A Sparse Representation-Based Sample Pseudo-Labeling Method for Hyperspectral Image Classification
Abstract
1. Introduction
2. Related Work
2.1. Intrinsic Image Decomposition of Hyperspectral Images
2.2. Sparse Representation Classification of Hyperspectral Images
3. Proposed Method
3.1. Feature Extraction of Hyperspectral Images
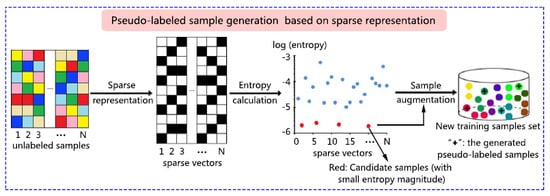
3.2. Candidate Sample Selection Based on Sparse Representation
3.3. Pseudo-Label Assignment for Candidate Samples
3.4. Classification Model Optimization Using Pseudo-Labeled Samples
3.5. Pseudo-Code of the Proposed SRSPL Method
| Algorithm 1: Sparse Representation-based Sample Pseudo-Labeling (SRSPL) for Hyperspectral Image |
| Classification |
| Input: Hyperspectral image ; the initial labeled samples set ; the hyper- |
| spectral samples set other than the training samples and test samples: |
| Output: Classification map |
| 1: Reduce the dimension and noise of based on averaging image fusion (Equation (7)) to obtain the |
| dimensionally reduced image . |
| 2: According to Equation (8), partition the into several subgroups of adjacent bands, denoted as . |
| 3: According to Equations (9) and (10), obtain the spectral reflectance components of each , (through the |
| optimization-based IID method) and combined to obtain the resulting IID features . |
| 4: For each Do |
| (1) According to Equation (11) and (12), solve the sparse representation coefficient of each sample |
| based on the reflectance component and the overcomplete dictionary . |
| (2) According to Equation (13), calculate the information entropy of each to discriminating the |
| purity of each sample . |
| 5:End For |
| 6: Sort the samples corresponding to these reflectance components in ascending order according to their |
| entropy magnitudes. |
| 7: Selected the first T samples as the candidate samples set . |
| 8: According to Equation (14), assign the pseudo-label for each sample in set , to obtain the pseudo- |
| labeled samples set . |
| 9: Combine the initial labeled samples set and the pseudo-labeled samples set to the new |
| labeled samples set . |
| 10: Classify the spectral reflectance components with the extended random walker (ERW) classifier and the |
| new labeled samples set to obtain the final classification map . |
| 11:Return |
4. Experiment
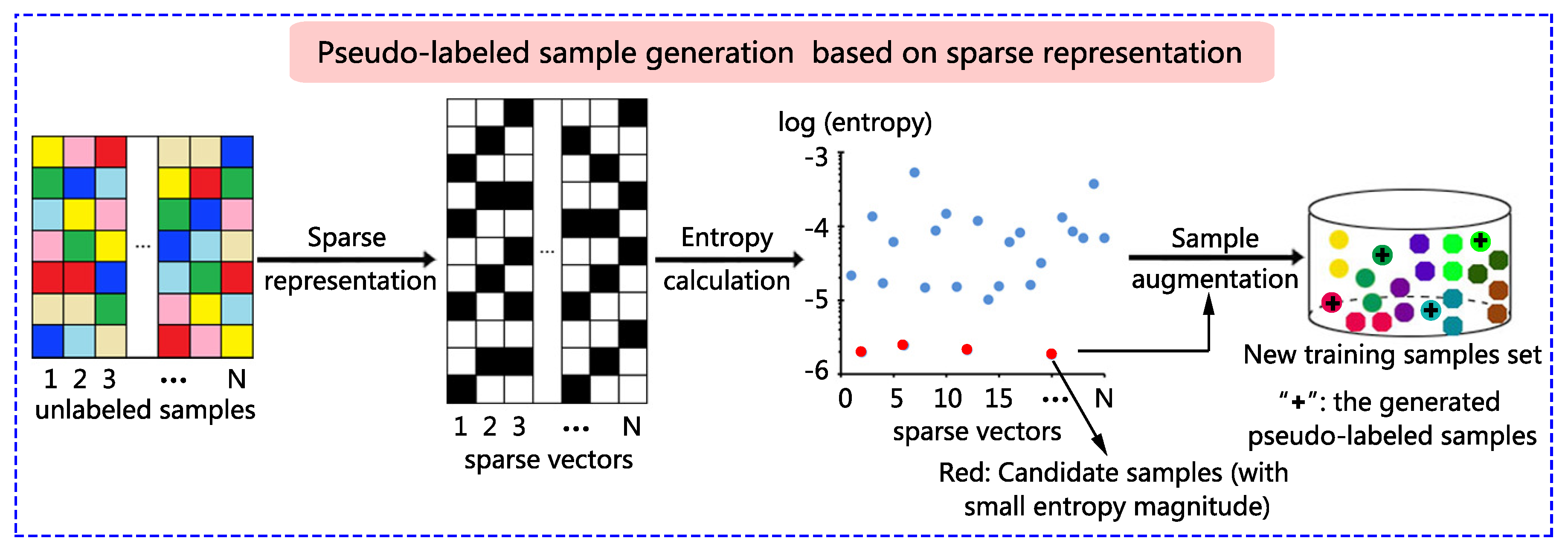
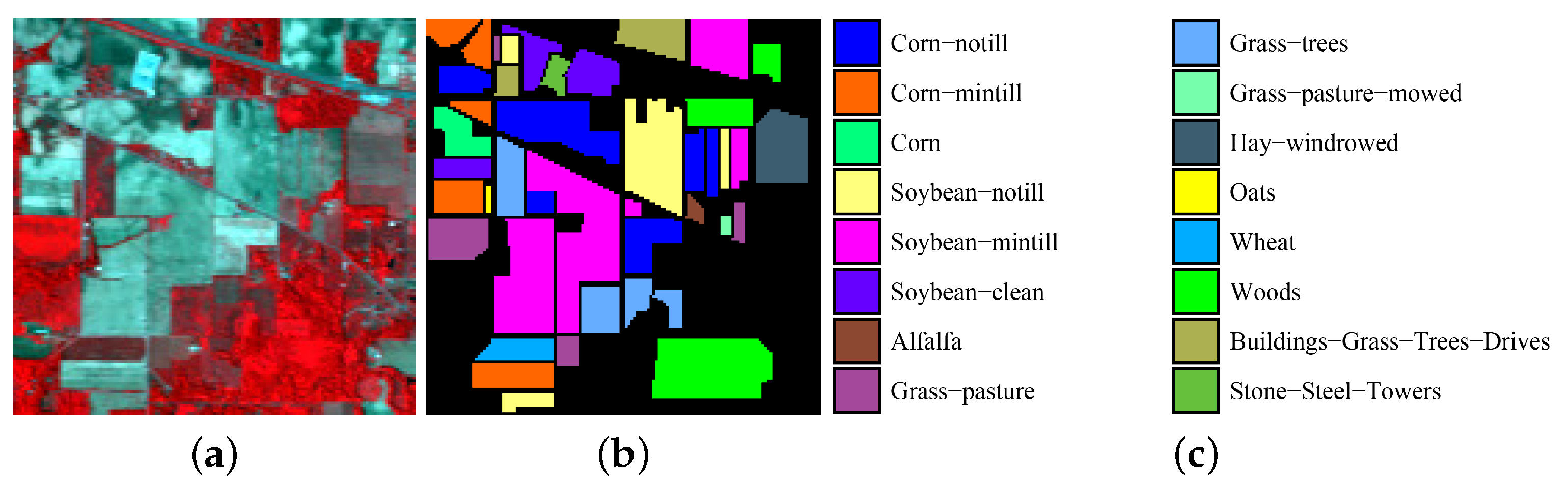
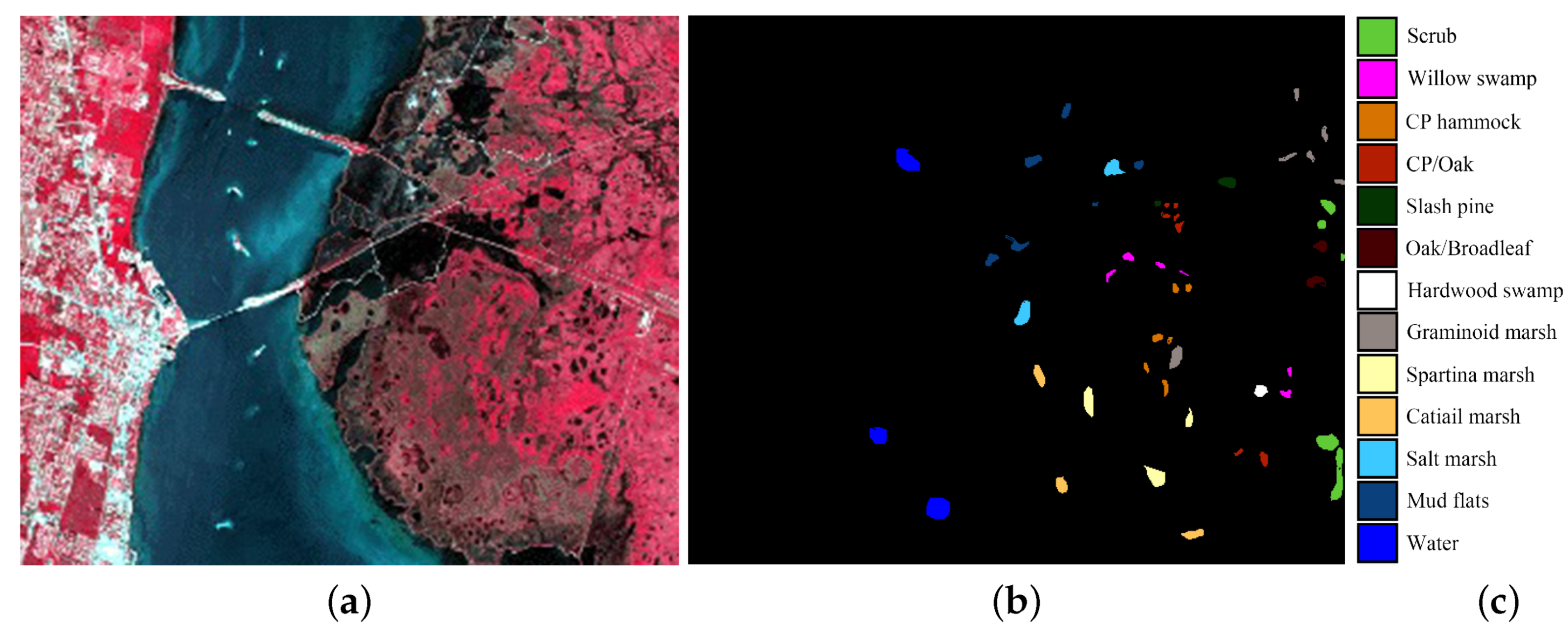
4.1. Experimental Data Sets
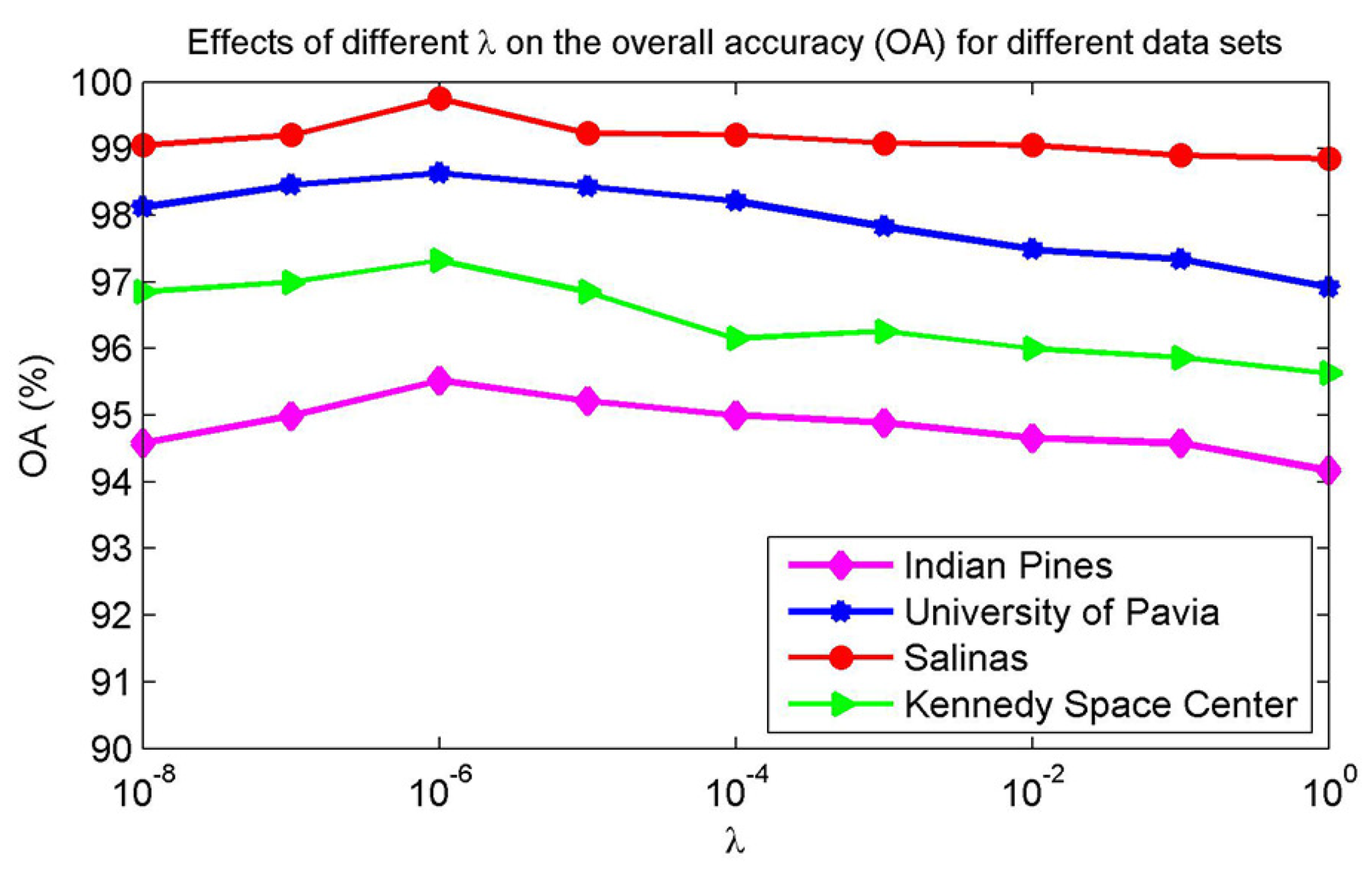
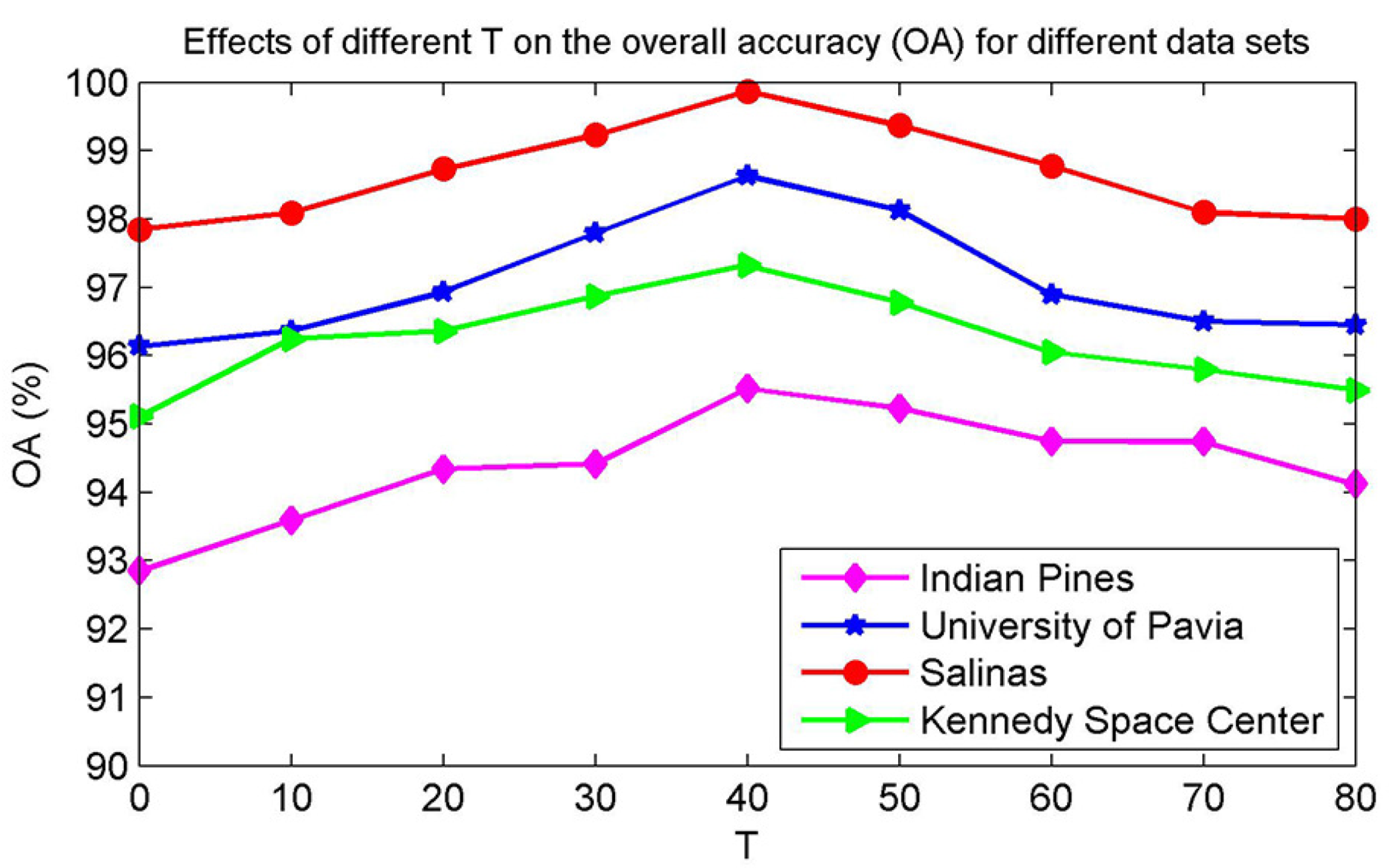
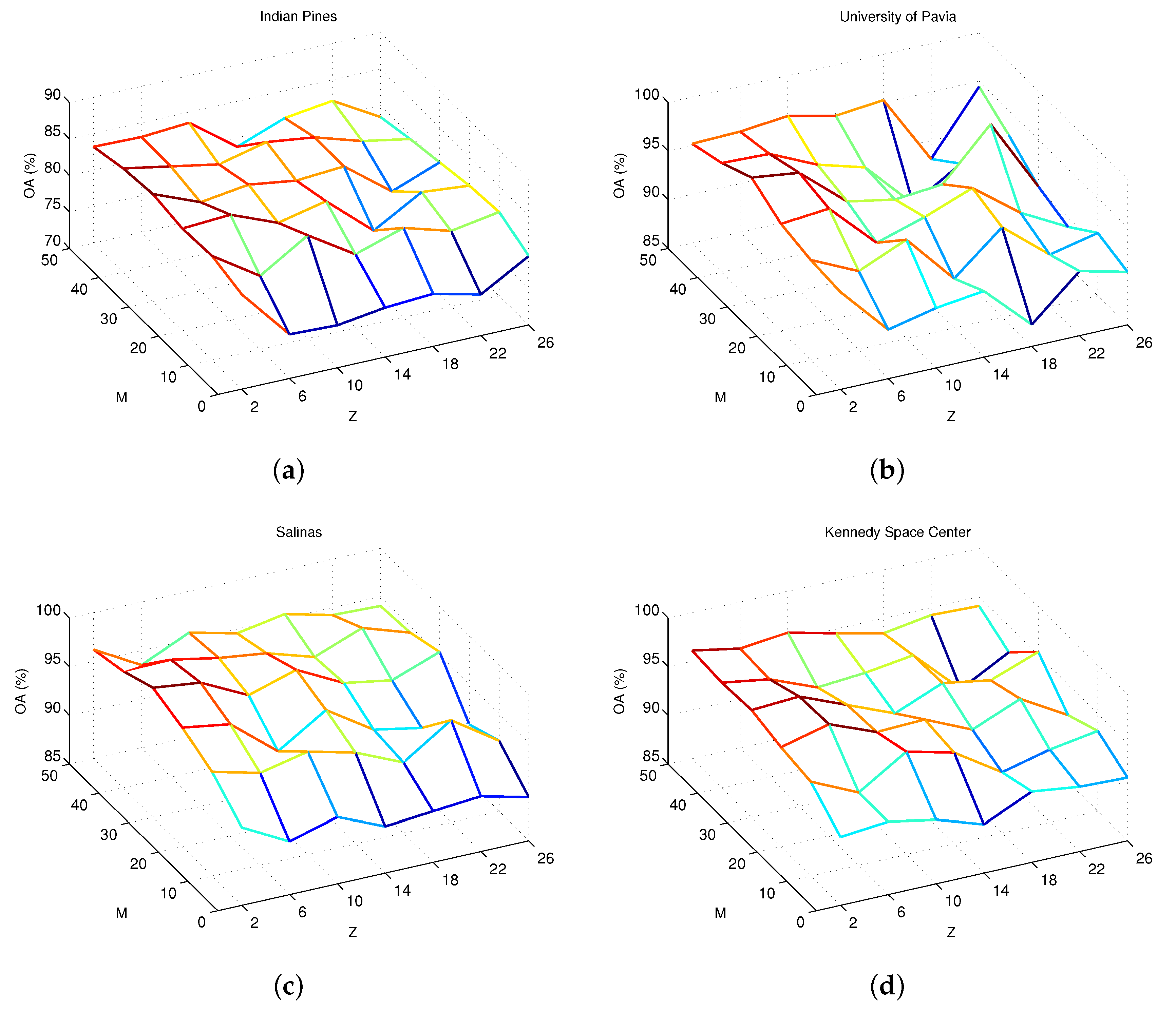
4.2. Parameter Analysis
4.3. Performance Evaluation
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kang, X.; Li, C.; Li, S.; Lin, H. Classification of hyperspectral images by Gabor filtering based deep network. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 11, 1166–1178. [Google Scholar] [CrossRef]
