The Use of Remotely Sensed Data and Polish NFI Plots for Prediction of Growing Stock Volume Using Different Predictive Methods
Abstract
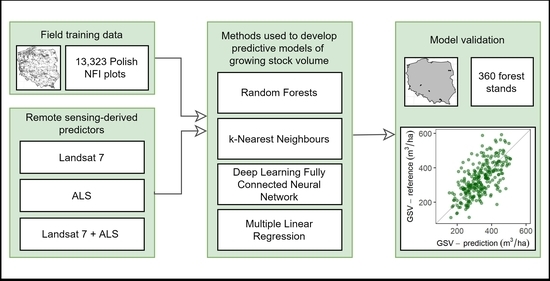
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. Polish National Forest Inventory Data
2.3. Landsat Images
2.4. Airborne Laser Scanning Point Clouds
2.5. Extraction of Predictor Variables
2.6. Validation Data
2.7. Predictive Methods
2.7.1. Random Forests
2.7.2. k-Nearest Neighbours
2.7.3. Deep Learning Fully Connected Neural Network
2.7.4. Multiple Linear Regression
2.8. Models Training and Validation
3. Results
3.1. Set of Predictor Variables
3.2. Optimization Results
3.3. Performance Assessment
3.4. Performance Assessment per Dominant Tree Species
4. Discussion
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kangas, A.; Astrup, R.; Breidenbach, J.; Fridman, J.; Gobakken, T.; Korhonen, K.T.; Maltamo, M.; Nilsson, M.; Nord-Larsen, T.; Næsset, E.; et al. Remote sensing and forest inventories in Nordic countries—Roadmap for the future. Scand. J. For. Res. 2018, 7581, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Tomppo, E.; Gschwantner, T.; Lawrence, M.; McRoberts, R.E. National Forest Inventories: Pathways for Common Reporting; Springer: Dordrecht, The Netherlands, 2010; ISBN 9789048132324. [Google Scholar]
- Gschwantner, T.; Lanz, A.; Vidal, C.; Bosela, M.; Di Cosmo, L.; Fridman, J.; Gasparini, P.; Kuliešis, A.; Tomter, S.; Schadauer, K. Comparison of methods used in European National Forest Inventories for the estimation of volume increment: Towards harmonisation. Ann. For. Sci. 2016. [Google Scholar] [CrossRef] [Green Version]
- Næsset, E. Area-Based Inventory in Norway—From Innovation to an Operational Reality. In Forestry Applications of Airborne Laser Scanning: Concepts and Case Studies; Springer: Dordrecht, The Netherlands, 2014; pp. 215–240. [Google Scholar]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Wulder, M.A.; Bater, C.W.; Coops, N.C.; Hilker, T.; White, J.C. The role of LiDAR in sustainable forest management. For. Chron. 2008, 84, 807–826. [Google Scholar] [CrossRef] [Green Version]
- Socha, J.; Hawryło, P.; Pierzchalski, M.; Stereńczak, K.; Krok, G.; Wężyk, P.; Tymińska-Czabańska, L. An allometric area-based approach—A cost-effective method for stand volume estimation based on ALS and NFI data. For. Int. J. For. Res. 2019, 93, 344–358. [Google Scholar] [CrossRef]
- Kurz, W.A.; Dymond, C.C.; White, T.M.; Stinson, G.; Shaw, C.H.; Rampley, G.J.; Smyth, C.; Simpson, B.N.; Neilson, E.T.; Trofymow, J.A.; et al. CBM-CFS3: A model of carbon-dynamics in forestry and land-use change implementing IPCC standards. Ecol. Model. 2009, 220, 480–504. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Varhola, A.; Vastaranta, M.; Coops, N.C.; Cook, B.D.; Pitt, D.G.; Woods, M. A Best Practices Guide for Generating Forest Inventory Attributes from Airborne Laser Scanning Data Using an Area-Based Approach; Technical Report FI-X-010; The Canadian Wood Fibre Centre: Victoria, BC, Canada, 2013. [Google Scholar]
- Næsset, E. Estimating timber volume of forest stands using airborne laser scanner data. Remote Sens. Environ. 1997, 61, 246–253. [Google Scholar] [CrossRef]
- Brach, M.; Stereńczak, K.; Bolibok, L.; Kwaśny, Ł.; Krok, G.; Laszkowski, M. Impacts of forest spatial structure on variation of the multipath phenomenon of navigation satellite signals. Folia For. Pol. 2019, 61, 3–21. [Google Scholar] [CrossRef] [Green Version]
