Predicting WNV Circulation in Italy Using Earth Observation Data and Extreme Gradient Boosting Model
Abstract
:1. Introduction
2. Materials and Methods
2.1. WNV Circulation Dataset (Ground Truth Data)
2.2. EO Products (LSTD, LSTN, NDVI, SSM): Sources and Preparation
2.3. Modelling
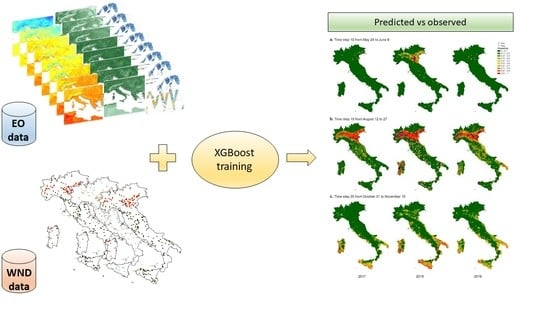
3. Results
3.1. WNV Dataset (Ground Truth Data)
3.2. EO Products (LSTD, LSTN, NDVI, SSM): Sources and Preparation
3.3. Modelling
4. Discussion
- The model classifies entomological positive cases better than birds. The worst classification in birds could be due to the fact that the coordinates used represent the place of death, rather than the place of infection and working at 250 m resolution, this aspect can affect also the classification of resident birds of target species. However, it should be noted that only 16 of the 67 bird cases had all the 3 × 3 pixels negative; in the other cases, at least one pixel was predicted as positive, showing a potential risk for the area. In addition, from the epidemiological point of view the detection of WNV in mosquito pools is clearly the best predictor, in time and space, of virus transmission. The distance of flight range can affect the correct location of virus exposure and infection in birds; as well, active movements for riding or other services can influence the exact estimation of the place of infection for horses.
- As far as the negatives are concerned, it is important to notice that only a consistent and frequent monitoring over the same area can be considered satisfying to define a true negative. In the Italian context, this occurs essentially for the entomological subset, where we have evidence of a positivity and the corresponding negativity in the previous period for the same place. In our model, we can distinguish the observed negatives in two groups (pseudo-absence in space and negatives in time, i.e., the entomological subset) and we found more accurate results in predicting negatives in time rather than pseudo-absence in space. The pseudo negatives, created randomly outside the VCA, and used to train the model, do not guarantee the real absence of the virus. The results of the model, however, show us that climatic and environmental conditions favourable to the spread of WNV can also occur in areas where the virus was not detected during 2019, although sporadically detected in the past.
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Komar, N. West Nile viral encephalitis. Rev. Sci. Tech. Int. Off. Epizoot. 2000, 19, 166–176. [Google Scholar] [CrossRef] [PubMed]
- Ecdc: West Nile Virus Infection. Available online: https://www.ecdc.europa.eu/en/west-nile-virus-infection (accessed on 23 July 2020).
- Autorino, G.L.; Battisti, A.; Deubel, V.; Ferrari, G.; Forletta, R.; Giovannini, A.; Lelli, R.; Murri, S.; Scicluna, M.T. West Nile virus Epidemic in Horses, Tuscany Region, Italy. Emerg. Infect. Dis. 2002, 8, 1372–1378. [Google Scholar] [CrossRef]
- Savini, G.; Monaco, F.; Calistri, P.; Lelli, R. Phylogenetic analysis of West Nile virus isolated in Italy in 2008. Eurosurveillance 2008, 13, 19048. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, C.; Napoli, C.; Venturi, G.; Pupella, S.; Lombardini, L.; Calistri, P.; Monaco, F.; Cagarelli, R.; Angelini, P.; Bellini, R.; et al. West Nile virus transmission: Results from the integrated surveillance system in Italy, 2008 to 2015. Eurosurveillance 2016, 21, 30340. [Google Scholar] [CrossRef] [Green Version]
- Savini, G.; Capelli, G.; Monaco, F.; Polci, A.; Russo, F.; Di Gennaro, A.; Marini, V.; Teodori, L.; Montarsi, F.; Pinoni, C.; et al. Evidence of West Nile virus lineage 2 circulation in Northern Italy. Vet. Microbiol. 2012, 158, 267–273. [Google Scholar] [CrossRef]
- Monaco, F.; Goffredo, M.; Briguglio, P.; Pinoni, C.; Polci, A.; Iannetti, S.; Marruchella, G.; Di Francesco, G.; Di Gennaro, A.P.; Pais, M.; et al. The 2011 West Nile disease outbreak in Sardinia region, Italy. Vet. Ital. 2015, 51, 5–16. [Google Scholar] [CrossRef]
- West Nile Disease. Available online: https://westnile.izs.it/j6_wnd/docMinisteriale?annoDocumento=2016 (accessed on 23 July 2020).