- Pontius, J.; Martin, M.; Plourde, L.; Hallett, R. Ash decline assessment in emerald ash borer-infested regions: A test of tree-level, hyperspectral technologies. Remote Sens. Environ. 2008, 112, 2665–2676. [Google Scholar] [CrossRef]
- Imani, M. Manifold structure preservative for hyperspectral target detection. Adv. Space Res. 2018, 61, 2510–2520. [Google Scholar] [CrossRef]
- Cao, X.; Zhou, F.; Xu, L.; Meng, D.; Xu, Z.; Paisley, J. Hyperspectral image classification with Markov random fields and a convolutional neural network. IEEE Trans. Image Process. 2018, 27, 2354–2367. [Google Scholar] [CrossRef]
- Sharma, S.; Buddhiraju, K.M. Spatial–spectral ant colony optimization for hyperspectral image classification. Int. J. Remote Sens. 2018, 39, 2702–2717. [Google Scholar] [CrossRef]
- Hughes, G. On the mean accuracy of statistical pattern recognizers. IEEE Trans. Inf. Theory 1968, 14, 55–63. [Google Scholar] [CrossRef]
- Xia, J.; Du, P.; He, X.; Chanussot, J. Hyperspectral remote sensing image classification based on rotation forest. IEEE Geosci. Remote Sens. Lett. 2013, 11, 239–243. [Google Scholar] [CrossRef]
- Mohanty, R.; Happy, S.; Routray, A. Spatial–Spectral Regularized Local Scaling Cut for Dimensionality Reduction in Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2018, 16, 932–936. [Google Scholar] [CrossRef]
- Shamsolmoali, P.; Zareapoor, M.; Yang, J. Convolutional neural network in network (CNNiN): Hyperspectral image classification and dimensionality reduction. IET Image Process. 2018, 13, 246–253. [Google Scholar] [CrossRef]
- Fang, B.; Li, Y.; Zhang, H.; Chan, J. Semi-supervised deep learning classification for hyperspectral image based on dual-strategy sample selection. Remote Sens. 2018, 10, 574. [Google Scholar] [CrossRef]
- Cui, B.; Xie, X.; Hao, S.; Cui, J.; Lu, Y. Semi-supervised classification of hyperspectral images based on extended label propagation and rolling guidance filtering. Remote Sens. 2018, 10, 515. [Google Scholar] [CrossRef]
- Liu, B.; Yu, X.; Zhang, P.; Yu, A.; Fu, Q.; Wei, X. Supervised deep feature extraction for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 56, 1909–1921. [Google Scholar] [CrossRef]
- He, N.; Paoletti, M.E.; Haut, J.M.; Fang, L.; Li, S.; Plaza, A.; Plaza, J. Feature extraction with multiscale covariance maps for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2018, 57, 755–769. [Google Scholar] [CrossRef]
- Medjahed, S.A.; Ouali, M. Band selection based on optimization approach for hyperspectral image classification. Egyptian J. Remote Sens. Space Sci. 2018, 21, 413–418. [Google Scholar] [CrossRef]
- Cui, B.; Xie, X.; Ma, X.; Ren, G.; Ma, Y. Superpixel-based extended random walker for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 3233–3243. [Google Scholar] [CrossRef]
- Jamshidpour, N.; Homayouni, S.; Safari, A. Graph-based semi-supervised hyperspectral image classification using spatial information. In Proceedings of the Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Los Angeles, LA, USA, 21–24 August 2016; pp. 1–4. [Google Scholar]