- Stereńczak, K.; Lisańczuk, M.; Parkitna, K.; Mitelsztedt, K.; Mroczek, P.; Miścicki, S. The influence of number and size of sample plots on modelling growing stock volume based on airborne laser scanning. Drewno 2018, 61. [Google Scholar] [CrossRef]
- Gobakken, T.; Næsset, E. Assessing effects of positioning errors and sample plot size on biophysical stand properties derived from airborne laser scanner data. Can. J. For. Res. 2009, 39, 1036–1052. [Google Scholar] [CrossRef]
- Mauro, F.; Valbuena, R.; Manzanera, J.A.; García-Abril, A. Influence of global navigation satellite system errors in positioning inventory plots for treeheight distribution studies. Can. J. For. Res. 2011, 41, 11–23. [Google Scholar] [CrossRef]
- Tomppo, E. Satellite image-based National Forest Inventory of Finland. In Proceedings of the ISPRS, COMISSION VII, Mid-Term Symposium Global and Environmental Monitoring, Techniques and Impacts, Victoria, BC, Canada, 17–21 September 1990; pp. 419–424. [Google Scholar]
- Chirici, G.; Giannetti, F.; McRoberts, R.E.; Travaglini, D.; Pecchi, M.; Maselli, F.; Chiesi, M.; Corona, P. Wall-to-wall spatial prediction of growing stock volume based on Italian National Forest Inventory plots and remotely sensed data. Int. J. Appl. Earth Obs. Geoinf. 2020, 84, 101959. [Google Scholar] [CrossRef]
- Nilsson, M.; Nordkvist, K.; Jonzén, J.; Lindgren, N.; Axensten, P.; Wallerman, J.; Egberth, M.; Larsson, S.; Nilsson, L.; Eriksson, J.; et al. A nationwide forest attribute map of Sweden predicted using airborne laser scanning data and field data from the National Forest Inventory. Remote Sens. Environ. 2017, 194, 447–454. [Google Scholar] [CrossRef]
- Hollaus, M.; Dorigo, W.; Wagner, W.; Schadauer, K.; Höfle, B.; Maier, B. Operational wide-area stem volume estimation based on airborne laser scanning and national forest inventory data. Int. J. Remote Sens. 2009, 30, 5159–5175. [Google Scholar] [CrossRef]
- Nord-Larsen, T.; Schumacher, J. Estimation of forest resources from a country wide laser scanning survey and national forest inventory data. Remote Sens. Environ. 2012, 119, 148–157. [Google Scholar] [CrossRef]
- CSO. Statistical Yearbook of Forestry; Statistics Poland: Warsaw, Poland, 2019. (In Polish)
- SFIC. Forests in Poland; The State Forests Information Centre: Warsaw, Poland, 2018. [Google Scholar]
- NFI National Forest Inventory. Wielkoobszarowa Inwentaryzacja Stanu Lasów, Wyniki za Okres 2014–2018; Biuro Urządzania Lasu i Geodezji Leśnej: Warsaw, Poland, 2018. (In Polish) [Google Scholar]
- Forest Europe. State of Europe’s Forests. In Proceedings of the Ministerial Conference on the Protection of Forests in Europe, Madrid, Spain, 20–21 October 2015. [Google Scholar]
- ICP ICP Forests Manual. Manual on Methods and Criteria for Harmonized Sampling, Assessment, Monitoring and Analysis of the Effects of Air Pollution on Forests; Thünen Institute Forest Ecosystems: Eberswalde, Germany, 2016. [Google Scholar]
- Bruchwald, A.; Rymer-Dudzińska, T.; Dudek, A.; Michalak, K.; Wróblewski, L.; Zasada, M. Wzory empiryczne do określania wysokości i pierśnicowej liczby kształtu grubizny drzewa. Sylwan 2000, 144, 5–12. [Google Scholar]
- Gschwantner, T.; Alberdi, I.; Balázs, A.; Bauwens, S.; Bender, S.; Borota, D.; Bosela, M.; Bouriaud, O.; Cañellas, I.; Donis, J.; et al. Harmonisation of stem volume estimates in European National Forest Inventories. Ann. For. Sci. 2019, 76. [Google Scholar] [CrossRef] [Green Version]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Schmidt, G.; Jenkerson, C.B.; Masek, J.; Vermote, E.; Gao, F. Landsat Ecosystem Disturbance Adaptive Processing System (LEDAPS) Algorithm Description; U.S. Geological Survey: Reston, VA, USA, 2013.
