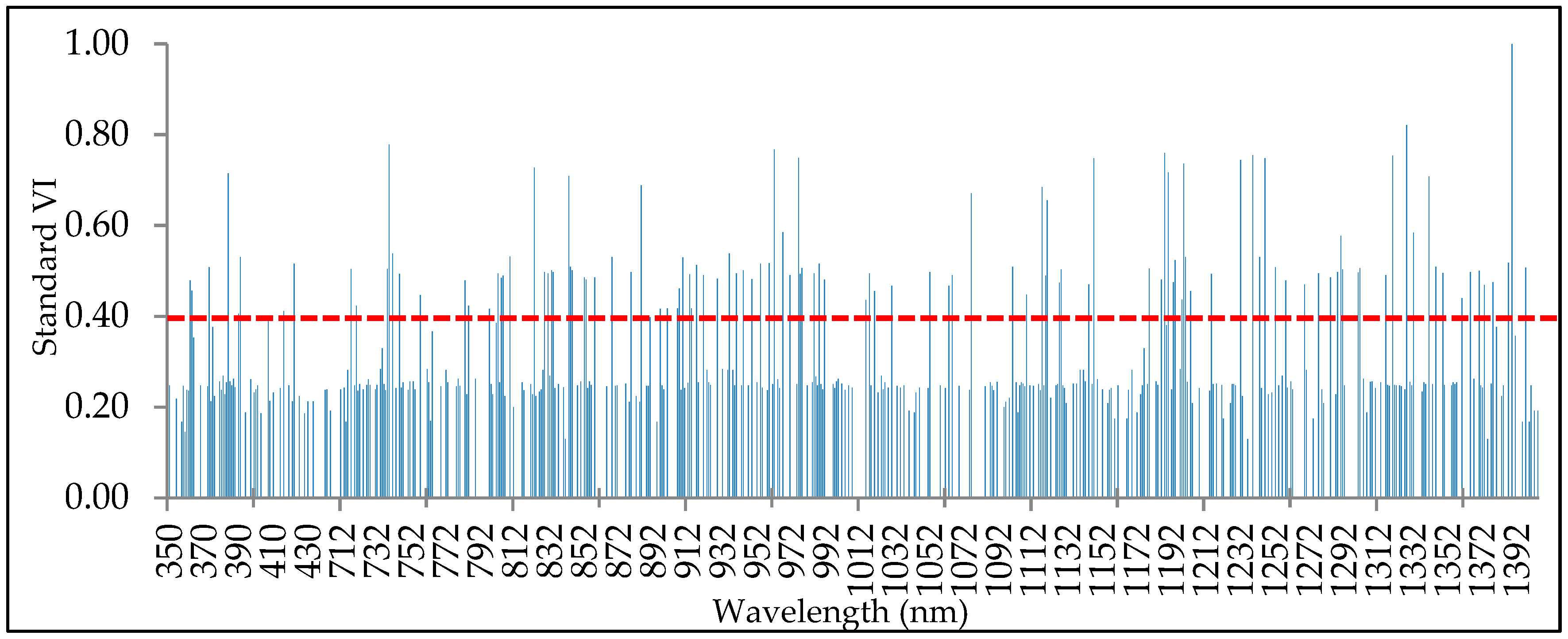
2.3.1. ANOVA–RFC–PCA Method
2.3.2. PLS
2.3.3. Accuracy Assessment
3. Results
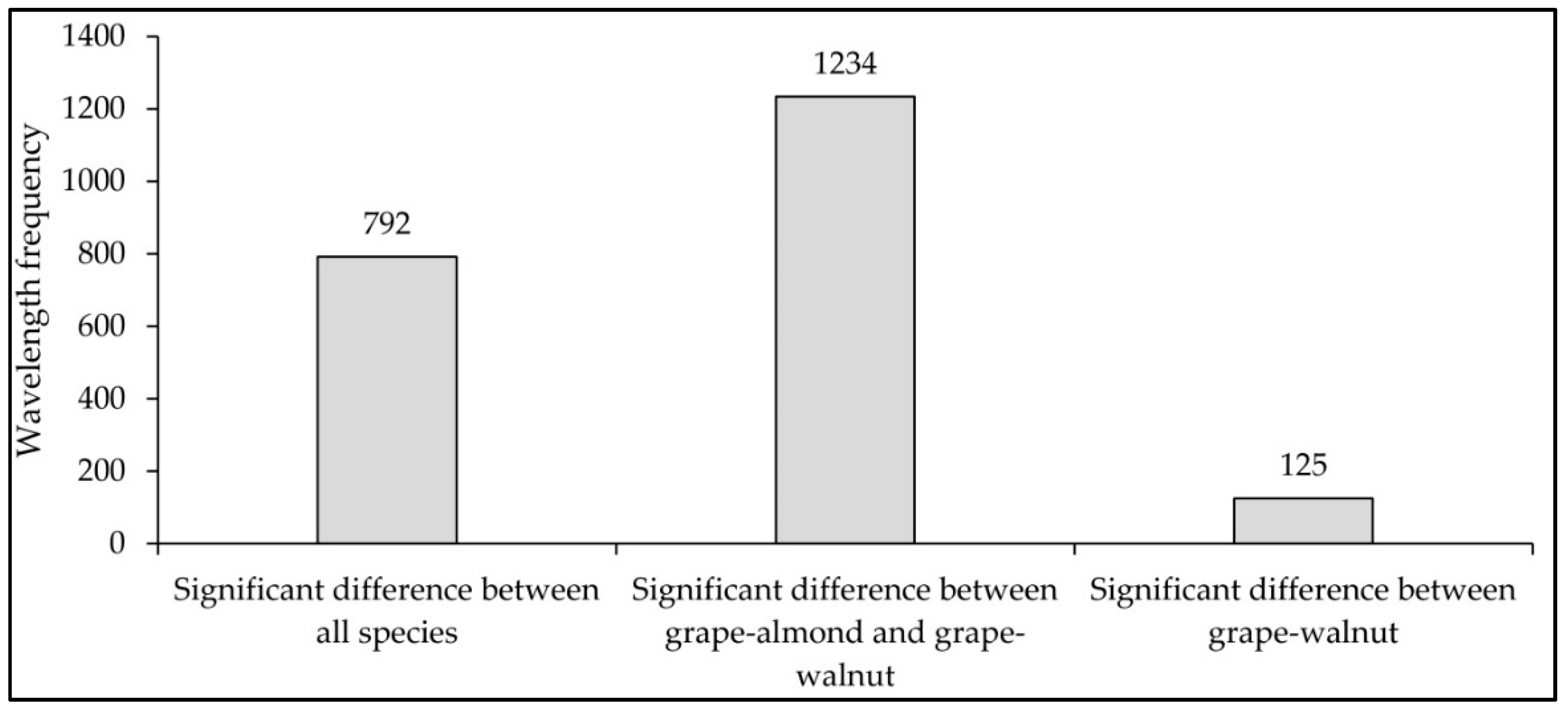
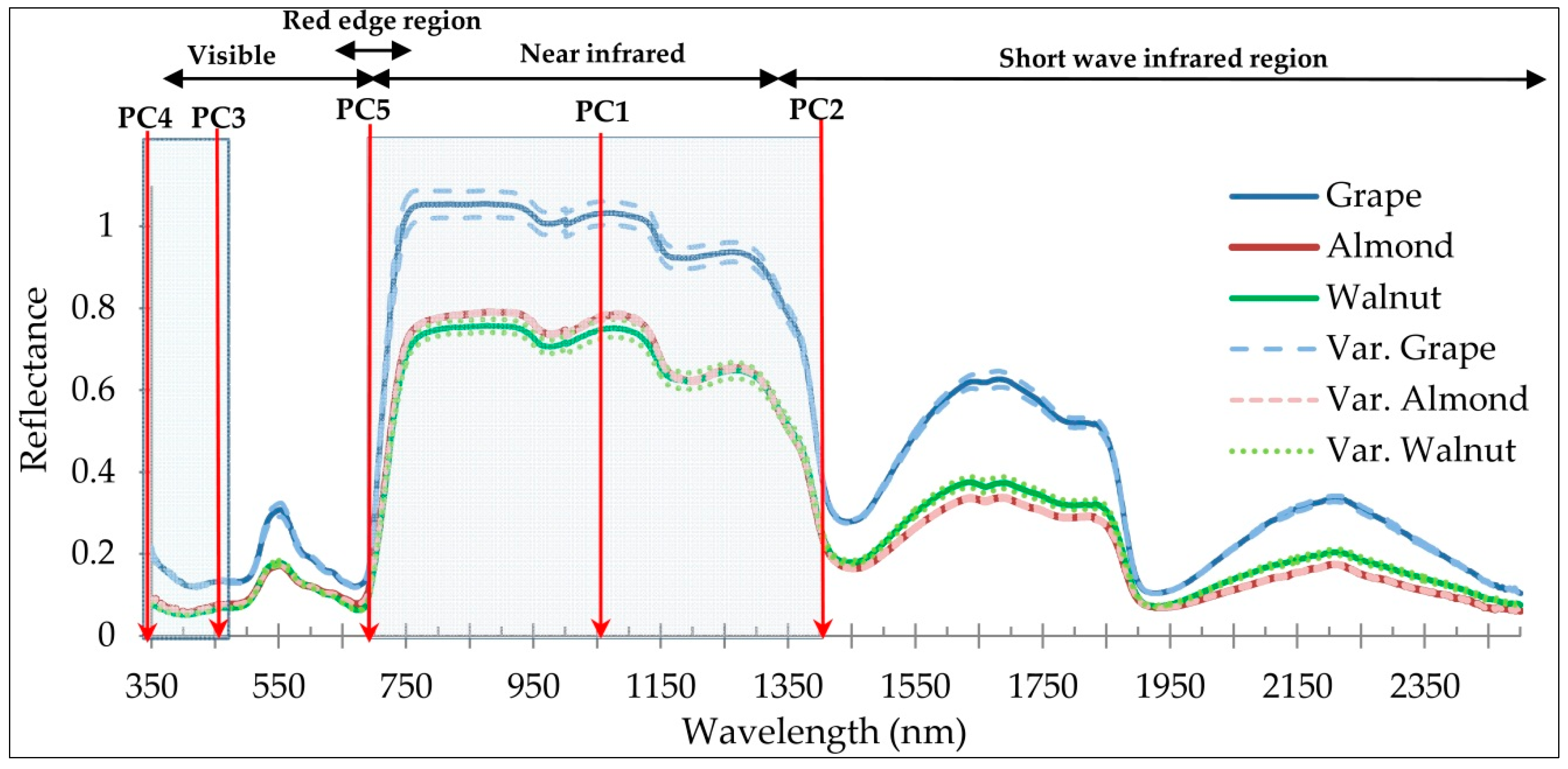
3.1. First Method: ANOVA–RFC–PCA
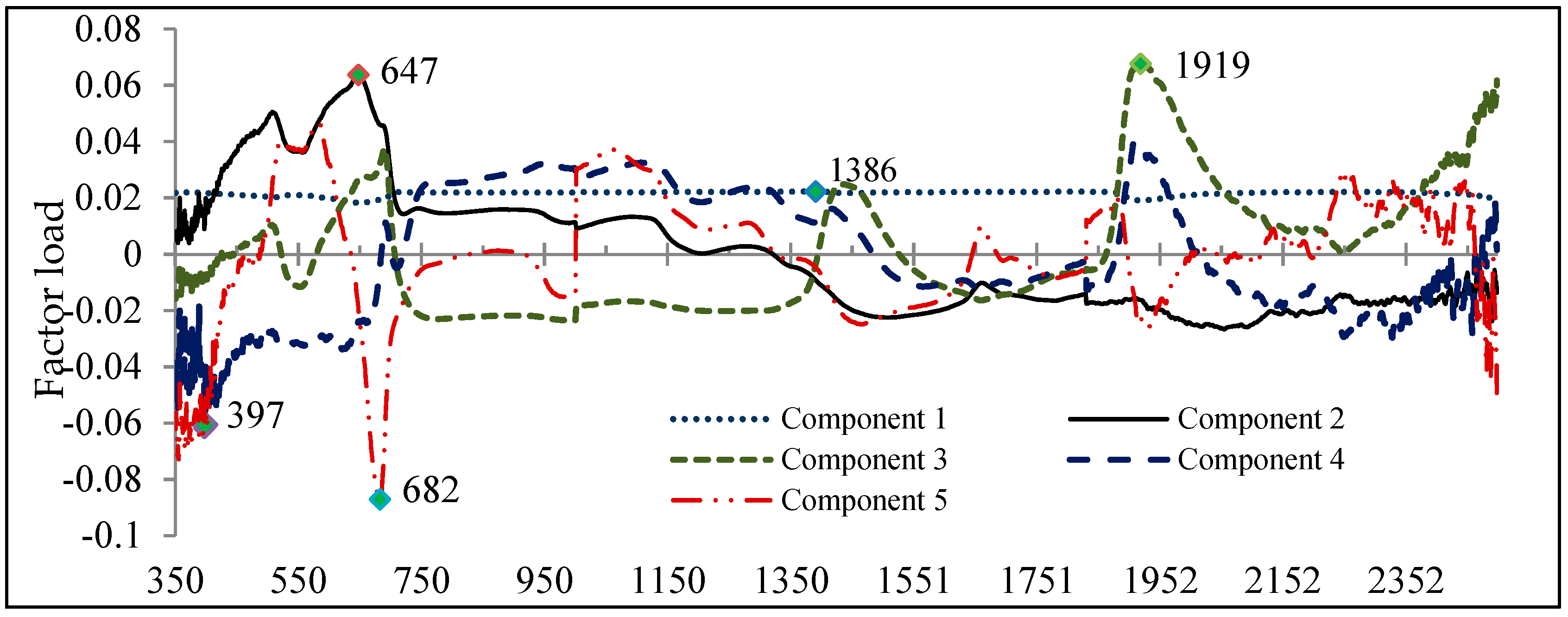
3.2. Second Method: PLS
3.3. Accuracy Assessment
4. Discussion
5. Conclusions and Recommendations
- The combination of ANOVA, RFC, and PCA can reduce the complex dimension of hyperspectral remote sensing data.
- The near infrared and red edge regions have played important roles in the introduction of optimal wavelengths for discriminating of plant species, which indicates the sensitivity and applicability of these spectral regions for discrimination targets.
- Similarity has been observed in the spectral behavior of walnut and almond species, and fewer wavelengths were able to discriminate these two species. While spectral behavior of grapes leaves was more distinctly separated from walnuts and almonds, and more wavelengths had the potential for separating grapes from almonds and walnuts.
- The PLS method showed superior and easier potential for discriminating the species as opposed to ANOVA–RFC–PCA approach.