- Haut, J.M.; Paoletti, M.E.; Plaza, J.; Li, J.; Plaza, A. Active learning with convolutional neural networks for hyperspectral image classification using a new bayesian approach. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6440–6461. [Google Scholar] [CrossRef]
- Wang, Z.; Du, B.; Zhang, L.; Zhang, L.; Jia, X. A novel semisupervised active-learning algorithm for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3071–3083. [Google Scholar] [CrossRef]
- Li, J.; Xi, B.; Li, Y.; Du, Q.; Wang, K. Hyperspectral classification based on texture feature enhancement and deep belief networks. Remote Sens. 2018, 10, 396. [Google Scholar] [CrossRef]
- Zhou, S.; Chen, Q.; Wang, X. Fuzzy deep belief networks for semi-supervised sentiment classification. Neurocomputing 2014, 131, 312–322. [Google Scholar] [CrossRef]
- Sun, B.; Kang, X.; Li, S.; Benediktsson, J.A. Random-walker-based collaborative learning for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2016, 55, 212–222. [Google Scholar] [CrossRef]
- Imani, M.; Ghassemian, H. Adaptive expansion of training samples for improving hyperspectral image classification performance. In Proceedings of the Iranian Conference on Electrical Engineering (ICEE), Mashhad, Iran, 14–16 May 2013; pp. 1–6. [Google Scholar]
- Wu, H.; Prasad, S. Semi-supervised deep learning using pseudo labels for hyperspectral image classification. IEEE Trans. Image Process. 2017, 27, 1259–1270. [Google Scholar] [CrossRef] [PubMed]
- Ma, D.; Tang, P.; Zhao, L. SiftingGAN: Generating and Sifting Labeled Samples to Improve the Remote Sensing Image Scene Classification Baseline In Vitro. IEEE Geosci. Remote Sens. Lett. 2019, 16, 1046–1050. [Google Scholar] [CrossRef]
- Prasad, S.; Labate, D.; Cui, M.; Zhang, Y. Morphologically decoupled structured sparsity for rotation-invariant hyperspectral image analysis. IEEE Trans. Geosci. Remote Sens. 2017, 55, 4355–4366. [Google Scholar] [CrossRef]
- Prasad, S.; Labate, D.; Cui, M.; Zhang, Y. Rotation invariance through structured sparsity for robust hyperspectral image classification. In Proceedings of the International Conference on Acoustics, Speech and Signal Processing (ICASSP), New Orleans, LA, USA, 5–9 March 2017; pp. 6205–6209. [Google Scholar]
- Li, J.; Zhang, H.; Zhang, L. Efficient superpixel-level multitask joint sparse representation for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2015, 53, 5338–5351. [Google Scholar]
- Tu, B.; Zhang, X.; Kang, X.; Zhang, G.; Wang, J.; Wu, J. Hyperspectral image classification via fusing correlation coefficient and joint sparse representation. IEEE Geosci. Remote Sens. Lett. 2018, 15, 340–344. [Google Scholar] [CrossRef]
- Chen, Y.; Nasrabadi, N.M.; Tran, T.D. Hyperspectral image classification via kernel sparse representation. IEEE Trans. Geosci. Remote Sens. 2012, 51, 217–231. [Google Scholar] [CrossRef]
- Gan, L.; Xia, J.; Du, P.; Chanussot, J. Multiple feature kernel sparse representation classifier for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5343–5356. [Google Scholar] [CrossRef]