- Zhu, Z.; Woodcock, C.E. Object-based cloud and cloud shadow detection in Landsat imagery. Remote Sens. Environ. 2012, 118, 83–94. [Google Scholar] [CrossRef]
- Roussel, J.-R.; Auty, D. lidR: Airborne LiDAR Data Manipulation and Visualization for Forestry Applications. R Package Version 2.2.2. Available online: https://CRAN.R-project.org/package=lidR2020 (accessed on 28 January 2020).
- Woods, M.; Lim, K.; Treitz, P. Predicting forest stand variables from LiDAR data in the Great Lakes—St. Lawrence forest of Ontario. For. Chron. 2008, 84, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Li, I.Z.W.; Xin, X.P.; Huan, T.A.N.G.; Fan, Y.A.N.G.; Chen, B.R.; Zhang, B.H. Estimating grassland LAI using the Random Forests approach and Landsat imagery in the meadow steppe of Hulunber, China. J. Integr. Agric. 2017, 16, 286–297. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Baccini, A.; Goetz, S.J.; Walker, W.S.; Laporte, N.T.; Sun, M.; Sulla-Menashe, D.; Hackler, J.; Beck, P.S.A.; Dubayah, R.; Friedl, M.A.; et al. Estimated carbon dioxide emissions from tropical deforestation improved by carbon-density maps. Nat. Clim. Chang. 2012, 2, 182–185. [Google Scholar] [CrossRef]
- Evans, J.S.; Cushman, S.A. Gradient modeling of conifer species using random forests. Landsc. Ecol. 2009, 24, 673–683. [Google Scholar] [CrossRef]
- Falkowski, M.J.; Evans, J.S.; Martinuzzi, S.; Gessler, P.E.; Hudak, A.T. Characterizing forest succession with lidar data: An evaluation for the Inland Northwest, USA. Remote Sens. Environ. 2009, 113, 946–956. [Google Scholar] [CrossRef] [Green Version]
- Houghton, R.A. Balancing the Global Carbon Budget. Annu. Rev. Earth Planet. Sci. 2007, 35, 313–347. [Google Scholar] [CrossRef] [Green Version]
- Stumpf, A.; Kerle, N. Object-oriented mapping of landslides using Random Forests. Remote Sens. Environ. 2011, 115, 2564–2577. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăgu, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Chirici, G.; Mura, M.; McInerney, D.; Py, N.; Tomppo, E.O.; Waser, L.T.; Travaglini, D.; McRoberts, R.E. A meta-analysis and review of the literature on the k-Nearest Neighbors technique for forestry applications that use remotely sensed data. Remote Sens. Environ. 2016, 176, 282–294. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016. [Google Scholar]
- Chollet, F. Deep Learning with Python; Manning Publications Co.: New York, NY, USA, 2018. [Google Scholar]
- Mura, M.; McRoberts, R.E.; Chirici, G.; Marchetti, M. Statistical inference for forest structural diversity indices using airborne laser scanning data and the k-Nearest Neighbors technique. Remote Sens. Environ. 2016, 186, 678–686. [Google Scholar] [CrossRef]
- McRoberts, R.E. Estimating forest attribute parameters for small areas using nearest neighbors techniques. For. Ecol. Manag. 2012, 272, 3–12. [Google Scholar] [CrossRef]
- Efron, B.; Tibshirani, R.J. An Introduction to the Bootstrap; Chapman & Hall/CRC: Boca Raton, FL, USA, 1994. [Google Scholar]
