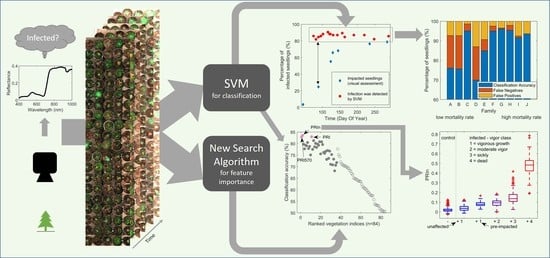
Using Hyperspectral Imagery to Detect an Invasive Fungal Pathogen and Symptom Severity in Pinus strobiformis Seedlings of Different Genotypes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Set-Up
2.2. Vigor Assessments of Seedlings Inoculated with White Pine Blister Rust
2.3. Hyperspectral Image Acquisition
2.4. Hyperspectral Image Processing
2.5. Hyperspectral Target Extraction
2.6. Hyperspectral Features
2.7. Classification
2.8. Statistical Analysis to Assess Feature Importance
3. Results
3.1. Visual Assessments of White Pine Blister Rust
3.2. Classification Results
3.2.1. White Pine Blister Rust Infection Identification Detected by Hyperspectral Imaging
3.2.2. Classification Accuracy of Infection per Family
3.2.3. Classification into Vigor Class
3.3. Hyperspectral Feature Importance
3.3.1. Feature Identification for Infection Detection
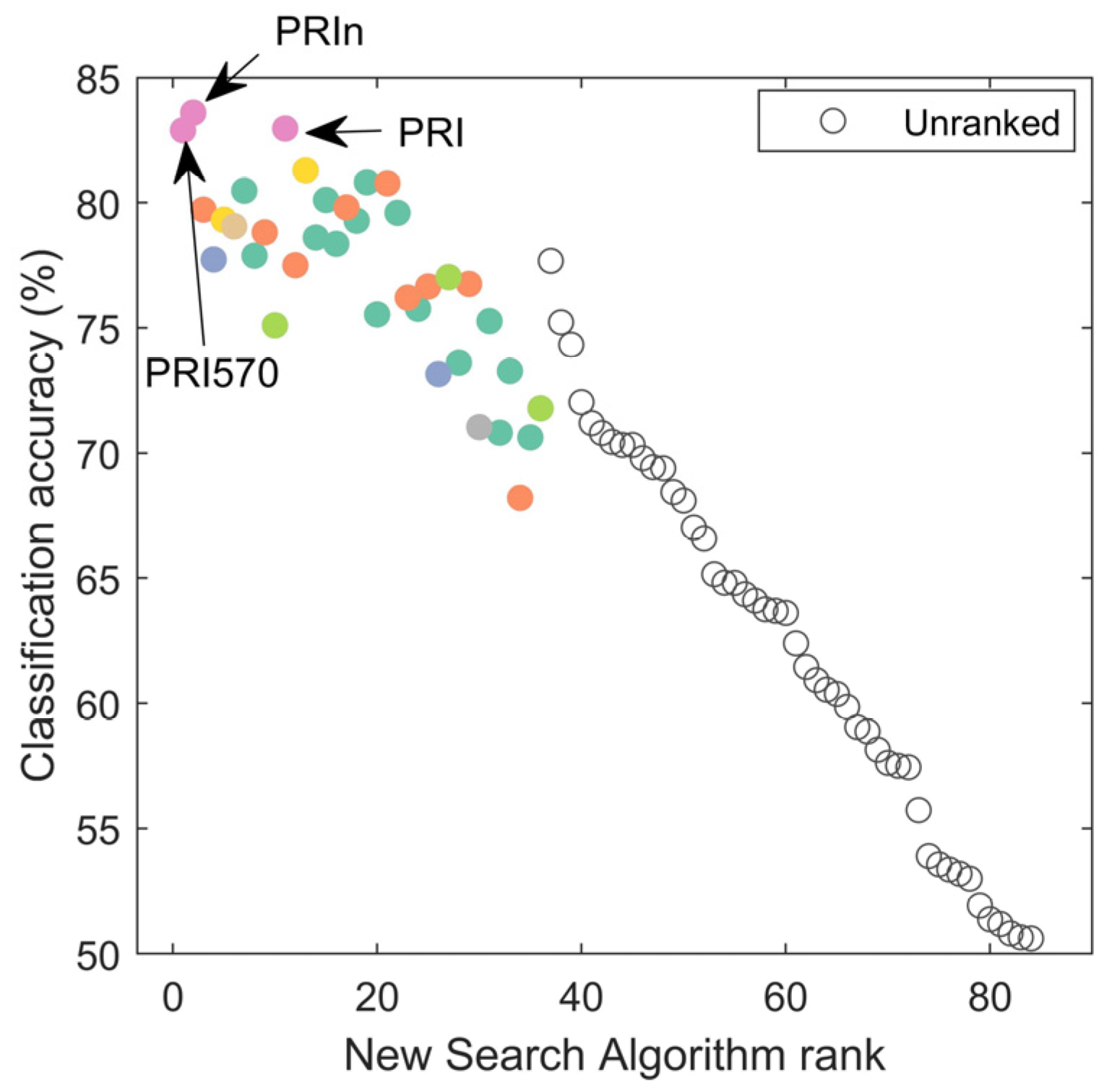
3.3.2. Feature Identification Using ‘New Search Algorithm’
3.3.3. New Search Algorithm Applied to All Features
3.3.4. Feature Identification for Family Separation
3.3.5. Feature Identification for Vigor Classes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Parmesan, C.; Yohe, G. A globally coherent fingerprint of climate change impacts across natural systems. Nature 2003, 421, 37–42. [Google Scholar] [CrossRef] [PubMed]
- Trumbore, S.; Brando, P.; Hartmann, H. Forest health and global change. Science 2015, 349, 814–818. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shirk, A.J.; Cushman, S.A.; Waring, K.M.; Wehenkel, C.A.; Leal-Sáenz, A.; Toney, C.; Lopez-Sanchez, C.A. Southwestern white pine (Pinus strobiformis) species distribution models project a large range shift and contraction due to regional climatic changes. For. Ecol. Manag. 2018, 411, 176–186. [Google Scholar] [CrossRef]
- Chakraborty, S.; Tiedemann, A.V.; Teng, P.S. Climate change: Potential impact on plant diseases. Environ. Pollut. 2000, 108, 317–326. [Google Scholar] [CrossRef]
- National Academies of Sciences and Medicine. Forest Health and Biotechnology: Possibilities and Considerations; National Academies Press: Washington, DC, USA, 2019; ISBN 978-0-309-48288-2. [Google Scholar]
- Fei, S.; Morin, R.S.; Oswalt, C.M.; Liebhold, A.M. Biomass losses resulting from insect and disease invasions in US forests. Proc. Natl. Acad. Sci. USA 2019, 116, 17371–17376. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kuska, M.; Wahabzada, M.; Leucker, M.; Dehne, H.-W.; Kersting, K.; Oerke, E.-C.; Steiner, U.; Mahlein, A.-K. Hyperspectral phenotyping on the microscopic scale: Towards automated characterization of plant-pathogen interactions. Plant Methods 2015, 11, 28. [Google Scholar] [CrossRef] [Green Version]
- Sniezko, R.A.; Koch, J. Breeding trees resistant to insects and diseases: Putting theory into application. Biol. Invasions 2017, 19, 3377–3400. [Google Scholar] [CrossRef]
- Sniezko, R.A.; Johnson, J.S.; Savin, D.P. Assessing the durability, stability, and usability of genetic resistance to a non-native fungal pathogen in two pine species. Plants People Planet 2020, 2, 57–68. [Google Scholar] [CrossRef] [Green Version]
