Deep Relation Network for Hyperspectral Image Few-Shot Classification
Abstract
:1. Introduction
- The RN-FSC method is proposed to carry out classification on the new HSI with only a few labeled samples. The RN-FSC method has the ability to learn how to learn through meta-learning on the source HSI data set, so it can accurately classify the new HSI;
- The network model containing the feature learning module and relation learning module is designed for HSI classification. Specifically, 3D convolution is utilized for feature extraction to make full use of spatial–spectral information in HSI, and the 2D convolution layer and fully connected layer are utilized to approximate the relationship between sample features in an abstract nonlinear approach;
- Experiments are conducted on three well-known HSI data sets, which demonstrate that the proposed method can outperform conventional semisupervised methods and the semisupervised deep learning model with a few labeled samples.
2. HSI Few-Shot Classification
2.1. Definition of Few-Shot Classification
2.2. Task-Based Learning Strategy
2.3. HSI Few-Shot Classification
- (1)
- In the first phase, learning tasks are built on the source data set, and the model performs meta-learning;
- (2)
- In the second phase, learning tasks are built on the fine-tuning data set, and the model performs few-shot learning;
- (3)
- In the third phase, the entire fine-tuning data set is regarded as the support set, and the testing data set is regarded as the query set, so as to build tasks for HSI classification.
3. The Designed Relation Network Model
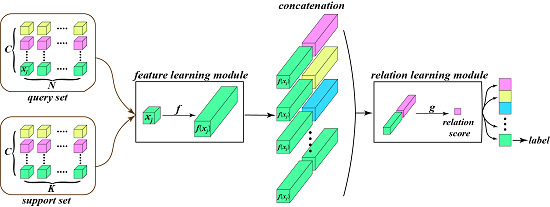
3.1. Model Overview
3.2. The Feature Learning Module
3.3. The Relation Learning Module
3.4. The Ability of Learning to Learn
- (1)
- Learning processGeneral deep learning models are trained based on the unique correspondence between data and labels and can only be trained in one specific task space at a time. However, the proposed method is task-based learning at any phase. The model focuses not on the specific classification task but on the learning ability with many different tasks;
- (2)
- ScalabilityThe proposed method performs meta-learning on the source data set to extract the transferable feature knowledge and cultivate the ablitity of learning to learn. From the perspective of knowledge transfer, the richer the categories in the source data set, the stronger the acquired learning ability, which is consistent with the human learning experience. Therefore, we can appropriately extend the source data set to enhance the generalization ability of the model;
- (3)
- Core mechanismThe proposed method is not to learn how to classify a specific data set, but to learn a deep metric space with the help of many tasks from different data sets, in which relation learning is performed by comparison. In a data-driven way, this metric space is nonlinear and transferrable. By comparing the similarity between the support samples and the query samples in the deep metric space, the classification is realized indirectly.
4. Experiments and Discussion
4.1. Experimental Data Sets
4.1.1. Source Data Sets
4.1.2. Target Data Sets
4.2. Experimental Setup
- (1)
- The number of convolutional layers has an important influence on the classification results. From NO.4 to NO.1, the number of convolutional layers in the feature learning module increases gradually, and the corresponding classification accuracy increases first and then decreases gradually. This indicates that the appropriate number of convolutional layers can obtain the best classification results, while too much or too little will reduce the effect of feature learning. In addition, a comparison between NO.2 and NO.5 can also verify a similar conclusion;
- (2)
- By comparing NO.2 and NO.6 network settings, it can be found that the convolution in the relation learning module can effectively improve the classification accuracy by 3.57%. The convolution is mainly used to extract cross-channel cascaded features and reduce the dimension of concatenations, which is conducive to relation learning;
- (3)
- The experimental results of NO.7 setting show that the classification effect of only applying the fully connected layer in the relational learning module is very poor, which directly proves the importance of the convolutional layer in relation learning.