- Jebadurai, J.; Jebadurai, I.J.; Paulraj, G.J.L.; Samuel, N.E. Super-resolution of digital images using CNN with leaky ReLU. Int. J. Recent Technol. Eng. 2019, 8, 210–212. [Google Scholar] [CrossRef]
- Huk, M. Stochastic Optimization of Contextual Neural Networks with RMSprop BT—Intelligent Information and Database Systems; Nguyen, N.T., Jearanaitanakij, K., Selamat, A., Trawiński, B., Chittayasothorn, S., Eds.; Springer International Publishing: Cham, Switzerland, 2020; pp. 343–352. [Google Scholar]
- Miścicki, S.; Stereńczak, K. Określanie miąższości i zagęszczenia drzew w drzewostanach centralnej Polski na podstawie danych lotniczego skanowania laserowego w dwufazowej metodzie inwentaryzacji zasobów drzewnych. Leśne Pr. Badaw. 2013, 74, 127–136. [Google Scholar] [CrossRef] [Green Version]
- Guyon, I.; Elisseeff, A. An introduction to variable and feature selection. J. Mach. Learn. Res. 2003, 3, 1157–1182. [Google Scholar] [CrossRef] [Green Version]
- Straub, C.; Weinacker, H.; Koch, B. A comparison of different methods for forest resource estimation using information from airborne laser scanning and CIR orthophotos. Eur. J. For. Res. 2010, 129, 1069–1080. [Google Scholar] [CrossRef]
- Ayrey, E.; Hayes, D.J.; Kilbride, J.B.; Fraver, S.; Kershaw, J.A.; Cook, B.D.; Weiskittel, A.R. Synthesizing Disparate LiDAR and Satellite Datasets through Deep Learning to Generate Wall-to-Wall Regional Forest Inventories. BioRxiv 2019, 580514. [Google Scholar] [CrossRef]
- Ayrey, E.; Hayes, D.J. The use of three-dimensional convolutional neural networks to interpret LiDAR for forest inventory. Remote Sens. 2018, 10, 649. [Google Scholar] [CrossRef] [Green Version]
- McRoberts, R.E.; Chen, Q.; Walters, B.F.; Kaisershot, D.J. The effects of global positioning system receiver accuracy on airborne laser scanning-assisted estimates of aboveground biomass. Remote Sens. Environ. 2018, 207, 42–49. [Google Scholar] [CrossRef]
- Packalen, P.; Strunk, J.; Packalen, T.; Maltamo, M.; Mehtätalo, L. Resolution dependence in an area-based approach to forest inventory with airborne laser scanning. Remote Sens. Environ. 2019, 224, 192–201. [Google Scholar] [CrossRef]






| Dominant Tree Species | Number of Plots | Percentage of Plots (%) | Minimum GSV (m3/ha) | Mean GSV (m3/ha) | Maximum GSV (m3/ha) | Standard Deviation of GSV (m3/ha) |
|---|---|---|---|---|---|---|
| Scots pine | 8334 | 62.6 | 0.2 | 300.7 | 935.1 | 139.6 |
| silver birch | 904 | 6.8 | 0.3 | 192.8 | 708.4 | 119.8 |
| Norway spruce | 731 | 5.5 | 0.4 | 309.3 | 1069.0 | 200.8 |
| European beech | 708 | 5.3 | 0.1 | 351.8 | 1452.8 | 220.6 |
| sessile oak | 680 | 5.1 | 0.2 | 296.0 | 1237.3 | 186.0 |
| other species | 663 | 5.0 | 0.2 | 243.4 | 1347.1 | 174.0 |
| common alder | 577 | 4.3 | 0.7 | 270.9 | 908.3 | 165.5 |
| silver fir | 459 | 3.4 | 2.7 | 368.0 | 1338.0 | 217.5 |
| European larch | 267 | 2.0 | 0.3 | 253.0 | 795.1 | 181.0 |
| Maximum Absolute Difference between NFI and ALS (Years) | Number of Plots | Percentage of Plots (%) |