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kent, M. Vegetation Description and Data Analysis: A Practical Approach; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar]
- Thenkabail, P.S.; Lyon, J.G. Hyperspectral Remote Sensing of Vegetation; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar]
- Li, X.; Liu, X.; Liu, M.; Wang, C.; Xia, X. A hyperspectral index sensitive to subtle changes in the canopy chlorophyll content under arsenic stress. Int. J. Appl. Earth Obs. Geoinf. 2015, 36, 41–53. [Google Scholar] [CrossRef]
- Damm, A.; Paul-Limoges, E.; Haghighi, E.; Simmer, C.; Morsdorf, F.; Schneider, F.D.; van der Tol, C.; Migliavacca, M.; Rascher, U. Remote sensing of plant-water relations: An overview and future perspectives. J. Plant Physiol. 2018, 227, 3–19. [Google Scholar] [CrossRef] [PubMed]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: A review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar]
- Song, X.-P.; Potapov, P.; Adusei, B.; King, L.; Khan, A.; Krylov, A.; Di Bella, C.M.; Pickens, A.; Stehman, S.V.; Hansen, M. National-scale crop type mapping and area estimation using multi-resolution remote sensing and field survey. In Proceedings of the AGU Fall Meeting Abstracts, San Francisco, CA, USA, 12–16 December 2016. [Google Scholar]
- Zarco-Tejada, P.J.; González-Dugo, M.; Fereres, E. Seasonal stability of chlorophyll fluorescence quantified from airborne hyperspectral imagery as an indicator of net photosynthesis in the context of precision agriculture. Remote Sens. Environ. 2016, 179, 89–103. [Google Scholar] [CrossRef]
- Fagan, M.; DeFries, R.; Sesnie, S.; Arroyo-Mora, J.; Soto, C.; Singh, A.; Townsend, P.; Chazdon, R. Mapping species composition of forests and tree plantations in Northeastern Costa Rica with an integration of hyperspectral and multitemporal Landsat imagery. Remote Sens. 2015, 7, 5660–5696. [Google Scholar]
- Lopatin, J.; Fassnacht, F.E.; Kattenborn, T.; Schmidtlein, S. Mapping plant species in mixed grassland communities using close range imaging spectroscopy. Remote Sens. Environ. 2017, 201, 12–23. [Google Scholar] [CrossRef]
- Tesfamichael, S.G.; Newete, S.W.; Adam, E.; Dubula, B. Field spectroradiometer and simulated multispectral bands for discriminating invasive species from morphologically similar cohabitant plants. GISci. Remote Sens. 2018, 55, 417–436. [Google Scholar]
- Cochrane, M. Using vegetation reflectance variability for species level classification of hyperspectral data. Int. J. Remote Sens. 2000, 21, 2075–2087. [Google Scholar] [CrossRef]
- Laurin, G.V.; Puletti, N.; Hawthorne, W.; Liesenberg, V.; Corona, P.; Papale, D.; Chen, Q.; Valentini, R. Discrimination of tropical forest types, dominant species, and mapping of functional guilds by hyperspectral and simulated multispectral Sentinel-2 data. Remote Sens. Environ. 2016, 176, 163–176. [Google Scholar]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar]
- Schmidt, K.; Skidmore, A. Spectral discrimination of vegetation types in a coastal wetland. Remote Sens. Environ. 2003, 85, 92–108. [Google Scholar] [CrossRef]
- Jia, K.; Wu, B.; Tian, Y.; Zeng, Y.; Li, Q. Vegetation classification method with biochemical composition estimated from remote sensing data. Int. J. Remote Sens. 2011, 32, 9307–9325. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E.; Kellner, J.R.; Wright, S.J. Operational tree species mapping in a diverse tropical forest with airborne imaging spectroscopy. PLoS ONE 2015, 10, e0118403. [Google Scholar] [CrossRef] [PubMed]
- Prospere, K.; McLaren, K.; Wilson, B. Plant species discrimination in a tropical wetland using in situ hyperspectral data. Remote Sens. 2014, 6, 8494–8523. [Google Scholar] [CrossRef] [Green Version]
- Mirzaei, M.; Marofi, S.; Abbasi, M.; Solgi, E.; Karimi, R.; Verrelst, J. Scenario-based discrimination of common grapevine varieties using in-field hyperspectral data in the western of Iran. Int. J. Appl. Earth Obs. Geoinf. 2019, 80, 26–37. [Google Scholar] [CrossRef]