4.3. Comparison and Analysis
- (1)
- In general, the performance of the traditional SVM classifier is better than that of the supervised deep learning model. Deep learning models need sufficient training samples for parameter optimization. However, in the HSI few-shot classification problem, limited labeled samples cannot provide guarantee for enough training, so the performance of supervised deep learning models is worse than that of SVM. For example, the OA of SVM is 6.04% higher than that of Res-3D-CNN on the Salinas data set;
- (2)
- By comparing SVM and semisupervised SVM, Res-3D-CNN, and other semisupervised deep models, it can be found that the classification performance of the methods trained with only the labeled samples is poor. In this case, the semisupervised method can further improve the classification accuracy by utilizing the information of unlabeled samples;
- (3)
- The classification performance of the semisupervised deep model is always better than that of the traditional semisupervised SVM. Deep learning models can extract more discriminative features from labeled and unlabeled samples by building an end-to-end hierarchical framework, so they can obtain better classification results;
- (4)
- Compared with other methods, RN-FSC has the best classification performance, with the highest OA, AA, and in all target data sets. The OA of RN-FSC is about 8.5%, 5%, and 6% higher than DCGAN+SEMI and GCN, which have similar performances on the three data sets. The most significant difference between RN-FSC and other methods is that other methods only perform training and classification on specific target data sets, while RN-FSC performs meta-learning on the collected source data sets through a large number of different tasks. Therefore, when processing new target data sets, RN-FSC has stronger generalization ability and can obtain better classification results with only a few labeled samples;
- (5)
- For the classes that other methods do not recognize accurately, RN-FSC can obtain better results, such as Bricks, Bare Soil and Gravel in UP, and Corn_senesced_green_weeds, Fallow in Salinas. Benefitting from meta-learning and network design, RN-FSC can acquire the ability to learn how to learn in the form of comparison. By comparing similarities between samples in the deep metric space, RN-FSC can take advantage of more abstract features. Therefore, RN-FSC can accurately recognize the uneasily distinguished classes.
4.4. Influence of the Number of Labeled Samples
4.5. Exploration on the Effectiveness of Meta-Learning
4.6. Execution Time Analysis
4.7. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sun, S.; Zhong, P.; Xiao, H.; Liu, F.; Wang, R. An active learning method based on SVM classifier for hyperspectral images classification. In Proceedings of the 7th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Tokyo, Japan, 2–5 June 2015; pp. 1–4. [Google Scholar] [CrossRef]
- Yuemei, R.; Yanning, Z.; Wei, W.; Lei, L. A spectral-spatial hyperspectral data classification approach using random forest with label constraints. In Proceedings of the IEEE Workshop on Electronics, Computer and Applications, Ottawa, ON, Canada, 8–9 May 2014; pp. 344–347. [Google Scholar] [CrossRef]
- Agarwal, A.; El-Ghazawi, T.; El-Askary, H.; Le-Moigne, J. Efficient Hierarchical-PCA Dimension Reduction for Hyperspectral Imagery. In Proceedings of the IEEE International Symposium on Signal Processing and Information Technology, Cairo, Egypt, 15–18 December 2007; pp. 353–356. [Google Scholar] [CrossRef]