|---|---|---|
| 1 | 5209 | 39.1 |
| 2 | 4246 | 31.9 |
| 3 | 2588 | 19.4 |
| 4 | 1092 | 8.2 |
| 5 | 188 | 1.4 |
| Description of Predictor Variables | Acronyms |
|---|---|
| Bottom of atmosphere reflectance of Landsat 7 ETM+ spectral bands | B1_30, B2_30, B_3_30, B4_30, B5_30, B6_30, B7_30 |
| Mean value of point heights (m) | zmean |
| Maximum value of point heights (m) | zmax |
| Standard deviation of point heights (m) | zsd |
| Skewness of point heights | Zskew |
| Kurtosis of point heights | Zkurt |
| Percentile values of point heights: 5th, 10th, 15th, …, 95th (m) | zq5, zq10, zq15, …, zq95 |
| Entropy calculated as a normalized Shannon vertical complexity index | zentropy |
| Percentage of all returns above 2 m (%) | pzabove2 |
| Percentage of all returns above zmean (%) | pzabovezmean |
| Cumulative percentage of returns from nine height layers. The height measurements were divided into 10 equal intervals according to Reference [31] (%) | zpcum1, zpcum2, zcum3, …, zpcum9 |
| Dominant Tree Species | Number of Stands | Percentage of Stands (%) | Minimum GSV (m3/ha) | Mean GSV (m3/ha) | Maximum GSV (m3/ha) | Standard Deviation of GSV (m3/ha) |
|---|---|---|---|---|---|---|
| Scots pine | 270 | 75.0 | 111.0 | 332.6 | 593.0 | 97.4 |
| silver fir | 29 | 8.1 | 223.0 | 387.7 | 629.0 | 97.4 |
| European beech | 22 | 6.1 | 158.0 | 336.0 | 590.0 | 116.3 |
| sessile oak | 20 | 5.6 | 174.0 | 348.0 | 473.0 | 86.8 |
| other species | 19 | 5.3 | 190.0 | 277.6 | 479.0 | 77.7 |
| Model | Hyperparameter | Values | Tuning Method |
|---|---|---|---|
| KNN | K | 1–40 | Random Search |
| Minkowski distance | Euclidean distance, Manhattan distance | ||
| Distance metrics | Unweighted, Weighted, Inverse, Reciprocal | ||
| RF | ntree | 100–500 | Random Search |
| mtry | 2—number of predictors | ||
| tdepth | 2–10 | ||
| DL | Hidden layers | Trial-and-error | |
| Nodes | |||
| Optimizer | |||
| Learning Rate | |||
| Dropout layers | |||
| Dropout percentage | |||
| Activation functions | |||
| Regularization layers | |||
| L1 regularization | |||
| L2 regularization | |||
| Predictive Approach | Predictors | Configurations | ||
|---|---|---|---|---|
| RF | mtry | tdepth | ntree | |
| Landsat | 2 | 9 | 500 | |
| ALS | 2 | 6 | 500 | |
| Landsat+ALS | 2 | 3 | 500 | |
| k-NN | kmax | distance | kernel | |
| Landsat | 36 | Manhattan | unweighted | |
| ALS | 37 | Manhattan | inverse | |
| Landsat+ALS | 35 | Manhattan | inverse | |
| Model | Predictors | R2 | RMSE (m3/ha) | RMSE% | Bias (m3/ha) | Bias% |
|---|---|---|---|---|---|---|
| DL | Landsat | 0.01 | 106.24 | 31.69 | 40.39 | 12.03 |
| DL | ALS | 0.39 | 81.43 | 24.30 | −11.64 | −3.48 |
| DL | Landsat+ALS | 0.38 | 80.98 | 24.16 | −7.31 | −2.2 |
| KNN | Landsat | 0.04 | 104.54 | 31.19 | 39.94 | 11.9 |
| KNN | ALS | 0.38 | 83.24 | 24.85 | −16.77 | −5.02 |
| KNN | Landsat+ALS | 0.38 | 83.19 | 24.83 | −16.94 | −5.06 |
| LM | Landsat | 0.05 | 104.92 | 31.3 | 38 | 11.3 |