- Rischbeck, P.; Elsayed, S.; Mistele, B.; Barmeier, G.; Heil, K.; Schmidhalter, U. Data fusion of spectral, thermal and canopy height parameters for improved yield prediction of drought stressed spring barley. Eur. J. Agron. 2016, 78, 44–59. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.; Camino, C.; Beck, P.; Calderon, R.; Hornero, A.; Hernández-Clemente, R.; Kattenborn, T.; Montes-Borrego, M.; Susca, L.; Morelli, M. Previsual symptoms of Xylella fastidiosa infection revealed in spectral plant-trait alterations. Nat. Plants 2018, 4, 432. [Google Scholar] [CrossRef]
- Clevers, J.G.; Kooistra, L.; Schaepman, M.E. Estimating canopy water content using hyperspectral remote sensing data. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, 119–125. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Peng, Y.; Viña, A.; Arkebauer, T.; Schepers, J.S. Efficiency of chlorophyll in gross primary productivity: A proof of concept and application in crops. J. Plant Physiol. 2016, 201, 101–110. [Google Scholar] [CrossRef] [Green Version]
- Koch, B. Status and future of laser scanning, synthetic aperture radar and hyperspectral remote sensing data for forest biomass assessment. ISPRS J. Photogramm. Remote Sens. 2010, 65, 581–590. [Google Scholar] [CrossRef]
- Cordon, G.; Lagorio, M.G.; Paruelo, J.M. Chlorophyll fluorescence, photochemical reflective index and normalized difference vegetative index during plant senescence. J. Plant Physiol. 2016, 199, 100–110. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guo, B.-B.; Qi, S.-L.; Heng, Y.-R.; Duan, J.-Z.; Zhang, H.-Y.; Wu, Y.-P.; Feng, W.; Xie, Y.-X.; Zhu, Y.-J. Remotely assessing leaf N uptake in winter wheat based on canopy hyperspectral red-edge absorption. Eur. J. Agron. 2017, 82, 113–124. [Google Scholar] [CrossRef]
- Zhang, M.; Qin, Z.; Liu, X.; Ustin, S.L. Detection of stress in tomatoes induced by late blight disease in California, USA, using hyperspectral remote sensing. Int. J. Appl. Earth Obs. Geoinf. 2003, 4, 295–310. [Google Scholar] [CrossRef]
- Fedenko, V.S.; Shemet, S.A.; Landi, M. UV–vis spectroscopy and colorimetric models for detecting anthocyanin-metal complexes in plants: An overview of in vitro and in vivo techniques. J. Plant Physiol. 2017, 212, 13–28. [Google Scholar] [CrossRef] [PubMed]
- Liu, M.; Liu, X.; Ding, W.; Wu, L. Monitoring stress levels on rice with heavy metal pollution from hyperspectral reflectance data using wavelet-fractal analysis. Int. J. Appl. Earth Obs. Geoinf. 2011, 13, 246–255. [Google Scholar] [CrossRef]
- Mirzaei, M.; Verrelst, J.; Marofi, S.; Abbasi, M.; Azadi, H. Eco-Friendly Estimation of Heavy Metal Contents in Grapevine Foliage Using In-Field Hyperspectral Data and Multivariate Analysis. Remote Sens. 2019, 11, 2731. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.A.; Merzlyak, M.N. Signature analysis of leaf reflectance spectra: Algorithm development for remote sensing of chlorophyll. J. Plant Physiol. 1996, 148, 494–500. [Google Scholar] [CrossRef]
- Goswami, S.; Matharasi, K. Development of a Web-Based Vegetation Spectral Library (VSL) for Remote Sensing Research and Applications; PeerJ PrePrints: San Diego, CA, USA, 2015; pp. 2167–9843. [Google Scholar]
- Jiménez, M.; Díaz-Delgado, R. Towards a standard plant species spectral library protocol for vegetation mapping: A case study in the Shrubland of Doñana National Park. ISPRS Int. J. Geo-Inf. 2015, 4, 2472–2495. [Google Scholar] [CrossRef] [Green Version]
- Gomez, C.; Rossel, R.A.V.; McBratney, A.B. Soil organic carbon prediction by hyperspectral remote sensing and field vis-NIR spectroscopy: An Australian case study. Geoderma 2008, 146, 403–411. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O. Spectral discrimination of papyrus vegetation (Cyperus papyrus L.) in swamp wetlands using field spectrometry. ISPRS J. Photogramm. Remote Sens. 2009, 64, 612–620. [Google Scholar] [CrossRef]
- Ye, X.; Sakai, K.; Sasao, A.; Asada, S.-I. Potential of airborne hyperspectral imagery to estimate fruit yield in citrus. Chemom. Intell. Lab. Syst. 2008, 90, 132–144. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectral and chemical analysis of tropical forests: Scaling from leaf to canopy levels. Remote Sens. Environ. 2008, 112, 3958–3970. [Google Scholar] [CrossRef]
- Yao, Z.; Sakai, K.; Ye, X.; Akita, T.; Iwabuchi, Y.; Hoshino, Y. Airborne hyperspectral imaging for estimating acorn yield based on the PLS B-matrix calibration technique. Ecol. Inform. 2008, 3, 237–244. [Google Scholar] [CrossRef]