- Falco, N.; Bruzzone, L.; Benediktsson, J.A. An ICA based approach to hyperspectral image feature reduction. In Proceedings of the IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 3470–3473. [Google Scholar] [CrossRef]
- Li, C.; Chu, H.; Kuo, B.; Lin, C. Hyperspectral image classification using spectral and spatial information based linear discriminant analysis. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium, Vancouver, BC, Canada, 4–29 July 2011; pp. 1716–1719. [Google Scholar] [CrossRef]
- Liao, W.; Pizurica, A.; Philips, W.; Pi, Y. A fast iterative kernel PCA feature extraction for hyperspectral images. In Proceedings of the IEEE International Conference on Image Processing, Hong Kong, China, 26–29 September 2010; pp. 1317–1320. [Google Scholar] [CrossRef]
- Chen, Y.; Qu, C.; Lin, Z. Supervised Locally Linear Embedding based dimension reduction for hyperspectral image classification. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium—IGARSS, Melbourne, Australia, 21–26 July 2013; pp. 3578–3581. [Google Scholar] [CrossRef]
- Gao, L.; Gu, D.; Zhuang, L.; Ren, J.; Yang, D.; Zhang, B. Combining t-Distributed Stochastic Neighbor Embedding With Convolutional Neural Networks for Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2019, 1–5. [Google Scholar] [CrossRef]
- Quesada-Barriuso, P.; Argüello, F.; Heras, D.B. Spectral–Spatial Classification of Hyperspectral Images Using Wavelets and Extended Morphological Profiles. IEEE J. Sel. Topics Appl. Earth Obs. Remote Sens. 2014, 7, 1177–1185. [Google Scholar] [CrossRef]
- Jia, S.; Hu, J.; Zhu, J.; Jia, X.; Li, Q. Three-Dimensional Local Binary Patterns for Hyperspectral Imagery Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 2399–2413. [Google Scholar] [CrossRef]
- Bau, T.C.; Sarkar, S.; Healey, G. Hyperspectral Region Classification Using a Three-Dimensional Gabor Filterbank. IEEE Trans. Geosci. Remote Sens. 2010, 48, 3457–3464. [Google Scholar] [CrossRef]
- Xu, Y.; Wu, Z.; Wei, Z. Markov random field with homogeneous areas priors for hyperspectral image classification. In Proceedings of the IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 3426–3429. [Google Scholar] [CrossRef]
- He, L.; Chen, X. A three-dimensional filtering method for spectral-spatial hyperspectral image classification. In Proceedings of the IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 2746–2748. [Google Scholar] [CrossRef]
- Casalino, G.; Gillis, N. Sequential dimensionality reduction for extracting localized features. Pattern Recognit. 2017, 63, 15–29. [Google Scholar] [CrossRef] [Green Version]
- Yin, J.; Li, S.; Zhu, H.; Luo, X. Hyperspectral Image Classification Using CapsNet With Well-Initialized Shallow Layers. IEEE Geosci. Remote Sens. Lett. 2019, 16, 1095–1099. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Du, B. Deep Learning for Remote Sensing Data: A Technical Tutorial on the State of the Art. IEEE Geosci. Remote Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Chen, Y.; Lin, Z.; Zhao, X.; Wang, G.; Gu, Y. Deep Learning-Based Classification of Hyperspectral Data. IEEE J. Sel.Topics Appl. Earth Obs. Remote Sens. 2014, 7, 2094–2107. [Google Scholar] [CrossRef]
- Mou, L.; Ghamisi, P.; Zhu, X.X. Deep Recurrent Neural Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef] [Green Version]
- Hang, R.; Liu, Q.; Hong, D.; Ghamisi, P. Cascaded Recurrent Neural Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 5384–5394. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Zhao, X.; Jia, X. Spectral–Spatial Classification of Hyperspectral Data Based on Deep Belief Network. IEEE J. Sel. Topics Appl. Earth Obs. Remote Sens. 2015, 8, 2381–2392. [Google Scholar] [CrossRef]