| LM | ALS | 0.37 | 82.72 | 24.69 | −11.7 | −3.51 |
| LM | Landsat+ALS | 0.37 | 82.57 | 24.64 | −12.01 | −3.6 |
| RF | Landsat | 0.07 | 102.10 | 30.45 | 36.56 | 10.89 |
| RF | ALS | 0.38 | 83.90 | 25.03 | −16.51 | −4.93 |
| RF | Landsat+ALS | 0.39 | 84.41 | 25.18 | −17.81 | −5.32 |
| Species | Number of Stands | Method | R2 | RMSE (m3/ha) | RMSE% | Bias (m3/ha) | Bias% |
|---|---|---|---|---|---|---|---|
| European beech | 22 | DL | 0.46 | 106.94 | 32.13 | −66.62 | −20.19 |
| KNN | 0.51 | 108.39 | 32.59 | −74.39 | −22.5 | ||
| LM | 0.53 | 110.19 | 33.11 | −78.55 | −23.71 | ||
| RF | 0.49 | 109.92 | 33.03 | −74.21 | −22.45 | ||
| Scots pine | 270 | DL | 0.42 | 76.02 | 22.86 | −2.80 | −0.86 |
| KNN | 0.42 | 78.6 | 23.64 | −12.08 | −3.65 | ||
| LM | 0.40 | 77.79 | 23.40 | −6.13 | −1.86 | ||
| RF | 0.42 | 80.00 | 24.06 | −13.18 | −3.98 | ||
| sessile oak | 20 | DL | 0.33 | 72.61 | 20.99 | −10.91 | −3.32 |
| KNN | 0.31 | 77.22 | 22.34 | −20.19 | −5.99 | ||
| LM | 0.36 | 73.38 | 21.24 | −22.78 | −6.73 | ||
| RF | 0.32 | 77.62 | 22.45 | −20.27 | −6.01 | ||
| silver fir | 29 | DL | 0.28 | 87.74 | 22.63 | 18.01 | 4.53 |
| KNN | 0.3 | 83.32 | 21.49 | 6.78 | 1.63 | ||
| LM | 0.29 | 84.08 | 21.68 | 12.81 | 3.18 | ||
| RF | 0.32 | 84.53 | 21.82 | 3.15 | 0.70 | ||
| other species | 19 | DL | 0.13 | 103.23 | 37.11 | −37.98 | −14.17 |
| KNN | 0.12 | 109.53 | 39.45 | −51.86 | −19.19 | ||
| LM | 0.14 | 108.51 | 39.04 | −45.55 | −16.91 | ||
| RF | 0.12 | 109.09 | 39.27 | −47.89 | −17.74 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hawryło, P.; Francini, S.; Chirici, G.; Giannetti, F.; Parkitna, K.; Krok, G.; Mitelsztedt, K.; Lisańczuk, M.; Stereńczak, K.; Ciesielski, M.; et al. The Use of Remotely Sensed Data and Polish NFI Plots for Prediction of Growing Stock Volume Using Different Predictive Methods. Remote Sens. 2020, 12, 3331. https://doi.org/10.3390/rs12203331
Hawryło P, Francini S, Chirici G, Giannetti F, Parkitna K, Krok G, Mitelsztedt K, Lisańczuk M, Stereńczak K, Ciesielski M, et al. The Use of Remotely Sensed Data and Polish NFI Plots for Prediction of Growing Stock Volume Using Different Predictive Methods. Remote Sensing. 2020; 12(20):3331. https://doi.org/10.3390/rs12203331
Chicago/Turabian StyleHawryło, Paweł, Saverio Francini, Gherardo Chirici, Francesca Giannetti, Karolina Parkitna, Grzegorz Krok, Krzysztof Mitelsztedt, Marek Lisańczuk, Krzysztof Stereńczak, Mariusz Ciesielski, and et al. 2020. "The Use of Remotely Sensed Data and Polish NFI Plots for Prediction of Growing Stock Volume Using Different Predictive Methods" Remote Sensing 12, no. 20: 3331. https://doi.org/10.3390/rs12203331
APA StyleHawryło, P., Francini, S., Chirici, G., Giannetti, F., Parkitna, K., Krok, G., Mitelsztedt, K., Lisańczuk, M., Stereńczak, K., Ciesielski, M., Wężyk, P., & Socha, J. (2020). The Use of Remotely Sensed Data and Polish NFI Plots for Prediction of Growing Stock Volume Using Different Predictive Methods. Remote Sensing, 12(20), 3331. https://doi.org/10.3390/rs12203331