- Liu, J.; Xiao, Z.; Chen, Y.; Yang, J. Spatial-spectral graph regularized kernel sparse representation for hyperspectral image classification. ISPRS Int. J. Geo-Inf. 2017, 6, 258. [Google Scholar] [CrossRef]
- Fang, L.; Wang, C.; Li, S.; Benediktsson, J.A. Hyperspectral image classification via multiple-feature-based adaptive sparse representation. IEEE Trans. Instrum. Meas. 2017, 66, 1646–1657. [Google Scholar] [CrossRef]
- Tong, F.; Tong, H.; Jiang, J.; Zhang, Y. Multiscale union regions adaptive sparse representation for hyperspectral image classification. Remote Sens. 2017, 9, 872. [Google Scholar] [CrossRef]
- Pan, B.; Shi, Z.; Xu, X. Multiobjective-based sparse representation classifier for hyperspectral imagery using limited samples. IEEE Trans. Geosci. Remote Sens. 2018, 57, 239–249. [Google Scholar] [CrossRef]
- Jian, M.; Jung, C. Class-discriminative kernel sparse representation-based classification using multi-objective optimization. IEEE Trans. Signal Process. 2013, 61, 4416–4427. [Google Scholar] [CrossRef]
- Sun, X.; Yang, L.; Zhang, B.; Gao, L.; Gao, J. An endmember extraction method based on artificial bee colony algorithms for hyperspectral remote sensing images. Remote Sens. 2015, 7, 16363–16383. [Google Scholar] [CrossRef]
- Kang, X.; Li, S.; Fang, L.; Benediktsson, J.A. Intrinsic image decomposition for feature extraction of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2014, 53, 2241–2253. [Google Scholar] [CrossRef]
- Tappen, M.F.; Freeman, W.T.; Adelson, E.H. Recovering intrinsic images from a single image. In Advances in Neural Information Processing Systems; Massachusetts Institute of Technology Press: Cambridge, MA, USA, 2003; pp. 1367–1374. [Google Scholar]
- Yacoob, Y.; Davis, L.S. Segmentation using appearance of mesostructure roughness. Int. J. Comput. Vis. 2009, 83, 248–273. [Google Scholar] [CrossRef]
- Shen, J.; Yang, X.; Li, X.; Jia, Y. Intrinsic image decomposition using optimization and user scribbles. IEEE Trans. Cybern. 2013, 43, 425–436. [Google Scholar] [CrossRef]
- Shen, J.; Yang, X.; Jia, Y.; Li, X. Intrinsic images using optimization. In Proceedings of the Conference on Computer Vision and Pattern Recognition (CVPR), Providence, RI, USA, 20–25 June 2011; pp. 3481–3487. [Google Scholar]
- Wright, J.; Yang, A.Y.; Ganesh, A.; Sastry, S.S.; Ma, Y. Robust face recognition via sparse representation. IEEE Trans. Pattern Anal. Mach. Intell. 2008, 31, 210–227. [Google Scholar] [CrossRef]
- Chen, Y.; Nasrabadi, N.M.; Tran, T.D. Hyperspectral image classification using dictionary-based sparse representation. IEEE Trans. Geosci. Remote Sens. 2011, 49, 3973–3985. [Google Scholar] [CrossRef]
- Tibshirani, R. Regression shrinkage and selection via the lasso: A retrospective. J. R. Stat. Soc. Ser. (Stat. Methodol.) 2011, 73, 273–282. [Google Scholar] [CrossRef]