- Showalter, D.N.; Raffa, K.F.; Sniezko, R.A.; Herms, D.A.; Liebhold, A.M.; Smith, J.A.; Bonello, P. Strategic development of tree resistance against forest pathogen and insect invasions in defense-free space. Front. Ecol. Evol. 2018, 6, 124. [Google Scholar] [CrossRef] [Green Version]
- Sniezko, R.A. Resistance breeding against nonnative pathogens in forest trees - Current successes in North America. Can. J. Plant Pathol. 2006, 28, S270–S279. [Google Scholar] [CrossRef]
- Kumar, A.; Lee, W.S.; Ehsani, R.J.; Albrigo, L.G.; Yang, C.; Mangan, R.L. Citrus greening disease detection using aerial hyperspectral and multispectral imaging techniques. J. Appl. Remote Sens. 2012, 6, 063542. [Google Scholar] [CrossRef]
- Devadas, R.; Lamb, D.W.; Simpfendorfer, S.; Backhouse, D. Evaluating ten spectral vegetation indices for identifying rust infection in individual wheat leaves. Precis. Agric. 2009, 10, 459–470. [Google Scholar] [CrossRef]
- López-López, M.; Calderón, R.; González-Dugo, V.; Zarco-Tejada, P.J.; Fereres, E. Early detection and quantification of almond red leaf blotch using high-resolution hyperspectral and thermal imagery. Remote Sens. 2016, 8, 276. [Google Scholar] [CrossRef] [Green Version]
- Calderón, R.; Navas-Cortés, J.A.; Lucena, C.; Zarco-Tejada, P.J. High-resolution airborne hyperspectral and thermal imagery for early detection of Verticillium wilt of olive using fluorescence, temperature and narrow-band. Remote Sens. Environ. 2013, 139, 231–245. [Google Scholar] [CrossRef]
- Calderón, R.; Navas-Cortés, J.A.; Zarco-Tejada, P.J. Early detection and quantification of Verticillium wilt in olive using hyperspectral and thermal imagery over large areas. Remote Sens. 2015, 7, 5584–5610. [Google Scholar] [CrossRef] [Green Version]
- Rumpf, T.; Mahlein, A.-K.; Steiner, U.; Oerke, E.-C.; Dehne, H.-W.; Plümer, L. Early detection and classification of plant diseases with support vector machines based on hyperspectral reflectance. Comput. Electron. Agric. 2010, 74, 91–99. [Google Scholar] [CrossRef]
- Mahlein, A.-K.; Steiner, U.; Dehne, H.-W.; Oerke, E.-C. Spectral signatures of sugar beet leaves for the detection and differentiation of diseases. Precis. Agric. 2010, 11, 413–431. [Google Scholar] [CrossRef]
- Gold, K.M.; Townsend, P.A.; Chlus, A.; Herrmann, I.; Couture, J.J.; Larson, E.R.; Gevens, A.J. Hyperspectral Measurements Enable Pre-Symptomatic Detection and Differentiation of Contrasting Physiological Effects of Late Blight and Early Blight in Potato. Remote Sens. 2020, 12, 286. [Google Scholar] [CrossRef] [Green Version]
- Leucker, M.; Wahabzada, M.; Kersting, K.; Peter, M.; Beyer, W.; Steiner, U.; Mahlein, A.-K.; Oerke, E.-C. Hyperspectral imaging reveals the effect of sugar beet QTLs on Cercospora leaf spot resistance. Funct. Plant Biol. 2017, 44, 1–9. [Google Scholar] [CrossRef]
- Alisaac, E.; Behmann, J.; Kuska, M.T.; Dehne, H.-W.; Mahlein, A.-K. Hyperspectral quantification of wheat resistance to Fusarium head blight: Comparison of two Fusarium species. Eur. J. Plant Pathol. 2018, 152, 869–884. [Google Scholar] [CrossRef]