- Barmeier, G.; Hofer, K.; Schmidhalter, U. Mid-season prediction of grain yield and protein content of spring barley cultivars using high-throughput spectral sensing. Eur. J. Agron. 2017, 90, 108–116. [Google Scholar] [CrossRef]
- Soriano-Disla, J.M.; Janik, L.J.; Forrester, S.T.; Grocke, S.F.; Fitzpatrick, R.W.; McLaughlin, M.J. The use of mid-infrared diffuse reflectance spectroscopy for acid sulfate soil analysis. Sci. Total Environ. 2019, 646, 1489–1502. [Google Scholar] [CrossRef]
- Diago, M.P.; Fernandes, A.M.; Millan, B.; Tardáguila, J.; Melo-Pinto, P. Identification of grapevine varieties using leaf spectroscopy and partial least squares. Comput. Electron. Agric. 2013, 99, 7–13. [Google Scholar] [CrossRef]
- Páscoa, R.; Lopo, M.; dos Santos, C.T.; Graça, A.; Lopes, J. Exploratory study on vineyards soil mapping by visible/near-infrared spectroscopy of grapevine leaves. Comput. Electron. Agric. 2016, 127, 15–25. [Google Scholar] [CrossRef]
- Nguyen, D.V.; Rocke, D.M. On partial least squares dimension reduction for microarray-based classification: A simulation study. Comput. Stat. Data Anal. 2004, 46, 407–425. [Google Scholar] [CrossRef]
- Li, F.; Mistele, B.; Hu, Y.; Chen, X.; Schmidhalter, U. Reflectance estimation of canopy nitrogen content in winter wheat using optimised hyperspectral spectral indices and partial least squares regression. Eur. J. Agron. 2014, 52, 198–209. [Google Scholar] [CrossRef]
- Preisner, O.; Lopes, J.A.; Menezes, J.C. Uncertainty assessment in FT-IR spectroscopy based bacteria classification models. Chemom. Intell. Lab. Syst. 2008, 94, 33–42. [Google Scholar] [CrossRef]
- Aneece, I.; Epstein, H. Identifying invasive plant species using field spectroscopy in the VNIR region in successional systems of north-central Virginia. Int. J. Remote Sens. 2017, 38, 100–122. [Google Scholar] [CrossRef]
- Mureriwa, N.; Adam, E.; Sahu, A.; Tesfamichael, S. Examining the spectral separability of Prosopis glandulosa from co-existent species using field spectral measurement and guided regularized random forest. Remote Sens. 2016, 8, 144. [Google Scholar] [CrossRef] [Green Version]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
- Christian, B.; Krishnayya, N. Classification of tropical trees growing in a sanctuary using Hyperion (EO-1) and SAM algorithm. Curr. Sci. 2009, 1601–1607. [Google Scholar]
- Dudley, K.L.; Dennison, P.E.; Roth, K.L.; Roberts, D.A.; Coates, A.R. A multi-temporal spectral library approach for mapping vegetation species across spatial and temporal phenological gradients. Remote Sens. Environ. 2015, 167, 121–134. [Google Scholar] [CrossRef]
- Okujeni, A.; Canters, F.; Cooper, S.D.; Degerickx, J.; Heiden, U.; Hostert, P.; Priem, F.; Roberts, D.A.; Somers, B.; van der Linden, S. Generalizing machine learning regression models using multi-site spectral libraries for mapping vegetation-impervious-soil fractions across multiple cities. Remote Sens. Environ. 2018, 216, 482–496. [Google Scholar] [CrossRef]
- Verrelst, J.; Schaepman, M.E.; Malenovský, Z.; Clevers, J.G. Effects of woody elements on simulated canopy reflectance: Implications for forest chlorophyll content retrieval. Remote Sens. Environ. 2010, 114, 647–656. [Google Scholar] [CrossRef] [Green Version]
- Schaepman, M.E.; Koetz, B.; Schaepman-Strub, G.; Itten, K.I. Spectrodirectional remote sensing for the improved estimation of biophysical and-chemical variables: Two case studies. Int. J. Appl. Earth Obs. Geoinf. 2005, 6, 271–282. [Google Scholar] [CrossRef]
- Mirzaei, M.; Marofi, S.; Solgi, E.; Abbasi, M.; Karimi, R.; Bakhtyari, H.R.R. Ecological and health risks of soil and grape heavy metals in long-term fertilized vineyards (Chaharmahal and Bakhtiari province of Iran). Environ. Geochem. Health 2019, 1–17. [Google Scholar] [CrossRef]
- Mirzaei, M.; Marofi, S.; Abbasi, M.; Karimi, R. Evaluation of heavy metal contamination ecological risk in a food-producing Ecosystem. J. Health Res. Commun. 2017, 3, 1–16. [Google Scholar]
- Klančnik, K.; Gaberščik, A. Leaf spectral signatures differ in plant species colonizing habitats along a hydrological gradient. J. Plant Ecol. 2015, 9, 442–450. [Google Scholar] [CrossRef]