- Nitanda, A. Stochastic proximal gradient descent with acceleration techniques. In Advances in Neural Information Processing Systems; Massachusetts Institute of Technology Press: Cambridge, MA, USA, 2014; pp. 1574–1582. [Google Scholar]
- Kang, X.; Li, S.; Fang, L.; Li, M.; Benediktsson, J.A. Extended random walker-based classification of hyperspectral images. IEEE Trans. Geosci. Remote. Sens. 2014, 53, 144–153. [Google Scholar] [CrossRef]
- Bouda, M.; Rousseau, A.N.; Gumiere, S.J.; Gagnon, P.; Konan, B.; Moussa, R. Implementation of an automatic calibration procedure for HYDROTEL based on prior OAT sensitivity and complementary identifiability analysis. Hydrol. Process. 2014, 28, 3947–3961. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Wang, L.; Hao, S.; Wang, Q.; Wang, Y. Semi-supervised classification for hyperspectral imagery based on spatial-spectral label propagation. ISPRS J. Photogramm. Remote Sens. 2014, 97, 123–137. [Google Scholar] [CrossRef]
- Wu, Y.; Li, J.; Gao, L.; Tan, X.; Zhang, B. Graphics processing unit–accelerated computation of the Markov random fields and loopy belief propagation algorithms for hyperspectral image classification. J. Appl. Remote Sens. 2015, 9, 097295. [Google Scholar] [CrossRef]
- Yang, X.; Ye, Y.; Li, X.; Lau, R.Y.; Zhang, X.; Huang, X. Hyperspectral image classification with deep learning models. IEEE Trans. Geosci. Remote. Sens. 2018, 56, 5408–5423. [Google Scholar] [CrossRef]
- Hang, R.; Liu, Q.; Hong, D.; Ghamisi, P. Cascaded recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote. Sens. 2019, 57, 5384–5394. [Google Scholar] [CrossRef]
- Farrell, M.D.; Mersereau, R.M. On the impact of PCA dimension reduction for hyperspectral detection of difficult targets. IEEE Geosci. Remote Sens. Lett. 2005, 2, 192–195. [Google Scholar] [CrossRef]
- Kang, X.; Li, S.; Benediktsson, J.A. Feature extraction of hyperspectral images with image fusion and recursive filtering. IEEE Trans. Geosci. Remote Sens. 2013, 52, 3742–3752. [Google Scholar] [CrossRef]

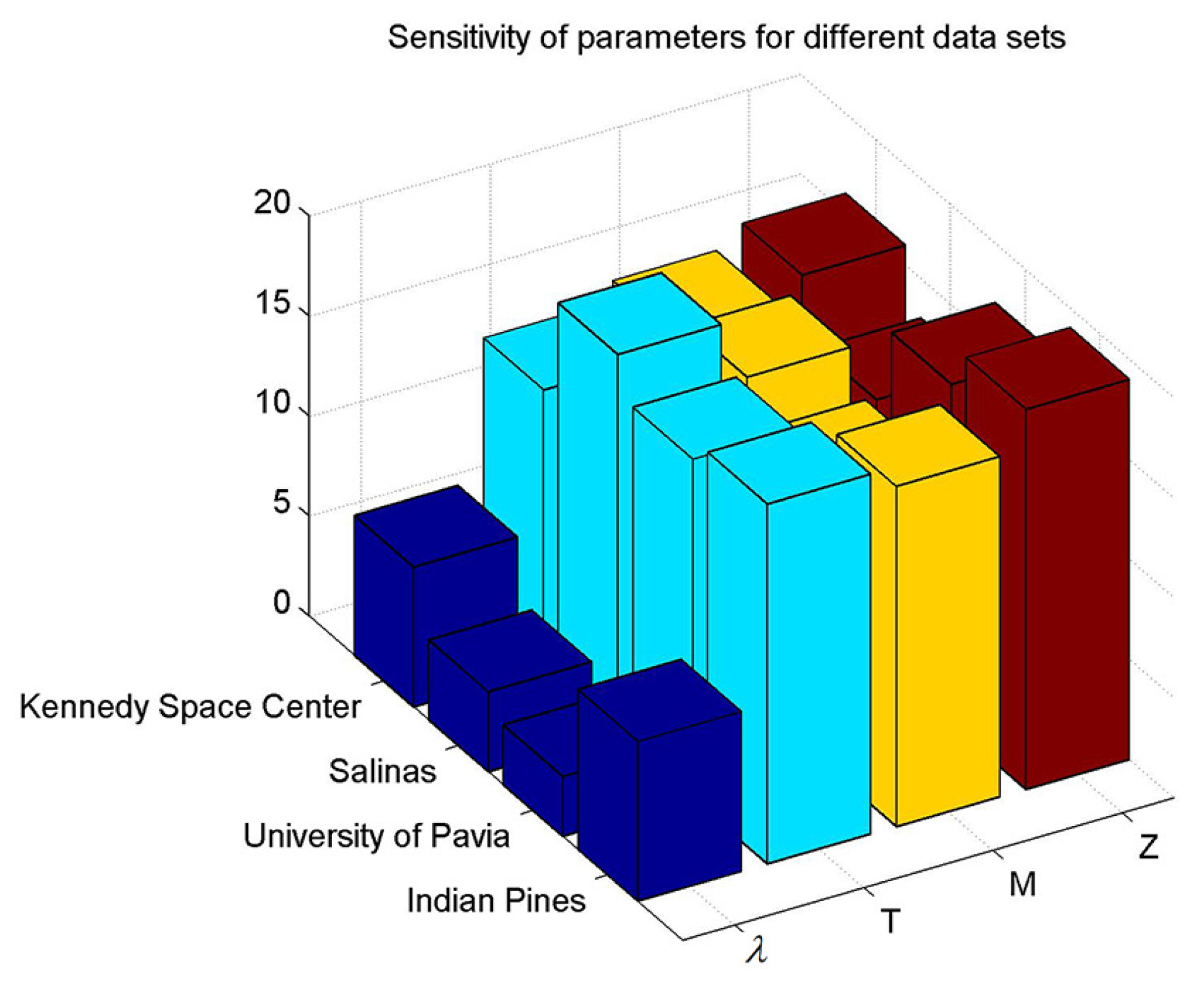
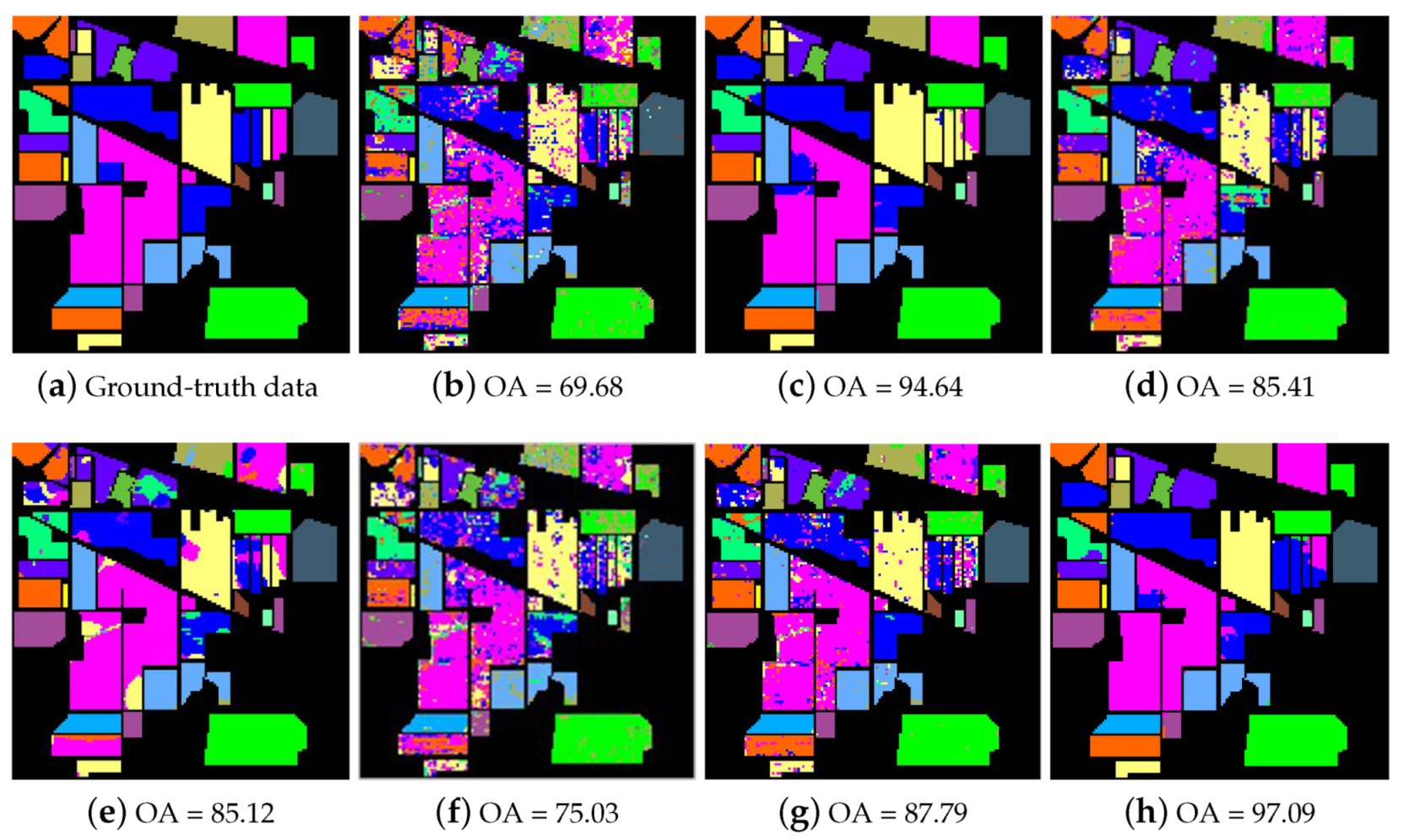


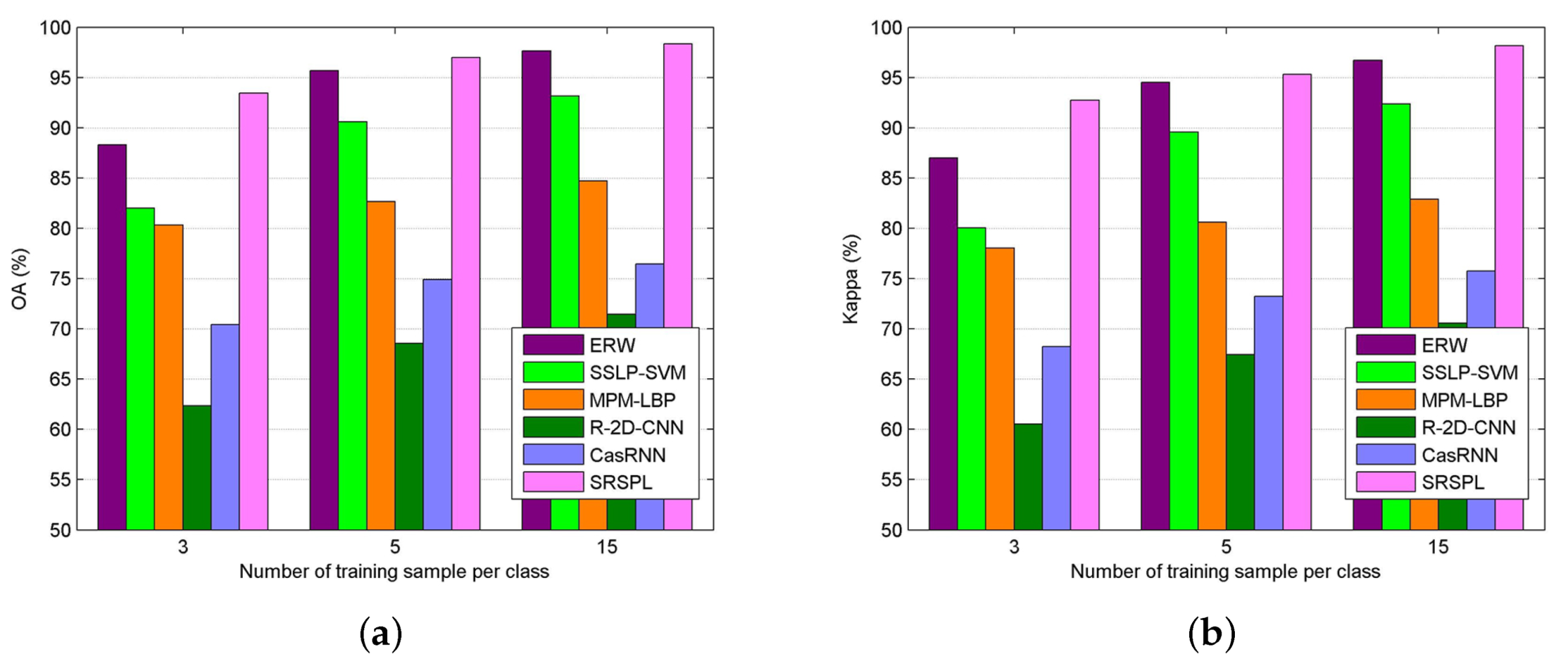
| Method | Metric | Number of Training Samples per Class | ||||
|---|---|---|---|---|---|---|
| 5 | 10 | 15 | 20 | 25 | ||
| OA | 45.31 ± 5.19 | 57.58 ± 2.98 | 63.56 ± 2.61 | 66.92 ± 1.47 | 69.68 ± 0.97 | |
| SVM | AA | 47.41 ± 3.71 | 55.19 ± 2.27 | 59.84 ± 2.21 | 62.13 ± 1.86 | 63.84 ± 0.91 |
| Kappa | 39.21 ± 5.43 | 52.52 ± 3.17 | 59.11 ± 2.82 | 62.80 ± 1.56 | 65.82 ± 1.08 | |
| OA | 81.97 ± 5.93 | 86.89 ± 3.45 | 90.13 ± 3.47 | 92.64 ± 1.74 | 94.92 ± 1.70 | |
| Without-IID | AA | 84.35 ± 6.40 | 89.07 ± 3.47 | 92.73 ± 1.23 | 95.63 ± 0.97 | 96.76 ± 0.95 |
| Kappa | 80.91 ± 6.50 | 85.12 ± 3.77 | 89.42 ± 3.91 | 91.59 ± 1.99 | 94.20 ± 1.94 | |
| OA | 76.08 ± 3.60 | 82.62 ± 2.90 | 86.80 ± 2.13 | 90.90 ± 1.83 | 93.53 ± 1.55 | |
| With-PCA | AA | 81.47 ± 4.20 | 80.47 ± 2.94 | 87.36 ± 3.51 | 91.02 ± 2.76 | 95.06 ± 0.79 |
| Kappa | 73.03 ± 3.91 | 80.40 ± 3.20 | 85.30 ± 2.39 | 89.78 ± 2.07 | 92.61 ± 1.77 | |
