Fusarium Wilt of Radish Detection Using RGB and Near Infrared Images from Unmanned Aerial Vehicles
Abstract
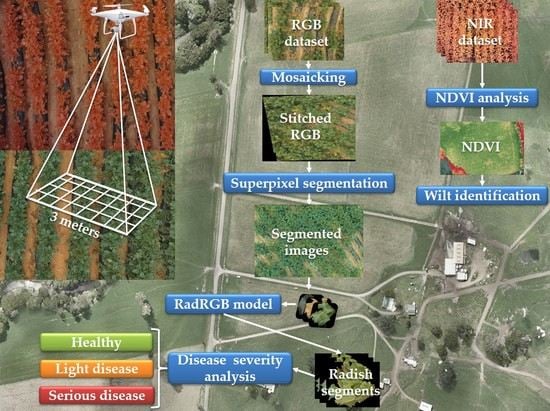
:1. Introduction
- Collect a high-resolution radish field dataset that contains both regular RGB images and multispectral NIR images.
- Introduce a customized CNN model to precisely classify radish, mulching film, and soil in RGB images.
- Apply a series of image processing methods to identify different stages of the wilt disease.
- Comprehensively compare the NIR-based approach and the conventional RGB-based approach.
2. Radish Field Dataset
2.1. Data Collection
2.2. Description of Training Data
3. Methodology
3.1. Radish Wilt Detection Using RGB Dataset
3.1.1. Mosaicking Technique (Stitching Technique)
3.1.2. Radish Field Segmentation
3.1.3. Detailed Implementation of the RadRGB Model
3.1.4. Disease Severity Analysis
| Algorithm 1: Wilt Severity Analysis |
| 0: Initialize count_seg_black, count_white equals to 0 and total_pixel equals to total number of pixels in img; 1: for in img do 2: if equals to 0 then 3: 1 4: end if 5: end for 6: adaptiveThreshold(img, blockSize, C) 7: for in bin_img do 8: if equals to 255 then 9: 10: end if 11: end for 12: |
3.2. Radish Wilt Detection Using NIR Dataset
4. Experimental Results
4.1. Experiments on RGB Dataset
4.2. Experiments on the NIR Dataset
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lee, G.-G.; Lee, H.-W.; Lee, J.-H. Greenhouse gas emission reduction effect in the transportation sector by urban agriculture in Seoul, Korea. Landsc. Urban Plan. 2015, 140, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Ha, J.G.; Moon, H.; Kwak, J.T.; Hassan, S.I.; Dang, L.M.; Lee, O.N.; Park, H.Y. Deep convolutional neural network for classifying Fusarium wilt of radish from unmanned aerial vehicles. J. Appl. Remote Sens. 2017, 11, 042621. [Google Scholar] [CrossRef]
- Dang, L.M.; Hassan, S.I.; Suhyeon, I.; Sangaiah, A.K.; Mehmood, I.; Rho, S.; Seo, S.; Moon, H.; Syed, I.H. UAV based wilt detection system via convolutional neural networks. Sustain. Comput. Inform. Syst. 2018. [Google Scholar] [CrossRef] [Green Version]
- Drapikowska, M.; Drapikowski, P.; Borowiak, K.; Hayes, F.; Harmens, H.; Dziewiątka, T.; Byczkowska, K. Application of novel image base estimation of invisible leaf injuries in relation to morphological and photosynthetic changes of Phaseolus vulgaris L. exposed to tropospheric ozone. Atmos. Pollut. Res. 2016, 7, 1065–1071. [Google Scholar] [CrossRef]
- Khirade, S.D.; Patil, A. Plant disease detection using image processing. In Proceedings of the 2015 International Conference on Computing Communication Control and Automation, Maharashtra, India, 26–27 February 2015. [Google Scholar]
- Singh, V.; Misra, A.K. Detection of plant leaf diseases using image segmentation and soft computing techniques. Inf. Process. Agric. 2017, 4, 41–49. [Google Scholar] [CrossRef] [Green Version]