- Lausch, A.; Erasmi, S.; King, D.J.; Magdon, P.; Heurich, M. Understanding forest health with Remote sensing-Part II-A review of approaches and data models. Remote Sens. 2017, 9, 129. [Google Scholar] [CrossRef] [Green Version]
- Conklin, D.A.; Fairweather, M.L.; Ryerson, D.E.; Geils, B.W.; Vogler, D.R. White pines, blister rust, and management in the Southwest. USDA For. Serv. Southwest Reg. R3-Fh-09-01 2009, 16, 3. [Google Scholar]
- Sniezko, R.A.; Mahalovich, M.F.; Schoettle, A.W.; Vogler, D.R. Past and current investigations of the genetic resistance to Cronartium ribicola in high-elevation five-needle pines. In The Future of High-Elevation, Five-Needle White Pines in Western North America. Proc High Five Symp RMRS-P-63; Keane, R.E., Tomback, D.F., Murray, M.P., Smith, C.M., Eds.; USDA Forest Service Rocky Mountain research Station: Fort Collins, CO, USA, 2011; pp. 246–264. [Google Scholar]
- King, J.N.; David, A.; Noshad, D.; Smith, J. A review of genetic approaches to the management of blister rust in white pines. For. Pathol. 2010, 40, 292–313. [Google Scholar] [CrossRef]
- Kinloch, B.B. Forest Pathology for the Last Century: A Retrospective and Directions for the Future White Pine Blister Rust in North America: Past and Prognosis. Phytopathology 2003, 93, 1044–1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Looney, C.E.; Waring, K.M. Patterns of forest structure, competition and regeneration in southwestern white pine (Pinus strobiformis) forests. For. Ecol. Manag. 2012, 286, 159–170. [Google Scholar] [CrossRef]
- Hoff, R.; Bingham, R.T.; McDonald, G.I. Relative blister rust resistance of white pines. Eur. J. For. Pathol. 1980, 10, 307–316. [Google Scholar] [CrossRef]
- Wyka, S.A.; Munck, I.A.; Brazee, N.J.; Broders, K.D. Response of eastern white pine and associated foliar, blister rust, canker and root rot pathogens to climate change. For. Ecol. Manag. 2018, 423, 18–26. [Google Scholar] [CrossRef]
- Kegley, A.J.; Sniezko, R.A. Variation in blister rust resistance among 226 Pinus monticola and 217 P. lambertiana seedling families in the Pacific Northwest. In Breeding and Genetic Resources of Five-Needle Pines: Growth, Adaptability and Pest Resistance; Sniezko, R.A., Samman, S., Schlarbaum, S.E., Kriebel, H.B., Eds.; USDA Forest Service, Rocky Mountain Research Station RMPS-P-32: Fort Collins, CO, USA, 1998; pp. 209–225. [Google Scholar]
- Sniezko, R.A.; Kegley, A.J.; Danchok, R. White pine blister rust resistance in North American, Asian and European species-results from artificial inoculartion trials in Oregon. Ann. For. Res. 2008, 51, 53–66. [Google Scholar]
- Lopez Alcala, J.M.; Haagsma, M.; Udell, C.J.; Selker, J.S. HyperRail: Modular, 3D Printed, 1-100 meter, Programmable, and Low-cost Linear Motion Control System for Imaging and Sensor Suites. HardwareX 2019, 6, e00081. [Google Scholar] [CrossRef]
- Smith, G.M.; Milton, E.J. The use of the empirical line method to calibrate remotely sensed data to reflectance. Int. J. Remote Sens. 1999, 20, 2653–2662. [Google Scholar] [CrossRef]