| OA | 82.47 ± 2.30 | 88.62 ± 2.91 | 92.80 ± 2.13 | 94.90 ± 1.66 | 96.67 ± 1.21 | |
| With-IFRF | AA | 89.68 ± 0.91 | 90.47 ± 0.94 | 93.36 ± 2.03 | 95.02 ± 2.63 | 97.83 ± 0.72 |
| Kappa | 80.12 ± 2.56 | 89.40 ± 3.23 | 92.97 ± 1.96 | 93.78 ± 2.80 | 96.19 ± 1.39 | |
| OA | 72.30 ± 4.38 | 84.87 ± 4.41 | 90.02 ± 1.30 | 92.94 ± 1.86 | 94.64 ± 1.17 | |
| ERW | AA | 83.32 ± 2.41 | 91.48 ± 2.30 | 94.39 ± 0.94 | 96.18 ± 1.00 | 97.14 ± 0.62 |
| Kappa | 68.96 ± 4.69 | 82.95 ± 4.86 | 88.69 ± 1.46 | 91.99 ± 2.10 | 93.91 ± 1.33 | |
| OA | 64.84 ± 1.43 | 76.05 ± 1.44 | 80.79 ± 0.73 | 82.13 ± 0.44 | 85.41 ± 0.26 | |
| SSLP-SVM | AA | 65.96 ± 2.38 | 78.07 ± 0.98 | 82.08 ± 0.64 | 82.25 ± 0.68 | 86.87 ± 0.35 |
| Kappa | 60.63 ± 1.49 | 73.17 ± 1.60 | 78.38 ± 0.80 | 79.90 ± 0.49 | 83.49 ± 0.29 | |
| OA | 59.36 ± 5.80 | 72.83 ± 3.99 | 78.15 ± 2.51 | 81.92 ± 2.17 | 85.12 ± 2.18 | |
| MPM-LBP | AA | 72.10 ± 3.61 | 83.61 ± 1.27 | 87.34 ± 0.92 | 90.33 ± 1.63 | 91.82 ± 1.09 |
| Kappa | 54.39 ± 6.31 | 69.50 ± 4.25 | 75.38 ± 2.75 | 79.61 ± 2.42 | 83.14 ± 2.42 | |
| OA | 51.63 ± 3.81 | 59.83 ± 3.79 | 64.15 ± 2.18 | 70.92 ± 1.07 | 75.03 ± 1.18 | |
| R-2D-CNN | AA | 52.01 ± 3.16 | 58.61 ± 2.72 | 65.43 ± 1.21 | 69.33 ± 1.36 | 76.82 ± 1.39 |
| Kappa | 50.93 ± 3.13 | 60.50 ± 2.52 | 63.82 ± 1.57 | 71.61 ± 1.24 | 74.19 ± 1.43 | |
| OA | 60.57 ± 4.51 | 70.25 ± 2.18 | 75.22 ± 2.23 | 81.43 ± 1.69 | 87.79 ± 1.50 | |
| CasRNN | AA | 58.44 ± 3.12 | 72.86 ± 1.87 | 76.59 ± 1.74 | 83.66 ± 1.87 | 88.94 ± 1.21 |
| Kappa | 60.92 ± 3.58 | 69.34 ± 2.74 | 74.54 ± 2.05 | 80.49 ± 1.72 | 86.62 ± 1.74 | |
| OA | 84.73 ± 1.23 | 90.90 ± 1.25 | 93.88 ± 0.67 | 95.52 ± 0.39 | 97.09 ± 0.21 | |
| SRSPL | AA | 88.86 ± 1.51 | 93.97 ± 0.95 | 96.27 ± 0.58 | 97.04 ± 0.57 | 97.96 ± 0.39 |
| Kappa | 81.79 ± 1.45 | 89.64 ± 1.05 | 93.02 ± 0.72 | 94.88 ± 0.45 | 96.66 ± 0.27 | |
| Class | Training | Test | SVM | Without-IID | Different Feature Extraction Methods | Different Semi-Supervised Methods | Deep Learning Methods | SRSPL | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| With-PCA | With-IFRF | ERW | SSLP-SVM | MPM-LBP | R-2D-CNN | CasRNN | ||||||
| Asphalt | 20 | 6611 | 92.99 ± 2.27 | 90.92 ± 2.21 | 70.23 ± 6.45 | 99.84 ± 0.03 | 86.36 ± 6.78 | 96.22 ± 0.64 | 90.39 ± 8.38 | 79.34 ± 5.38 | 83.52 ± 6.33 | 98.93 ± 1.33 |
| Meadows | 20 | 18,629 | 90.78 ± 2.36 | 99.99 ± 0.58 | 80.84 ± 4.31 | 90.03 ± 3.61 | 98.32 ± 2.67 | 97.65 ± 0.20 | 92.95 ± 11.81 | 79.96 ± 3.61 | 71.37 ± 5.81 | 99.48 ± 0.57 |
| Gravel | 20 | 2079 | 55.61 ± 6.89 | 99.86 ± 0.07 | 98.98 ± 0.74 | 98.97 ± 0.03 | 96.34 ± 5.82 | 71.05 ± 1.69 | 85.25 ± 8.21 | 78.88 ± 6.12 | 64.51 ± 10.21 | 99.95 ± 0.03 |
| Trees | 20 | 3044 | 67.48 ± 10.20 | 85.81 ± 2.07 | 89.01 ± 5.77 | 90.70 ± 3.45 | 78.92 ± 4.07 | 81.88 ± 4.50 | 89.48 ± 5.91 | 73.91 ± 4.48 | 85.43 ± 3.42 | 91.86 ± 1.66 |
| Metal Sheets | 20 | 1325 | 94.27 ± 3.49 | 100.00 ± 0.00 | 99.31 ± 0.16 | 99.77 ± 0.02 | 99.66 ± 0.27 | 96.95 ± 1.91 | 98.56 ± 0.66 | 98.25 ± 0.20 | 98.55 ± 0.47 | 99.62 ± 0.20 |