- Dang, L.M.; Piran, J.; Han, D.; Min, K.; Moon, H. A Survey on Internet of Things and Cloud Computing for Healthcare. Electronics 2019, 8, 768. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Reddy, K.N.; Fletcher, R.S.; Pennington, D. UAV low-altitude remote sensing for precision weed management. Weed Technol. 2018, 32, 2–6. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B.; et al. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Wang, H.; Dang, L.M.; Sadeghi-Niaraki, A.; Moon, H. Crop pest recognition in natural scenes using convolutional neural networks. Comput. Electron. Agric. 2020, 169, 105174. [Google Scholar] [CrossRef]
- Zhao, J.; Zhang, X.; Gao, C.; Qiu, X.; Tian, Y.; Zhu, Y.; Cao, W. Rapid Mosaicking of Unmanned Aerial Vehicle (UAV) Images for Crop Growth Monitoring Using the SIFT Algorithm. Remote Sens. 2019, 11, 1226. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Ji, Q. Robust facial landmark detection under significant head poses and occlusion. In Proceedings of the IEEE International Conference on Computer Vision, Santiago, Chile, 7–13 December 2015. [Google Scholar]
- Redmon, J.; Farhadi, A. Yolov3: An Incremental Improvement. Retrieved Sept. 2018, 17, 1–6. [Google Scholar]
- Ren, S.; He, K.; Girshick, R.; Sun, J. Faster R-CNN: Towards real-time object detection with region proposal networks. In Proceedings of the Advances in Neural Information Processing Systems 2015, Montreal, QC, Canada, 7–12 December 2015. [Google Scholar]
- Li, Y.; Hou, X.; Koch, C.; Rehg, J.M.; Yuille, A.L. The secrets of salient object segmentation. In Proceedings of the 27th IEEE Conference on Computer Vision and Pattern Recognition, Columbus, OH, USA, 24–27 June 2014. [Google Scholar]
- Ren, X.; Malik, J. Learning a classification model for segmentation. In Proceedings of the Ninth IEEE International Conference on Computer Vision, Nice, France, 13–16 October 2003. [Google Scholar]
- Felzenszwalb, P.F.; Huttenlocher, D.P. Efficient graph-based image segmentation. Int. J. Comput. Vis. 2004, 59, 167–181. [Google Scholar] [CrossRef]
- Comaniciu, D.; Meer, P. Mean shift: A robust approach toward feature space analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef] [Green Version]
- Shi, J.; Malik, J. Normalized cuts and image segmentation. IEEE Trans. Pattern Anal. Mach. Intell. 2000, 22, 888–905. [Google Scholar] [CrossRef] [Green Version]
- Vedaldi, A.; Soatto, S. Quick shift and kernel methods for mode seeking. In Proceedings of the European Conference on Computer Vision, Marseille, France, 12–18 October 2008. [Google Scholar]
- Patil, J.K.; Kumar, R. Analysis of content based image retrieval FO–R plant leaf diseases using color, shape and texture features. Eng. Agric. Environ. Food 2017, 10, 69–78. [Google Scholar] [CrossRef]
- Dubey, S.R.; Jalal, A.S. Apple disease classification using color, texture and shape features from images. Signal Image Video Process. 2016, 10, 819–826. [Google Scholar] [CrossRef]
- Rançon, F.; Bombrun, L.; Keresztes, B.; Germain, C. Comparison of SIFT Encoded and Deep Learning Features for the Classification and Detection of Esca Disease in Bordeaux Vineyards. Remote Sens. 2019, 11, 1. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Wang, H.; Huang, W.; You, Z. Plant diseased leaf segmentation and recognition by fusion of superpixel, K-means and PHOG. Optik 2018, 157, 866–872. [Google Scholar] [CrossRef]