- Thenkabail, P.P.S.; Smith, R.B.; Pauw, E. De Hyperspectral vegetation indices and their relationships with agricultural crop characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Serrano, L.; Peñuelas, J.; Ustin, S.L. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: Decomposing biochemical from structural signals. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Carter, G.A. Ratios of leaf reflectances in narrow wavebands as indicators of plant stress. Remote Sens. 1994, 15, 697–703. [Google Scholar] [CrossRef]
- Dixit, L.; Ram, S. Quantitative analysis by derivative electronic spectroscopy. Appl. Spectrosc. Rev. 1985, 21, 311–418. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Bradley, A.E. The use of the area under the ROC curve in the evaluation of machine learning algorithms. Pattern Recognit. 1997, 30, 1145–1159. [Google Scholar] [CrossRef] [Green Version]
- Bhattacharyya, A. On a measure of divergence between two statistical populations defined by their probability distributions. Bull. Calcutta Math. Soc. 1943, 35, 99–109. [Google Scholar]
- Haagsma, M.; Page, G.F.M.; Johnson, J.S. Hyperspectral Imagery of Pinus Strobiformis Infected with Fungal Pathogen, Version 1; Oregon State University: Corvallis, OR, USA, 2020. [Google Scholar] [CrossRef]
- Chappelle, E.W.; Kim, M.S.; McMurtrey III, J.E. Ratio analysis of reflectance spectra (RARS): An algorithm for the remote estimation of the concentrations of chlorophyll a, chlorophyll b, and carotenoids in soybean. Remote Sens. Environ. 1992, 39, 239–247. [Google Scholar] [CrossRef]
- Gamon, J.A.; Peñuelas, J.; Field, C.B. A narrow-waveband spectral index that tracks diurnal changes in photosynthetic efficiency. Remote Sens. Environ. 1992, 41, 35–44. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; González-Dugo, V.; Williams, L.E.; Suárez, L.; Berni, J.A.J.; Goldhamer, D.; Fereres, E. A PRI-based water stress index combining structural and chlorophyll effects: Assessment using diurnal narrow-band airborne imagery and the CWSI thermal index. Remote Sens. Environ. 2013, 138, 38–50. [Google Scholar] [CrossRef]
- Rossini, M.; Fava, F.; Cogliati, S.; Meroni, M.; Marchesi, A.; Panigada, C.; Giardino, C.; Busetto, L.; Migliavacca, M.; Amaducci, S.; et al. Assessing canopy PRI from airborne imagery to map water stress in maize. Isprs J. Photogramm. Remote Sens. 2013, 86, 168–177. [Google Scholar] [CrossRef]
- Hernández-Clemente, R.; Navarro-Cerrillo, R.M.; Suárez, L.; Morales, F.; Zarco-Tejada, P.J. Assessing structural effects on PRI for stress detection in conifer forests. Remote Sens. Environ. 2011, 115, 2360–2375. [Google Scholar] [CrossRef]
- Merzlyak, M.N.; Gitelson, A.A.; Chivkunova, O.B.; Rakitin, V.Y. Non-destructive optical detection of leaf senescence and fruit ripening. Physiol. Plant. 1999, 106, 135–141. [Google Scholar] [CrossRef] [Green Version]
- Neuwirthová, E.; Lhotáková, Z.; Albrechtová, J. The effect of leaf stacking on leaf reflectance and vegetation indices measured by contact probe during the season. Sensors 2017, 17, 1202. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.; Merzlyak, M. Signature Analysis of Leaf Reflectance Spectra: Algorithm Development for Remote Sensing of Chlorophyll. J. Plant Physiol. 1996, 148, 494–500. [Google Scholar] [CrossRef]