- Calzolari, M.; Pautasso, A.; Montarsi, F.; Albieri, A.; Bellini, R.; Bonilauri, P.; Defilippo, F.; Lelli, D.; Moreno, A.; Chiari, M.; et al. West Nile Virus Surveillance in 2013 via Mosquito Screening in Northern Italy and the Influence of Weather on Virus Circulation. PLoS ONE 2015, 10, e0140915. [Google Scholar] [CrossRef] [Green Version]
- Riccardo, F.; Monaco, F.; Bella, A.; Savini, G.; Russo, F.; Cagarelli, R.; Dottori, M.; Rizzo, C.; Venturi, G.; Luca, M.D.; et al. An early start of West Nile virus seasonal transmission: The added value of One Heath surveillance in detecting early circulation and triggering timely response in Italy, June to July 2018. Eurosurveillance 2018, 23, 1800427. [Google Scholar] [CrossRef] [Green Version]
- West Nile Disease. Available online: https://westnile.izs.it/j6_wnd/docMinisteriale?annoDocumento=2020 (accessed on 29 July 2020).
- Calistri, P.; Giovannini, A.; Hubalek, Z.; Ionescu, A.; Monaco, F.; Savini, G.; Lelli, R. Epidemiology of West Nile in Europe and in the Mediterranean Basin. Open Virol. J. 2010, 4, 29–37. [Google Scholar] [CrossRef] [Green Version]
- Rappole, J.H.; Hubalek, Z. Migratory birds and West Nile virus. J. Appl. Microbiol. 2003, 94, 47–58. [Google Scholar] [CrossRef]
- Marini, G.; Rosà, R.; Pugliese, A.; Rizzoli, A.; Rizzo, C.; Russo, F.; Montarsi, F.; Capelli, G. West Nile virus transmission and human infection risk in Veneto (Italy): A modelling analysis. Sci. Rep. 2018, 8, 14005. [Google Scholar] [CrossRef]
- Montecino-Latorre, D.; Barker, C.M. Overwintering of West Nile virus in a bird community with a communal crow roost. Sci. Rep. 2018, 8, 6088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chevalier, V.; Tran, A.; Durand, B. Predictive Modeling of West Nile Virus Transmission Risk in the Mediterranean Basin: How Far from Landing? Int. J. Environ. Res. Public Health 2014, 11, 67–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tran, A.; Sudre, B.; Paz, S.; Rossi, M.; Desbrosse, A.; Chevalier, V.; Semenza, J.C. Environmental predictors of West Nile fever risk in Europe. Int. J. Health Geogr. 2014, 13, 26. [Google Scholar] [CrossRef] [Green Version]
- Paz, S. Climate change impacts on West Nile virus transmission in a global context. Philos. Trans. R. Soc. B Biol. Sci. 2015, 370, 20130561. [Google Scholar] [CrossRef]
- Paz, S.; Semenza, J.C. Environmental Drivers of West Nile Fever Epidemiology in Europe and Western Asia—A Review. Int. J. Environ. Res. Public Health 2013, 10, 3543–3562. [Google Scholar] [CrossRef] [Green Version]
- Marcantonio, M.; Rizzoli, A.; Metz, M.; Rosà, R.; Marini, G.; Chadwick, E.; Neteler, M. Identifying the Environmental Conditions Favouring West Nile Virus Outbreaks in Europe. PLoS ONE 2015, 10, e0121158. [Google Scholar] [CrossRef] [Green Version]
- Paz, S.; Malkinson, D.; Green, M.S.; Tsioni, G.; Papa, A.; Danis, K.; Sirbu, A.; Ceianu, C.; Katalin, K.; Ferenczi, E.; et al. Permissive Summer Temperatures of the 2010 European West Nile Fever Upsurge. PLoS ONE 2013, 8, e56398. [Google Scholar] [CrossRef] [Green Version]
- Moirano, G.; Gasparrini, A.; Acquaotta, F.; Fratianni, S.; Merletti, F.; Maule, M.; Richiardi, L. West Nile Virus infection in Northern Italy: Case-crossover study on the short-term effect of climatic parameters. Environ. Res. 2018, 167, 544–549. [Google Scholar] [CrossRef]