- Mohanty, S.P.; Hughes, D.P.; Salathé, M. Using deep learning for image-based plant disease detection. Front. Plant Sci. 2016, 7, 1419. [Google Scholar] [CrossRef] [Green Version]
- Nguyen, T.N.; Lee, S.; Nguyen-Xuan, H.; Lee, J. A novel analysis-prediction approach for geometrically nonlinear problems using group method of data handling. Comput. Methods Appl. Mech. Eng. 2019, 354, 506–526. [Google Scholar] [CrossRef]
- Nguyen, T.N.; Nguyen-Xuan, H.; Lee, J. A novel data-driven nonlinear solver for solid mechanics using time series forecasting. Finite Elem. Anal. Des. 2020, 171, 103377. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436. [Google Scholar] [CrossRef] [PubMed]
- Kerkech, M.; Hafiane, A.; Canals, R. Deep leaning approach with colorimetric spaces and vegetation indices for vine diseases detection in UAV images. Comput. Electron. Agric. 2018, 155, 237–243. [Google Scholar] [CrossRef]
- Albetis, J.; Duthoit, S.; Guttler, F.; Jacquin, A.; Goulard, M.; Poilvé, H.; Féret, J.-B.; Dedieu, G. Detection of Flavescence dorée grapevine disease using unmanned aerial vehicle (UAV) multispectral imagery. Remote Sens. 2017, 9, 308. [Google Scholar] [CrossRef] [Green Version]
- Dash, J.P.; Watt, M.S.; Pearse, G.D.; Heaphy, M.; Dungey, H.S. Assessing very high resolution UAV imagery for monitoring forest health during a simulated disease outbreak. ISPRS J. Photogramm. Remote Sens. 2017, 131, 1–14. [Google Scholar] [CrossRef]
- Barbedo, J.G.A. A Review on the Use of Unmanned Aerial Vehicles and Imaging Sensors for Monitoring and Assessing Plant Stresses. Drones 2019, 3, 40. [Google Scholar] [CrossRef] [Green Version]
- Wierzbicki, D.; Kedzierski, M.; Fryskowska, A.; Jasinski, J. Quality Assessment of the Bidirectional Reflectance Distribution Function for NIR Imagery Sequences from UAV. Remote Sens. 2018, 10, 1348. [Google Scholar] [CrossRef] [Green Version]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Gandhi, G.M.; Parthiban, S.; Thummalu, N.; Christy, A. NDVI: Vegetation change detection using remote sensing and GIS—A case study of Vellore District. Procedia Comput. Sci. 2015, 57, 1199–1210. [Google Scholar] [CrossRef] [Green Version]
- De Castro, A.I.; Ehsani, R.; Ploetz, R.C.; Crane, J.H.; Buchanon, S. Detection of laurel wilt disease in avocado using low altitude aerial imaging. PLoS ONE 2015, 10, e0124642. [Google Scholar] [CrossRef]
- Brown, M. Automatic Panoramic Image Stitching Using Invariant Features. Int. J. Comput. Vis. 2016, 74.1, 59–73. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Chen, J. Superpixel segmentation using linear spectral clustering. In Proceedings of the 28th IEEE Conference on Computer Vision and Pattern Recognition, Boston, MA, USA, 7–12 June 2015. [Google Scholar]
- Li, Y.; Yuan, Y. Convergence analysis of two-layer neural networks with relu activation. In Proceedings of the Advances in Neural Information Processing Systems, Long Beach, CA, USA, 4–9 December 2017; pp. 597–607. [Google Scholar]