- Daughtry, C.S.T.; Walthall, C.L.; Kim, M.S.; Brown de Colstoun, E.; McMurtrey III, J.E. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Wang, H.; Chen, J.; Lin, H.; Yuan, D. Research on effectiveness of hyperspectral data on identifying rice of different genotypes. Remote Sens. Lett. 2010, 1, 223–229. [Google Scholar] [CrossRef]









| PREDICTED (HSI Model) | Probability of Detection | ||||||
|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | |||
| TRUE (visual assessment) | 0 | 73.6 | 9.0 | 1.0 | 1.4 | 0.0 | 86.6% |
| 1 | 9.6 | 21.2 | 3.2 | 1.2 | 0.0 | 60.2% | |
| 2 | 1.0 | 4.2 | 5.2 | 4.4 | 0.0 | 35.1% | |
| 3 | 0.0 | 2.6 | 4.6 | 12.4 | 0.4 | 62.0% | |
| 4 | 0.0 | 0.0 | 0.0 | 1.2 | 13.6 | 91.9% | |
| precision | 87.4% | 57.3% | 37.1% | 60.2% | 97.1% | OA = 74.2% | |
| AUC | 0.926 | 0.782 | 0.856 | 0.926 | 0.998 | κ = 0.62 | |
| PREDICTED (HSI Model) | Probability of Detection | |||||
|---|---|---|---|---|---|---|
| 0 | 1 | 2 & 3 | 4 | |||
| TRUE (visual assessment) | 0 | 73.8 | 9.0 | 2.2 | 0.0 | 86.8% |
| 1 | 9.4 | 20.8 | 5.0 | 0.0 | 59.1% | |
| 2 & 3 | 1.2 | 6.0 | 27.2 | 0.4 | 78.2% | |
| 4 | 0.0 | 0.0 | 1.2 | 13.6 | 91.9% | |
| precision | 87.4% | 58.1% | 76.4% | 97.1% | OA = 79.7% | |
| AUC | 0.927 | 0.792 | 0.935 | 0.998 | κ = 0.69 | |
| Rank | ||||||
|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | |
| Overall | SR (578/526) | PRIn | SR (576/526) | PSRI | SR (582/522) | SR (531/700) |
| Early Time | SR (578/522) | PRIn | SR (569/526) | SR (533/564) | SR (582/522) | SR (526/575) |
| Late Time | SR (680/511) | PSRI | SR (678/506) | PRIn | SR (676/511) | SR (529/689) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Haagsma, M.; Page, G.F.M.; Johnson, J.S.; Still, C.; Waring, K.M.; Sniezko, R.A.; Selker, J.S. Using Hyperspectral Imagery to Detect an Invasive Fungal Pathogen and Symptom Severity in Pinus strobiformis Seedlings of Different Genotypes. Remote Sens. 2020, 12, 4041. https://doi.org/10.3390/rs12244041
Haagsma M, Page GFM, Johnson JS, Still C, Waring KM, Sniezko RA, Selker JS. Using Hyperspectral Imagery to Detect an Invasive Fungal Pathogen and Symptom Severity in Pinus strobiformis Seedlings of Different Genotypes. Remote Sensing. 2020; 12(24):4041. https://doi.org/10.3390/rs12244041
Chicago/Turabian StyleHaagsma, Marja, Gerald F. M. Page, Jeremy S. Johnson, Christopher Still, Kristen M. Waring, Richard A. Sniezko, and John S. Selker. 2020. "Using Hyperspectral Imagery to Detect an Invasive Fungal Pathogen and Symptom Severity in Pinus strobiformis Seedlings of Different Genotypes" Remote Sensing 12, no. 24: 4041. https://doi.org/10.3390/rs12244041
APA StyleHaagsma, M., Page, G. F. M., Johnson, J. S., Still, C., Waring, K. M., Sniezko, R. A., & Selker, J. S. (2020). Using Hyperspectral Imagery to Detect an Invasive Fungal Pathogen and Symptom Severity in Pinus strobiformis Seedlings of Different Genotypes. Remote Sensing, 12(24), 4041. https://doi.org/10.3390/rs12244041