- Mughees, A.; Tao, L. Multiple deep-belief-network-based spectral-spatial classification of hyperspectral images. Tsinghua Sci. Technol. 2019, 24, 183–194. [Google Scholar] [CrossRef]
- He, L.; Li, J.; Liu, C.; Li, S. Recent Advances on Spectral–Spatial Hyperspectral Image Classification: An Overview and New Guidelines. IEEE Trans. Geosci. Remote Sens. 2018, 56, 1579–1597. [Google Scholar] [CrossRef]
- Zhang, M.; Li, W.; Du, Q. Diverse Region-Based CNN for Hyperspectral Image Classification. IEEE Trans. Image Process. 2018, 27, 2623–2634. [Google Scholar] [CrossRef]
- Lee, H.; Kwon, H. Going Deeper With Contextual CNN for Hyperspectral Image Classification. IEEE Trans. Image Process. 2017, 26, 4843–4855. [Google Scholar] [CrossRef] [Green Version]
- Zhi, L.; Yu, X.; Liu, B.; Wei, X. A dense convolutional neural network for hyperspectral image classification. Remote Sens. Lett. 2019, 10, 59–66. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Y.; Zhang, Y.; Shen, Q. Spectral-spatial classification of hyperspectral imagery using a dual-channel convolutional neural network. Remote Sens. Lett. 2017, 8, 438–447. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Jiang, H.; Li, C.; Jia, X.; Ghamisi, P. Deep Feature Extraction and Classification of Hyperspectral Images Based on Convolutional Neural Networks. IEEE Trans. Geosci. Remote Sens. 2016, 54, 6232–6251. [Google Scholar] [CrossRef] [Green Version]
- Zhong, Z.; Li, J.; Luo, Z.; Chapman, M. Spectral–Spatial Residual Network for Hyperspectral Image Classification: A 3-D Deep Learning Framework. IEEE Trans. Geosci. Remote Sens. 2018, 56, 847–858. [Google Scholar] [CrossRef]
- Fang, B.; Li, Y.; Zhang, H.; Chan, J. Hyperspectral Images Classification Based on Dense Convolutional Networks with Spectral-Wise Attention Mechanism. Remote Sens. 2019, 11, 159. [Google Scholar] [CrossRef] [Green Version]
- Li, A.; Shang, Z. A new Spectral-Spatial Pseudo-3D Dense Network for Hyperspectral Image Classification. In Proceedings of the International Joint Conference on Neural Networks (IJCNN), Budapest, Hungary, 14–19 July 2019; pp. 1–7. [Google Scholar] [CrossRef]
- Xu, Q.; Xiao, Y.; Wang, D.; Luo, B. CSA-MSO3DCNN: Multiscale Octave 3D CNN with Channel and Spatial Attention for Hyperspectral Image Classification. Remote Sens. 2020, 12, 188. [Google Scholar] [CrossRef] [Green Version]
- Jamshidpour, N.; Aria, E.H.; Safari, A.; Homayouni, S. Adaptive Self-Learned Active Learning Framework for Hyperspectral Classification. In Proceedings of the 10th Workshop on Hyperspectral Imaging and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 24–26 September 2019; pp. 1–5. [Google Scholar] [CrossRef]
- Paoletti, M.E.; Haut, J.M.; Fernandez-Beltran, R.; Plaza, J.; Plaza, A.; Li, J.; Pla, F. Capsule Networks for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2019, 57, 2145–2160. [Google Scholar] [CrossRef]
- Fan, J.; Tan, H.L.; Toomik, M.; Lu, S. Spectral-spatial hyperspectral image classification using super-pixel-based spatial pyramid representation. In Image and Signal Processing for Remote Sensing XXII; Bruzzone, L., Bovolo, F., Eds.; International Society for Optics and Photonics, SPIE: San Francisco, CA, USA, 2016; Volume 10004, pp. 315–321. [Google Scholar] [CrossRef]
- Liu, B.; Yu, X.; Zhang, P.; Tan, X.; Yu, A.; Xue, Z. A semi-supervised convolutional neural network for hyperspectral image classification. Remote Sens. Lett. 2017, 8, 839–848. [Google Scholar] [CrossRef]