| Bare Soil | 20 | 5009 | 49.36 ± 7.80 | 92.25 ± 3.42 | 85.18 ± 7.13 | 98.42 ± 2.50 | 97.04 ± 3.62 | 66.64 ± 4.67 | 87.25 ± 10.38 | 80.94 ± 4.51 | 89.08 ± 5.25 | 98.49 ± 0.08 |
| Bitumen | 20 | 1310 | 53.58 ± 6.35 | 100.00 ± 0.00 | 99.19 ± 0.53 | 95.53 ± 1.85 | 99.42 ± 0.33 | 60.77 ± 3.59 | 97.94 ± 1.66 | 78.49 ± 2.92 | 91.13 ± 2.62 | 99.95 ± 0.09 |
| Bricks | 20 | 3662 | 76.30 ± 5.67 | 98.00 ± 1.83 | 98.14 ± 1.00 | 97.71 ± 2.85 | 97.34 ± 2.13 | 87.75 ± 1.46 | 88.32 ± 5.45 | 83.81 ± 2.45 | 93.54 ± 3.41 | 99.84 ± 3.07 |
| Shadows | 20 | 927 | 99.88 ± 0.09 | 98.05 ± 0.09 | 94.45 ± 1.48 | 98.88 ± 0.15 | 99.89 ± 0.09 | 99.95 ± 0.13 | 98.78 ± 1.58 | 94.27 ± 0.30 | 94.72 ± 1.78 | 99.03 ± 0.06 |
| OA | 75.49 ± 3.45 | 96.62 ± 1.35 | 83.49 ± 1.86 | 96.76 ± 1.15 | 94.86 ± 1.33 | 86.71 ± 1.44 | 87.40 ± 4.70 | 78.15 ± 1.47 | 80.86 ± 3.74 | 98.63 ± 0.31 | ||
| AA | 76.06 ± 1.98 | 96.42 ± 2.98 | 88.61 ± 2.56 | 96.54 ± 2.74 | 95.02 ± 3.25 | 88.55 ± 2.15 | 88.75 ± 2.05 | 79.19 ± 1.35 | 84.29 ± 4.53 | 98.34 ± 0.13 | ||
| Kappa | 69.06 ± 3.94 | 95.52 ± 1.72 | 79.88 ± 2.21 | 95.18 ± 3.73 | 93.19 ± 1.73 | 82.92 ± 1.75 | 87.62 ± 5.48 | 77.34 ± 1.81 | 77.93 ± 3.18 | 98.18 ± 0.41 | ||
| Class | Training | Test | SVM | Without-IID | Different Feature Extraction Methods | Different Semi-Supervised Methods | Deep Learning Methods | SRSPL | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| With-PCA | With-IFRF | ERW | SSLP-SVM | MPM-LBP | R-2D-CNN | CasRNN | ||||||
| Weeds 1 | 3 | 2006 | 86.74 ± 12.68 | 96.74 ± 12.68 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 99.87 ± 0.15 | 98.82 ± 0.80 | 88.83 ± 1.85 | 90.12 ± 1.28 | 100.00 ± 0.00 |
| Weeds 2 | 3 | 3723 | 99.08 ± 0.89 | 99.08 ± 0.89 | 100.00 ± 0.00 | 99.34 ± 0.04 | 99.95 ± 0.03 | 98.75 ± 0.24 | 99.40 ± 0.67 | 99.46 ± 0.37 | 99.20 ± 0.69 | 99.96 ± 0.06 |
| Fallow | 3 | 1973 | 77.08 ± 16.42 | 98.08 ± 1.42 | 100.00 ± 0.00 | 100.00 ± 0.00 | 100.00 ± 0.00 | 84.21 ± 2.16 | 90.98 ± 12.66 | 79.49 ± 2.66 | 81.88 ± 3.76 | 100.00 ± 0.00 |
| Fallow_P | 3 | 1391 | 96.94 ± 1.23 | 84.94 ± 1.23 | 96.81 ± 3.16 | 85.28 ± 6.84 | 86.28 ± 7.64 | 97.64 ± 0.12 | 99.47 ± 0.30 | 96.13 ± 0.46 | 97.48 ± 0.47 | 94.64 ± 2.84 |
| Fallow_S | 3 | 2675 | 90.08 ± 8.26 | 97.08 ± 1.26 | 97.36 ± 2.97 | 98.41 ± 0.25 | 99.41 ± 0.27 | 96.86 ± 3.22 | 97.94 ± 1.53 | 90.07 ± 1.35 | 92.97 ± 1.73 | 97.38 ± 6.17 |
| Stubble | 3 | 3956 | 99.11 ± 4.76 | 98.11 ± 1.76 | 98.00 ± 2.01 | 97.34 ± 1.58 | 97.72 ± 4.58 | 99.87 ± 0.25 | 97.75 ± 3.28 | 97.81 ± 1.78 | 97.89 ± 0.38 | 99.92 ± 0.06 |
| Celery | 3 | 3576 | 93.79 ± 4.17 | 98.79 ± 1.17 | 100.00 ± 0.00 | 90.20 ± 4.77 | 72.20 ± 24.77 | 98.95 ± 0.41 | 99.06 ± 0.84 | 94.91 ± 2.43 | 96.56 ± 1.64 | 99.35 ± 0.18 |
| Grapes | 3 | 11268 | 64.24 ± 7.87 | 94.24 ± 2.87 | 93.12 ± 2.18 | 99.15 ± 0.42 | 99.17 ± 0.92 | 72.05 ± 3.23 | 72.48 ± 18.65 | 70.84 ± 6.55 | 73.45 ± 7.67 | 99.70 ± 2.56 |
| Soil | 3 | 6200 | 96.20 ± 3.03 | 96.20 ± 2.03 | 98.38 ± 1.48 | 100.00 ± 0.00 | 100.00 ± 0.00 | 99.37 ± 0.17 | 99.21 ± 0.60 | 96.18 ± 1.06 | 97.41 ± 0.78 | 100.00 ± 0.00 |