- Vincenzi, S.; Porrello, A.; Buzzega, P.; Conte, A.; Ippoliti, C.; Candeloro, L.; Di Lorenzo, A.; Dondona, A.C.; Calderara, S. Spotting insects from satellites: Modeling the presence of Culicoides imicola through Deep CNNs. arXiv 2019, arXiv:1911.10024. [Google Scholar]
- Reichstein, M.; Camps-Valls, G.; Stevens, B.; Jung, M.; Denzler, J.; Carvalhais, N. Prabhat Deep learning and process understanding for data-driven Earth system science. Nature 2019, 566, 195–204. [Google Scholar] [CrossRef] [PubMed]
- VanderWaal, K.; Morrison, R.B.; Neuhauser, C.; Vilalta, C.; Perez, A.M. Translating Big Data into Smart Data for Veterinary Epidemiology. Front. Vet. Sci. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- Agany, D.D.M.; Pietri, J.E.; Gnimpieba, E.Z. Assessment of vector-host-pathogen relationships using data mining and machine learning. Comput. Struct. Biotechnol. J. 2020, 18, 1704–1721. [Google Scholar] [CrossRef]
- Kuo, F.-Y.; Wen, T.-H.; Sabel, C.E. Characterizing Diffusion Dynamics of Disease Clustering: A Modified Space–Time DBSCAN (MST-DBSCAN) Algorithm. Ann. Am. Assoc. Geogr. 2018, 108, 1168–1186. [Google Scholar] [CrossRef]
- Alobuia, W.M.; Missikpode, C.; Aung, M.; Jolly, P.E. Knowledge, Attitude, and Practices Regarding Vector-borne Diseases in Western Jamaica. Ann. Glob. Health 2015, 81, 654–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scavuzzo, J.M.; Trucco, F.; Espinosa, M.; Tauro, C.B.; Abril, M.; Scavuzzo, C.M.; Frery, A.C. Modeling Dengue vector population using remotely sensed data and machine learning. Acta Trop. 2018, 185, 167–175. [Google Scholar] [CrossRef] [Green Version]
- Sajana, T.; Narasingarao, M.R. Majority Voting Algorithm for Diagnosing of Imbalanced Malaria Disease. In Lecture Notes in Computational Vision and Biomechanics, Proceedings of the International Conference on ISMAC in Computational Vision and Bio-Engineering 2018 (ISMAC-CVB), Palladam, India, 16–17 May 2018; Pandian, D., Fernando, X., Baig, Z., Shi, F., Eds.; Springer International Publishing: Cham, Switzerland, 2019; pp. 31–40. [Google Scholar]
- Lim, S.; Tucker, C.S.; Kumara, S. An unsupervised machine learning model for discovering latent infectious diseases using social media data. J. Biomed. Inf. 2017, 66, 82–94. [Google Scholar] [CrossRef]
- Stilianakis, N.I.; Syrris, V.; Petroliagkis, T.; Pärt, P.; Gewehr, S.; Kalaitzopoulou, S.; Mourelatos, S.; Baka, A.; Pervanidou, D.; Vontas, J.; et al. Identification of Climatic Factors Affecting the Epidemiology of Human West Nile Virus Infections in Northern Greece. PLoS ONE 2016, 11, e0161510. [Google Scholar] [CrossRef]
- Conley, A.K.; Fuller, D.O.; Haddad, N.; Hassan, A.N.; Gad, A.M.; Beier, J.C. Modeling the distribution of the West Nile and Rift Valley Fever vector Culex pipiens in arid and semi-arid regions of the Middle East and North Africa. Parasites Vectors 2014, 7, 289. [Google Scholar] [CrossRef] [Green Version]
- Young, S.G.; Tullis, J.A.; Cothren, J. A remote sensing and GIS-assisted landscape epidemiology approach to West Nile virus. Appl. Geogr. 2013, 45, 241–249. [Google Scholar] [CrossRef]