- Wu, Y.; Mu, G.; Qin, C.; Miao, Q.; Ma, W.; Zhang, X. Semi-Supervised Hyperspectral Image Classification via Spatial-Regulated Self-Training. Remote Sens. 2020, 12, 159. [Google Scholar] [CrossRef] [Green Version]
- Kang, X.; Zhuo, B.; Duan, P. Semi-supervised deep learning for hyperspectral image classification. Remote Sens. Lett. 2019, 10, 353–362. [Google Scholar] [CrossRef]
- Li, W.; Wu, G.; Zhang, F.; Du, Q. Hyperspectral Image Classification Using Deep Pixel-Pair Features. IEEE Trans. Geosci. Remote Sens. 2017, 55, 844–853. [Google Scholar] [CrossRef]
- Liu, B.; Yu, X.; Zhang, P.; Yu, A.; Fu, Q.; Wei, X. Supervised Deep Feature Extraction for Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2018, 56, 1909–1921. [Google Scholar] [CrossRef]
- Xu, S.; Mu, X.; Chai, D.; Zhang, X. Remote sensing image scene classification based on generative adversarial networks. Remote Sens. Lett. 2018, 9, 617–626. [Google Scholar] [CrossRef]
- Qin, J.; Zhan, Y.; Wu, K.; Liu, W.; Yang, Z.; Yao, W.; Medjadba, Y.; Zhang, Y.; Yu, X. Semi-Supervised Classification of Hyperspectral Data for Geologic Body Based on Generative Adversarial Networks at Tianshan Area. In Proceedings of the IGARSS 2018—2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018; pp. 4776–4779. [Google Scholar] [CrossRef]
- Wang, H.; Tao, C.; Qi, J.; Li, H.; Tang, Y. Semi-Supervised Variational Generative Adversarial Networks for Hyperspectral Image Classification. In Proceedings of the IGARSS 2019—2019 IEEE International Geoscience and Remote Sensing Symposium, Yokohama, Japan, 28 July–2 August 2019; pp. 9792–9794. [Google Scholar] [CrossRef]
- Ren, M.; Triantafillou, E.; Ravi, S.; Snell, J.; Swersky, K.; Tenenbaum, J.; Larochelle, H.; Zemel, R. Meta-Learning for Semi-Supervised Few-Shot Classification. arXiv 2018, arXiv:1803.00676. [Google Scholar]
- Finn, C.; Abbeel, P.; Levine, S. Model-Agnostic Meta-Learning for Fast Adaptation of Deep Networks. arXiv 2017, arXiv:1703.03400. [Google Scholar]
- Andrychowicz, M.; Denil, M.; Gómez, S.; Hoffman, M.; Pfau, D.; Schaul, T.; Freitas, N. Learning to learn by gradient descent by gradient descent. arXiv 2016, arXiv:1606.04474. [Google Scholar]
- Li, Z.; Zhou, F.; Chen, F.; Li, H. Meta-SGD: Learning to Learn Quickly for Few Shot Learning. arXiv 2017, arXiv:1707.09835. [Google Scholar]
- Liang, H.; Fu, W.; Yi, F. A Survey of Recent Advances in Transfer Learning. In Proceedings of the IEEE 19th International Conference on Communication Technology (ICCT), Xi’an, China, 16–19 October 2019; pp. 1516–1523. [Google Scholar] [CrossRef]
- Liu, B.; Yu, X.; Yu, A.; Wan, G. Deep convolutional recurrent neural network with transfer learning for hyperspectral image classification. J. Appl. Remote Sens. 2018, 12, 1. [Google Scholar] [CrossRef]
- Sung, F.; Yang, Y.; Zhang, L.; Xiang, T.; Torr, P.; Hospedales, T. Learning to Compare: Relation Network for Few-Shot Learning. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, 18–23 June 2018; pp. 1199–1208. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Chen, Q.; Yan, S. Network In Network. arXiv 2013, arXiv:1312.4400. [Google Scholar]
- Sun, V.; Geng, X.; Chen, J.; Ji, L.; Tang, H.; Zhao, Y.; Xu, M. A robust and efficient band selection method using graph representation for hyperspectral imagery. Int. J. Remote Sens. 2016, 37, 4874–4889. [Google Scholar] [CrossRef]
- Glorot, X.; Bengio, Y. Understanding the difficulty of training deep feedforward neural networks. J. Mach. Learn. Res. Proc. Track 2010, 9, 249–256. [Google Scholar]