- Wang, H.; Li, Y.; Dang, L.M.; Ko, J.; Han, D.; Moon, H. Smartphone-based bulky waste classification using convolutional neural networks. Multimed. Tools Appl. 2020, 79, 1–21. [Google Scholar] [CrossRef]
- Yang, H.; Yang, X.; Heskel, M.A.; Sun, S.; Tang, J. Seasonal variations of leaf and canopy properties tracked by ground-based NDVI imagery in a temperate forest. Sci. Rep. 2017, 7, 1267. [Google Scholar] [CrossRef] [PubMed]













| Position (Latitude, Longitude) | Date | #Fields | Altitude (m) | #RGB Images | #NIR Images |
|---|---|---|---|---|---|
| 33°30′5.44″ N, 126°51′25.39″ E | 1 May 2018 | 2 | 3 | 627 | 406 |
| 7 | 434 | 267 | |||
| 15 | 207 | 187 | |||
| 2 May 2018 | 3 | 853 | 535 | ||
| 7 | 431 | 368 | |||
| 15 | 262 | 234 | |||
| Total number of images | 2814 | 1997 | |||
| RadRGB | ||
|---|---|---|
| Name | Structure | (Widths, Heights, Channels) |
| Input | 64 × 64 | |
| Convolution_1 | 7 × 7 | (58, 58, 32) |
| Convolution_2 | 5 × 5 | (54, 54, 64) |
| Maxpool_1 | 2 × 2 | (27, 27, 64) |
| Dropout_1 | Probability: 0.2 | |
| Convolution_3 | 3 × 3 | (25, 25, 128) |
| Maxpool_2 | 2 × 2 | (12, 12, 128) |
| Dropout_2 | Probability: 0.2 | |
| Convolution_4 | 3 × 3 | (10, 10, 256) |
| Maxpool_3 | 2 × 2 | (5, 5, 256) |
| Dropout_3 | Probability: 0.2 | |
| Convolution_5 | 3 × 3 | (3, 3, 512) |
| BatchNorm | (3, 3, 512) | |
| Dropout_4 | Probability: 0.5 | |
| Flatten | (4068) | |
| Dense | (3) | |
| Subset | Training Set | Testing Set | |
|---|---|---|---|
| Class | |||
| Radish | 510 | 90 | |
| Soil | 493 | 87 | |
| Plastic mulch | 442 | 78 | |
| Total | 1445 | 255 | |
| Radish | Soil | Plastic Mulch | |
|---|---|---|---|
| Radish | 87 | 3 | 1 |
| Soil | 1 | 84 | 2 |
| Plastic mulch | 2 | 0 | 75 |
| Accuracy (%) | 96.6 | 96.5 | 96.1 |
| Precision | 0.967 | 0.966 | 0.962 |
| Recall | 0.956 | 0.966 | 0.974 |
| F-measure | 0.961 | 0.966 | 0.968 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dang, L.M.; Wang, H.; Li, Y.; Min, K.; Kwak, J.T.; Lee, O.N.; Park, H.; Moon, H. Fusarium Wilt of Radish Detection Using RGB and Near Infrared Images from Unmanned Aerial Vehicles. Remote Sens. 2020, 12, 2863. https://doi.org/10.3390/rs12172863
Dang LM, Wang H, Li Y, Min K, Kwak JT, Lee ON, Park H, Moon H. Fusarium Wilt of Radish Detection Using RGB and Near Infrared Images from Unmanned Aerial Vehicles. Remote Sensing. 2020; 12(17):2863. https://doi.org/10.3390/rs12172863
Chicago/Turabian StyleDang, L. Minh, Hanxiang Wang, Yanfen Li, Kyungbok Min, Jin Tae Kwak, O. New Lee, Hanyong Park, and Hyeonjoon Moon. 2020. "Fusarium Wilt of Radish Detection Using RGB and Near Infrared Images from Unmanned Aerial Vehicles" Remote Sensing 12, no. 17: 2863. https://doi.org/10.3390/rs12172863
APA StyleDang, L. M., Wang, H., Li, Y., Min, K., Kwak, J. T., Lee, O. N., Park, H., & Moon, H. (2020). Fusarium Wilt of Radish Detection Using RGB and Near Infrared Images from Unmanned Aerial Vehicles. Remote Sensing, 12(17), 2863. https://doi.org/10.3390/rs12172863