| Corn | 3 | 3275 | 72.58 ± 19.06 | 97.58 ± 1.06 | 91.57 ± 6.35 | 97.21 ± 7.15 | 95.36 ± 7.15 | 89.21 ± 3.90 | 83.91 ± 10.82 | 75.06 ± 10.28 | 78.04 ± 9.12 | 97.36 ± 9.51 |
| Lettuce_4 | 3 | 1065 | 74.66 ± 19.37 | 96.66 ± 2.37 | 80.51 ± 6.47 | 98.76 ± 2.23 | 100.00 ± 0.00 | 83.41 ± 4.40 | 92.94 ± 4.87 | 77.27 ± 8.78 | 80.64 ± 6.76 | 99.98 ± 0.06 |
| Lettuce_5 | 3 | 1924 | 81.48 ± 10.07 | 98.48 ± 0.07 | 83.16 ± 12.54 | 99.60 ± 1.83 | 100.00 ± 0.00 | 92.60 ± 5.67 | 99.36 ± 1.97 | 83.55 ± 7.75 | 85.61 ± 5.67 | 100.00 ± 0.00 |
| Lettuce_6 | 3 | 913 | 78.58 ± 13.93 | 98.58 ± 0.93 | 98.31 ± 2.74 | 98.74 ± 0.47 | 97.88 ± 0.47 | 95.24 ± 0.66 | 97.17 ± 3.24 | 80.20 ± 6.24 | 82.80 ± 4.41 | 98.47 ± 0.27 |
| Lettuce_7 | 3 | 1067 | 83.59 ± 19.14 | 93.59 ± 2.14 | 97.24 ± 2.59 | 95.33 ± 2.21 | 95.35 ± 2.36 | 92.14 ± 4.74 | 94.76 ± 2.63 | 85.73 ± 7.36 | 88.78 ± 5.37 | 96.26 ± 0.68 |
| Vinyard_U | 3 | 7265 | 45.43 ± 6.21 | 97.43 ± 2.21 | 88.51 ± 8.64 | 99.37 ± 0.17 | 99.18 ± 0.05 | 58.67 ± 7.83 | 70.93 ± 25.10 | 71.81 ± 10.01 | 75.93 ± 9.70 | 98.92 ± 0.13 |
| Vinyard_T | 3 | 1804 | 83.43 ± 18.80 | 99.43 ± 0.80 | 100.00 ± 0.00 | 99.48 ± 0.89 | 100.00 ± 0.00 | 71.12 ± 4.44 | 76.88 ± 11.35 | 86.12 ± 6.35 | 89.06 ± 4.35 | 99.68 ± 0.79 |
| OA | 76.06 ± 2.97 | 95.88 ± 3.63 | 86.06 ± 2.97 | 97.65 ± 1.36 | 97.17 ± 2.23 | 83.30 ± 1.58 | 87.79 ± 3.26 | 78.12 ± 2.42 | 81.81 ± 3.89 | 98.69 ± 1.22 | ||
| AA | 82.69 ± 2.46 | 96.76 ± 2.64 | 90.69 ± 2.46 | 97.78 ± 1.69 | 96.62 ± 2.69 | 88.12 ± 0.78 | 92.57 ± 2.07 | 77.45 ± 2.04 | 82.34 ± 2.10 | 98.01 ± 1.44 | ||
| Kappa | 73.52 ± 3.22 | 94.59 ± 3.97 | 85.52 ± 3.22 | 98.00 ± 1.45 | 96.84 ± 2.49 | 81.46 ± 1.72 | 86.41 ± 3.63 | 79.47 ± 2.31 | 80.87 ± 3.38 | 98.54 ± 1.37 | ||
| Data Sets | Computational Times (s) | ||
|---|---|---|---|
| MPM-LBP | SSLP-SVM | SRSPL | |
| Indian Pines | 197.78 | 479.51 | 124.31 |
| University of Pavia | 604.35 | 164.09 | 240.14 |
| Salinas | 339.03 | 1592.75 | 202.14 |
| Kennedy Space Center | 939.519 | 122.20 | 72.32 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cui, B.; Cui, J.; Lu, Y.; Guo, N.; Gong, M. A Sparse Representation-Based Sample Pseudo-Labeling Method for Hyperspectral Image Classification. Remote Sens. 2020, 12, 664. https://doi.org/10.3390/rs12040664
Cui B, Cui J, Lu Y, Guo N, Gong M. A Sparse Representation-Based Sample Pseudo-Labeling Method for Hyperspectral Image Classification. Remote Sensing. 2020; 12(4):664. https://doi.org/10.3390/rs12040664
Chicago/Turabian StyleCui, Binge, Jiandi Cui, Yan Lu, Nannan Guo, and Maoguo Gong. 2020. "A Sparse Representation-Based Sample Pseudo-Labeling Method for Hyperspectral Image Classification" Remote Sensing 12, no. 4: 664. https://doi.org/10.3390/rs12040664
APA StyleCui, B., Cui, J., Lu, Y., Guo, N., & Gong, M. (2020). A Sparse Representation-Based Sample Pseudo-Labeling Method for Hyperspectral Image Classification. Remote Sensing, 12(4), 664. https://doi.org/10.3390/rs12040664