- Spectroradiometer, H. User’s Guide Version 4.05; Analytical Spectral Devices: Boulder, CO, USA, 2005. [Google Scholar]
- Kumar, L.; Schmidt, K.; Dury, S.; Skidmore, A. Review of hyperspectral remote sensing and vegetation science. In Imaging Spectrometry: Basic Principles and Prospective Applications; Kluwer Academic Press: Dordrecht, The Netherlands, 2001; pp. 111–155. [Google Scholar]
- Strobl, C.; Boulesteix, A.-L.; Kneib, T.; Augustin, T.; Zeileis, A. Conditional variable importance for random forests. BMC Bioinform. 2008, 9, 307. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Wang, Y.; Huang, H.-Y.; Zuo, Z.-T.; Wang, Y.-Z. Comprehensive quality assessment of Dendrubium officinale using ATR-FTIR spectroscopy combined with random forest and support vector machine regression. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 205, 637–648. [Google Scholar] [CrossRef] [PubMed]
- Mirzaei, M.; Jafari, A.; Gholamalifard, M.; Azadi, H.; Shooshtari, S.J.; Moghaddam, S.M.; Gebrehiwot, K.; Witlox, F. Mitigating environmental risks: Modeling the interaction of water quality parameters and land use cover. Land Use Policy 2019. [Google Scholar] [CrossRef]
- Shrestha, S.; Kazama, F. Assessment of surface water quality using multivariate statistical techniques: A case study of the Fuji river basin, Japan. Environ. Model. Softw. 2007, 22, 464–475. [Google Scholar] [CrossRef]
- Chen, S. Principal Component Analysis of Geochemical Data from the REE-Rich Maw Zone; Citeseer: Athabasca Basin, AB, Canada, 2015. [Google Scholar]
- Olawale, F.; Garwe, D. Obstacles to the growth of new SMEs in South Africa: A principal component analysis approach. Afr. J. Bus. Manag. 2010, 4, 729–738. [Google Scholar]
- Lehmann, J.; Große-Stoltenberg, A.; Römer, M.; Oldeland, J. Field spectroscopy in the VNIR-SWIR region to discriminate between Mediterranean native plants and exotic-invasive shrubs based on leaf tannin content. Remote Sens. 2015, 7, 1225–1241. [Google Scholar] [CrossRef] [Green Version]
- Mirzayi, M.; Riyahi Bakhtiyari, A.; Salman Mahini, A. Analysis of the physical and chemical quality of Mazandaran province (Iran) rivers using multivariate statistical methods. J. Maz. Univ. Med. Sci. 2014, 23, 41–52. [Google Scholar]
- Worley, B.; Powers, R. Multivariate analysis in metabolomics. Curr. Metabolomics 2013, 1, 92–107. [Google Scholar] [PubMed]
- Abbasi, M.; Darvishsefat, A.; Schaepman, M.; Sobhani, H.; Shirvany, N.; SHABESTANI, S. Spectral reflectance differences of Alnus subcordata, Quercus castaneifolia and Parrotia persica leaves based on nitrogen content using PLS regression. J. For. Wood Prod. 2012, 64, 399–417. [Google Scholar]
- Belton, P.S.; Colquhoun, I.J.; Kemsley, E.K.; Delgadillo, I.; Roma, P.; Dennis, M.J.; Sharman, M.; Holmes, E.; Nicholson, J.K.; Spraul, M. Application of chemometrics to the 1H NMR spectra of apple juices: Discrimination between apple varieties. Food Chem. 1998, 61, 207–213. [Google Scholar] [CrossRef]
- Vaiphasa, C.; Ongsomwang, S.; Vaiphasa, T.; Skidmore, A.K. Tropical mangrove species discrimination using hyperspectral data: A laboratory study. Estuar. Coast. Shelf Sci. 2005, 65, 371–379. [Google Scholar] [CrossRef]
- Belluco, E.; Camuffo, M.; Ferrari, S.; Modenese, L.; Silvestri, S.; Marani, A.; Marani, M. Mapping salt-marsh vegetation by multispectral and hyperspectral remote sensing. Remote Sens. Environ. 2006, 105, 54–67. [Google Scholar] [CrossRef]
- Raynolds, M.K.; Walker, D.A.; Epstein, H.E.; Pinzon, J.E.; Tucker, C.J. A new estimate of tundra-biome phytomass from trans-Arctic field data and AVHRR NDVI. Remote Sens. Lett. 2012, 3, 403–411. [Google Scholar] [CrossRef]
- Bratsch, S.; Epstein, H.; Buchhorn, M.; Walker, D. Differentiating among four Arctic tundra plant communities at Ivotuk, Alaska using field spectroscopy. Remote Sens. 2016, 8, 51. [Google Scholar] [CrossRef] [Green Version]
- Thenkabail, P.S.; Enclona, E.A.; Ashton, M.S.; Van Der Meer, B. Accuracy assessments of hyperspectral waveband performance for vegetation analysis applications. Remote Sens. Environ. 2004, 91, 354–376. [Google Scholar] [CrossRef]