- Parselia, E.; Kontoes, C.; Tsouni, A.; Hadjichristodoulou, C.; Kioutsioukis, I.; Magiorkinis, G.; Stilianakis, N.I. Satellite Earth Observation Data in Epidemiological Modeling of Malaria, Dengue and West Nile Virus: A Scoping Review. Remote Sens. 2019, 11, 1862. [Google Scholar] [CrossRef] [Green Version]
- Rochlin, I.; Turbow, D.; Gomez, F.; Ninivaggi, D.V.; Campbell, S.R. Predictive Mapping of Human Risk for West Nile Virus (WNV) Based on Environmental and Socioeconomic Factors. PLoS ONE 2011, 6, e23280. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salute, M. della Virus West Nile e Usutu, il Piano di Sorveglianza e Risposta. 2019. Available online: http://www.salute.gov.it/portale/news/p3_2_1_1_1.jsp?lingua=italiano&menu=notizie&p=dalministero&id=3701 (accessed on 2 September 2020).
- Mancini, G.; Montarsi, F.; Calzolari, M.; Capelli, G.; Dottori, M.; Ravagnan, S.; Lelli, D.; Chiari, M.; Santilli, A.; Quaglia, M.; et al. Mosquito species involved in the circulation of West Nile and Usutu viruses in Italy. Vet. Ital. 2017, 53, 97–110. [Google Scholar] [CrossRef] [PubMed]
- Colangeli, P.; Iannetti, S.; Cerella, A.; Ippoliti, C.; Di Lorenzo, A.; Santucci, U.; Simonetti, P.; Calistri, P.; Lelli, R. The national information system for the notification of animal diseases in Italy. Vet. Ital. 2011, 47, 303–312. [Google Scholar]
- Marini, G.; Calzolari, M.; Angelini, P.; Bellini, R.; Bellini, S.; Bolzoni, L.; Torri, D.; Defilippo, F.; Dorigatti, I.; Nikolay, B.; et al. A quantitative comparison of West Nile virus incidence from 2013 to 2018 in Emilia-Romagna, Italy. PLoS Negl. Trop. Dis. 2020, 14, e0007953. [Google Scholar] [CrossRef] [Green Version]
- Calistri, P.; Conte, A.; Monaco, F.; Goffredo, M.; Danzetta, M.L.; Sabatino, D.D.; Iapaolo, F.; Candeloro, L.; Ippoliti, C.; Mancini, G.; et al. Possible drivers for the increased West Nile virus transmission in Italy in 2018. Int. J. Infect. Dis. 2019, 79, 27–28. [Google Scholar] [CrossRef] [Green Version]
- Angenvoort, J.; Brault, A.C.; Bowen, R.A.; Groschup, M.H. West Nile viral infection of equids. Vet. Microbiol. 2013, 167, 168–180. [Google Scholar] [CrossRef] [Green Version]
- Bunning, M.L.; Bowen, R.A.; Cropp, C.B.; Sullivan, K.G.; Davis, B.S.; Komar, N.; Godsey, M.S.; Baker, D.; Hettler, D.L.; Holmes, D.A.; et al. Experimental infection of horses with West Nile virus. Emerg. Infect. Dis. 2002, 8, 380–386. [Google Scholar] [CrossRef]
- Komar, O.; Robbins, M.B.; Klenk, K.; Blitvich, B.J.; Marlenee, N.L.; Burkhalter, K.L.; Gubler, D.J.; Gonzálvez, G.; Peña, C.J.; Peterson, A.T.; et al. West Nile Virus Transmission in Resident Birds, Dominican Republic. Emerg. Infect. Dis. 2003, 9, 1299–1302. [Google Scholar] [CrossRef]
- Pérez-Ramírez, E.; Llorente, F.; Jiménez-Clavero, M.Á. Experimental Infections of Wild Birds with West Nile Virus. Viruses 2014, 6, 752–781. [Google Scholar] [CrossRef] [Green Version]
- Fortuna, C.; Remoli, M.E.; Di Luca, M.; Severini, F.; Toma, L.; Benedetti, E.; Bucci, P.; Montarsi, F.; Minelli, G.; Boccolini, D.; et al. Experimental studies on comparison of the vector competence of four Italian Culex pipiens populations for West Nile virus. Parasites Vectors 2015, 8, 463. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- EpiCentro La Sorveglianza dei Casi Umani di Infezione da West Nile viRus. Available online: https://www.epicentro.iss.it/westnile/bollettino (accessed on 29 July 2020).