- Mutanga, O.; Skidmore, A.K. Red edge shift and biochemical content in grass canopies. ISPRS J. Photogramm. Remote Sens. 2007, 62, 34–42. [Google Scholar] [CrossRef]
- Ham, J.; Chen, Y.; Crawford, M.M.; Ghosh, J. Investigation of the random forest framework for classification of hyperspectral data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 492–501. [Google Scholar] [CrossRef] [Green Version]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (RandomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Chan, J.C.-W.; Paelinckx, D. Evaluation of Random Forest and Adaboost tree-based ensemble classification and spectral band selection for ecotope mapping using airborne hyperspectral imagery. Remote Sens. Environ. 2008, 112, 2999–3011. [Google Scholar] [CrossRef]
- Asner, G.P. Biophysical and biochemical sources of variability in canopy reflectance. Remote Sens. Environ. 1998, 64, 234–253. [Google Scholar] [CrossRef]
- Galet, P. A Practical Ampelography; Cornell University Press: Ithaca, NY, USA, 1979. [Google Scholar]
- Cervera, M.-T.; Cabezas, J.A.; Sancha, J.; De Toda, F.M.; Martínez-Zapater, J.M. Application of AFLPs to the characterization of grapevine Vitis vinifera L. genetic resources. A case study with accessions from Rioja (Spain). Theor. Appl. Genet. 1998, 97, 51–59. [Google Scholar] [CrossRef] [Green Version]
- Johansen, K.; Raharjo, T.; McCabe, M. Using multi-spectral UAV imagery to extract tree crop structural properties and assess pruning effects. Remote Sens. 2018, 10, 854. [Google Scholar] [CrossRef] [Green Version]
| Wavelength | Principal Components | ||||
|---|---|---|---|---|---|
| PC1 | PC2 | PC3 | PC4 | PC5 | |
| 363 | 0.51 | 0.52 | 0.67 | 0.13 | −0.01 |
| 364 | 0.49 | 0.55 | 0.66 | 0.12 | −0.02 |
| 374 | 0.54 | 0.52 | 0.65 | 0.11 | −0.01 |
| 422 | 0.54 | 0.42 | 0.72 | −0.08 | 0.01 |
| 423 | 0.53 | 0.43 | 0.73 | −0.08 | −0.01 |
| 721 | 0.60 | 0.49 | 0.59 | −0.05 | 0.2 |
| 718 | 0.61 | 0.5 | 0.58 | −0.01 | 0.17 |
| 739 | 0.61 | 0.51 | 0.58 | −0.01 | 0.16 |
| 1053 | 0.72 | 0.52 | 0.47 | −0.05 | 0 |
| 1064 | 0.72 | 0.52 | 0.47 | −0.05 | 0 |
| 1077 | 0.72 | 0.51 | 0.47 | −0.05 | 0 |
| 1379 | 0.49 | 0.72 | 0.46 | −0.03 | 0.08 |
| 1388 | 0.50 | 0.72 | 0.46 | −0.02 | 0.08 |
| 1390 | 0.50 | 0.72 | 0.48 | −0.02 | 0.07 |
| Feature Selection Method | Selected Inputs | Train | Test |
|---|---|---|---|
| OAA % | OAA % | ||
| ANOVA–RFC–PCA | B363, B423, B721, B1064, B1388 | 100 | 95.6 |
| PLS | B397, B515, B647, B682, B1386, B1919 | 100 | 100 |
| Spectral Regions (nm) | Reference | Optimal Selected Wavelengths (nm) |
|---|---|---|
| Visible region (350–700) | This study | 363, 423 |
| Schmidt and Skidmore [14] | 404, 428 | |
| Adam and Mutanga [34] | No wavelength | |
| Aneece and Epstein [45] | 350 to 399 and 500 to 549 | |
| Red edge region (680–750) | This study | 721 |
| Schmidt and Skidmore [14] | No wavelength | |
| Vaiphasa et al. [71] | 720 | |
| Adam and Mutanga [34] | 745, 746 | |
| Lehmann et al. [66] | 675 to 710 | |
| Aneece and Epstein [45] | 700 to 749 | |
| Near infrared region (700–1300) | This study | 1064, 1388 |
| Schmidt and Skidmore [14] | 771 | |
| Adam and Mutanga [34] | 892, 932, 934, 958, 961, and 989 | |
| Aneece and Epstein [45] | 900 to 949 | |
| Short wave infrared (1300–2500) | This study | No wavelength |
| Schmidt and Skidmore [14] | 1398, 1803, and 2183 | |
| Adam and Mutanga [34] | No wavelength | |
| Lehmann et al. [66] | 1360 to 1450 and 1630 to 1740 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abbasi, M.; Verrelst, J.; Mirzaei, M.; Marofi, S.; Riyahi Bakhtiari, H.R. Optimal Spectral Wavelengths for Discriminating Orchard Species Using Multivariate Statistical Techniques. Remote Sens. 2020, 12, 63. https://doi.org/10.3390/rs12010063
Abbasi M, Verrelst J, Mirzaei M, Marofi S, Riyahi Bakhtiari HR. Optimal Spectral Wavelengths for Discriminating Orchard Species Using Multivariate Statistical Techniques. Remote Sensing. 2020; 12(1):63. https://doi.org/10.3390/rs12010063
Chicago/Turabian StyleAbbasi, Mozhgan, Jochem Verrelst, Mohsen Mirzaei, Safar Marofi, and Hamid Reza Riyahi Bakhtiari. 2020. "Optimal Spectral Wavelengths for Discriminating Orchard Species Using Multivariate Statistical Techniques" Remote Sensing 12, no. 1: 63. https://doi.org/10.3390/rs12010063