- West Nile Disease. Available online: https://westnile.izs.it/j6_wnd/wndItaliaPeriodici (accessed on 1 September 2020).
- Corbane, C.; Florczyk, A.; Pesaresi, M.; Politis, P.; Syrris, V. GHS-BUILT R2018A—GHS built-up grid, derived from Landsat, multitemporal (1975–1990–2000–2014). Eur. Comm. Joint Res. Cent. JRC Data Cat. 2018. [Google Scholar] [CrossRef]
- CLC 2012—Copernicus Land Monitoring Service. Available online: https://land.copernicus.eu/pan-european/corine-land-cover/clc-2012 (accessed on 29 July 2020).
- Stevens, D.L.; Olsen, A.R. Spatially Balanced Sampling of Natural Resources. J. Am. Stat. Assoc. 2004, 99, 262–278. [Google Scholar] [CrossRef]
- Theobald, D.M.; Stevens, D.L.; White, D.; Urquhart, N.S.; Olsen, A.R.; Norman, J.B. Using GIS to generate spatially balanced random survey designs for natural resource applications. Environ. Manag. 2007, 40, 134–146. [Google Scholar] [CrossRef]
- Wan, Z. New refinements and validation of the collection-6 MODIS land-surface temperature/emissivity product. Remote Sens. Environ. 2014, 140, 36–45. [Google Scholar] [CrossRef]
- R: The R Project for Statistical Computing. Available online: https://www.r-project.org/ (accessed on 27 July 2020).
- Didan, K. MOD13Q1 MODIS/Terra Vegetation Indices 16-Day L3 Global 250m SIN Grid V006. NASA EOSDIS Land Process. DAAC 2015. [Google Scholar] [CrossRef]
- Bauer-Marschallinger, B.; Freeman, V.; Cao, S.; Paulik, C.; Schaufler, S.; Stachl, T.; Modanesi, S.; Massari, C.; Ciabatta, L.; Brocca, L.; et al. Toward Global Soil Moisture Monitoring With Sentinel-1: Harnessing Assets and Overcoming Obstacles. IEEE Trans. Geosci. Remote Sens. 2019, 57, 520–539. [Google Scholar] [CrossRef]
- Gerber, F.; de Jong, R.; Schaepman, M.E.; Schaepman-Strub, G.; Furrer, R. Predicting Missing Values in Spatio-Temporal Remote Sensing Data. IEEE Trans. Geosci. Remote Sens. 2018, 56, 2841–2853. [Google Scholar] [CrossRef] [Green Version]
- Verdonschot, P.F.M.; Besse-Lototskaya, A.A. Flight distance of mosquitoes (Culicidae): A metadata analysis to support the management of barrier zones around rewetted and newly constructed wetlands. Limnologica 2014, 45, 69–79. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost: A Scalable Tree Boosting System. In KDD ’16: Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; Association for Computing Machinery: San Francisco, CA, USA, 2016; pp. 785–794. [Google Scholar]
- Nielsen, D. Tree Boosting with XGBoost. Nor. Univ. Sci. Technol. 2016. Available online: http://pzs.dstu.dp.ua/DataMining/boosting/bibl/Didrik.pdf (accessed on 18 September 2020).
- Bergstra, J.; Bengio, Y. Random Search for Hyper-Parameter Optimization. J. Mach. Learn. Res. 2012, 13, 281–305. [Google Scholar]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. Xgboost: Extreme Gradient Boosting. 2020. Available online: https://CRAN.R-project.org/package=xgboost (accessed on 18 September 2020).
- Kuhn, M.; Wing, J.; Weston, S.; Williams, A.; Keefer, C.; Engelhardt, A.; Cooper, T.; Mayer, Z.; Kenkel, B.; R Core Team; et al. Caret: Classification and Regression Training. 2020. Available online: https://CRAN.R-project.org/package=caret (accessed on 18 September 2020).
- Wickham, H.; François, R.; Henry, L.; Müller, K. RStudio Dplyr: A Grammar of Data Manipulation. 2020. Available online: https://CRAN.R-project.org/package=dplyr (accessed on 18 September 2020).
- Hijmans, R.J.; van Etten, J.; Sumner, M.; Cheng, J.; Baston, D.; Bevan, A.; Bivand, R.; Busetto, L.; Canty, M.; Forrest, D.; et al. Raster: Geographic Data Analysis and Modeling. 2020. Available online: https://CRAN.R-project.org/package=raster (accessed on 18 September 2020).
- Bivand, R.; Keitt, T.; Rowlingson, B.; Pebesma, E.; Sumner, M.; Hijmans, R.; Rouault, E.; Warmerdam, F.; Ooms, J.; Rundel, C. Rgdal: Bindings for the “Geospatial” Data Abstraction Library. 2020. Available online: https://CRAN.R-project.org/package=rgdal (accessed on 18 September 2020).
- Ooi, H.; Corporation, M.; Weston, S. doSNOW: Foreach Parallel Adaptor for the “Snow” Package. 2019. Available online: https://CRAN.R-project.org/package=doSNOW (accessed on 18 September 2020).
- McIntyre, K.M.; Setzkorn, C.; Hepworth, P.J.; Morand, S.; Morse, A.P.; Baylis, M. Systematic Assessment of the Climate Sensitivity of Important Human and Domestic Animals Pathogens in Europe. Sci. Rep. 2017, 7, 7134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ippoliti, C.; Candeloro, L.; Gilbert, M.; Goffredo, M.; Mancini, G.; Curci, G.; Falasca, S.; Tora, S.; Lorenzo, A.D.; Quaglia, M.; et al. Defining ecological regions in Italy based on a multivariate clustering approach: A first step towards a targeted vector borne disease surveillance. PLoS ONE 2019, 14, e0219072. [Google Scholar] [CrossRef] [PubMed]
- Conte, A.; Candeloro, L.; Ippoliti, C.; Monaco, F.; De Massis, F.; Bruno, R.; Di Sabatino, D.; Danzetta, M.L.; Benjelloun, A.; Belkadi, B.; et al. Spatio-Temporal Identification of Areas Suitable for West Nile Disease in the Mediterranean Basin and Central Europe. PLoS ONE 2015, 10, e0146024. [Google Scholar] [CrossRef] [PubMed]
- Calistri, P.; Ippoliti, C.; Candeloro, L.; Benjelloun, A.; El Harrak, M.; Bouchra, B.; Danzetta, M.L.; Di Sabatino, D.; Conte, A. Analysis of climatic and environmental variables associated with the occurrence of West Nile virus in Morocco. Prev. Vet. Med. 2013, 110, 549–553. [Google Scholar] [CrossRef]
- Savini, L.; Candeloro, L.; Perticara, S.; Conte, A. EpiExploreR: A Shiny Web Application for the Analysis of Animal Disease Data. Microorganisms 2019, 7, 680. [Google Scholar] [CrossRef] [Green Version]
- Bellini, R.; Calzolari, M.; Mattivi, A.; Tamba, M.; Angelini, P.; Bonilauri, P.; Albieri, A.; Cagarelli, R.; Carrieri, M.; Dottori, M.; et al. The experience of West Nile virus integrated surveillance system in the Emilia-Romagna region: Five years of implementation, Italy, 2009 to 2013. Eurosurveillance 2014, 19, 20953. [Google Scholar] [CrossRef] [Green Version]










| Year | Birds + | Mosquitoes + (−) | Equids + | Pseudo Absence (−) | Total + (−) |
|---|---|---|---|---|---|
| 2017 | 40 | 43 (90) | 47 | (186) | 130 (276) |
| 2018 | 216 | 137 (137) | 151 | (804) | 504 (941) |
| 2019 | 67 | 43 (86) | 8 | (150) | 118 (236) |
| Total | 323 | 223 (313) | 206 | (1140) | 752 (1453) |
| EO Product | Spatial Resolution | Temporal Resolution | Percentage of NoData Pixels in Source: Median (Min, Max) | Images in the Data-Cube (2016–2019) | ||
|---|---|---|---|---|---|---|
| Source | Model | Source | Model | |||
| MOD11A2 LSTD | 1 km | 250 m | 8 days | 8 days | 0.64% (0.24%, 38.10%) | 184 |
| MOD11A2 LSTN | 1 km | 8 days | 8 days | 1.05% (0.25%, 32.93%) | 184 | |
| MOD13Q1 NDVI | 250 m | 16 days | 16 days | 0.55% (0.41%, 1.70%) | 92 | |
| Copernicus SSM | 1 km | daily | 8 days | * 22.52% (22.3%, 68.25%) | 184 | |
| Observed Positive | Observed Negative | Total | |
|---|---|---|---|
| Predicted Positive | 771 | 230 | 1001 |
| Predicted Negative | 269 | 1818 | 2087 |
| Total | 1040 | 2048 | 3088 |
| Birds | Mosquitoes | Equids | Total | |
|---|---|---|---|---|
| Predicted Positive | 406 | 311 | 54 | 771 |
| Predicted Negative | 184 | 67 | 18 | 269 |
| Total | 590 | 378 | 72 | 1040 |
| Pseudo-Absence in Space | Negatives in Time | Total | |
|---|---|---|---|
| Predicted Positive | 208 | 22 | 230 |
| Predicted Negative | 1138 | 680 | 1818 |
| Total | 1346 | 702 | 2048 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Candeloro, L.; Ippoliti, C.; Iapaolo, F.; Monaco, F.; Morelli, D.; Cuccu, R.; Fronte, P.; Calderara, S.; Vincenzi, S.; Porrello, A.; et al. Predicting WNV Circulation in Italy Using Earth Observation Data and Extreme Gradient Boosting Model. Remote Sens. 2020, 12, 3064. https://doi.org/10.3390/rs12183064
Candeloro L, Ippoliti C, Iapaolo F, Monaco F, Morelli D, Cuccu R, Fronte P, Calderara S, Vincenzi S, Porrello A, et al. Predicting WNV Circulation in Italy Using Earth Observation Data and Extreme Gradient Boosting Model. Remote Sensing. 2020; 12(18):3064. https://doi.org/10.3390/rs12183064
Chicago/Turabian StyleCandeloro, Luca, Carla Ippoliti, Federica Iapaolo, Federica Monaco, Daniela Morelli, Roberto Cuccu, Pietro Fronte, Simone Calderara, Stefano Vincenzi, Angelo Porrello, and et al. 2020. "Predicting WNV Circulation in Italy Using Earth Observation Data and Extreme Gradient Boosting Model" Remote Sensing 12, no. 18: 3064. https://doi.org/10.3390/rs12183064
APA StyleCandeloro, L., Ippoliti, C., Iapaolo, F., Monaco, F., Morelli, D., Cuccu, R., Fronte, P., Calderara, S., Vincenzi, S., Porrello, A., D’Alterio, N., Calistri, P., & Conte, A. (2020). Predicting WNV Circulation in Italy Using Earth Observation Data and Extreme Gradient Boosting Model. Remote Sensing, 12(18), 3064. https://doi.org/10.3390/rs12183064