- Wang, L.; Hao, S.; Wang, Q.; Wang, Y. Semi-supervised classification for hyperspectral imagery based on spatial-spectral Label Propagation. ISPRS J. Photogramm. Remote Sens. 2014, 97, 123–137. [Google Scholar] [CrossRef]
- Liu, B.; Yu, X.; Zhang, P.; Tan, X. Deep 3D convolutional network combined with spatial-spectral features for hyperspectral image classification. Cehui Xuebao/Acta Geodaetica et Cartographica Sinica 2019, 48, 53–63. [Google Scholar] [CrossRef]
- Salimans, T.; Goodfellow, I.; Zaremba, W.; Cheung, V.; Radford, A.; Chen, X. Improved Techniques for Training GANs. arXiv 2016, arXiv:1606.03498. [Google Scholar]
- Kipf, T.; Welling, M. Semi-Supervised Classification with Graph Convolutional Networks. arXiv 2016, arXiv:1609.02907. [Google Scholar]
- Kang, X.; Li, S.; Benediktsson, J.A. Spectral-Spatial Hyperspectral Image Classification With Edge-Preserving Filtering. IEEE Transm Geoscim Remote Sens. 2014, 52, 2666–2677. [Google Scholar] [CrossRef]
- Zhong, S.; Chang, C.I.; Zhang, Y. Iterative Edge Preserving Filtering Approach to Hyperspectral Image Classification. IEEE Geosci. Remote Sens. Lett. 2018. [Google Scholar] [CrossRef]
- Zhong, S.; Chang, C.; Li, J.; Shang, X.; Chen, S.; Song, M.; Zhang, Y. Class Feature Weighted Hyperspectral Image Classification. IEEE J. Sel. Topics Appl. Earth Obs. Remote Sens. 2019, 12, 4728–4745. [Google Scholar] [CrossRef]















| Houston | Botswana | KSC | Chikusei | |
|---|---|---|---|---|
| Spatial size | ||||
| Spectral range | 380–1050 | 400–2500 | 400–2500 | 363–1018 |
| No. of bands | 144 | 145 | 176 | 128 |
| GSD | 2.5 | 30 | 18 | 2.5 |
| Sensor type | ITRES-CASI 1500 | EO-1 | AVIRIS | Hyperspec-VNIR-C |
| Areas | Houston | Botswana | Florida | Chikusei |
| No. of classes | 30 | 14 | 13 | 19 |
| Labeled samples | 15,029 | 3248 | 5211 | 77,592 |
| The Selected Bands | |
|---|---|
| Houston | 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 77 107 109 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 129 130 132 133 134 135 143 144 |
| Botswana | 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 88 110 111 112 113 114 115 116 117 118 119 120 121 122 123 124 125 126 127 128 137 138 139 140 141 142 143 144 145 |
| KSC | 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 28 29 31 32 33 35 36 37 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 95 101 120 132 143 144 145 146 147 148 149 150 151 155 167 175 176 |
| Chikusei | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 65 66 67 94 95 96 97 98 99 100 101 102 103 104 105 106 107 108 109 110 111 112 113 114 116 117 118 119 120 121 122 123 124 125 126 127 128 |
| UP | PC | Salinas | |
|---|---|---|---|
| Spatial size | |||
| Spectral range | 430–860 | 430–860 | 400–2500 |
| No. of bands | 103 | 102 | 204 |
| GSD | 1.3 | 1.3 | 3.7 |
| Sensor type | ROSIS | ROSIS | AVIRIS |
| Areas | Pavia | Pavia | California |
| No. of classes | 9 | 9 | 16 |
| Labeled samples | 42,776 | 148,152 | 54,129 |
| The Selected Bands | |
|---|---|
| UP | 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 103 |
| PC | 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 21 22 23 24 25 26 27 28 29 30 31 32 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 101 102 |
| Salinas | 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18 19 20 21 22 23 24 25 26 27 28 29 31 33 34 35 36 37 38 39 40 41 42 43 44 45 46 47 48 49 50 51 52 53 54 55 56 57 58 59 60 61 62 63 64 65 66 67 68 69 70 71 72 73 74 75 76 77 78 79 80 81 82 83 84 85 86 87 88 89 90 91 92 93 94 95 96 97 98 99 100 126 139 204 |
| K = 1, N = 19 | K = 5, N = 15 | K = 10, N = 10 | K = 15, N = 5 | |
|---|---|---|---|---|
| UP | ||||
| PC | ||||
| Salinas | 85.97 |
| No. | 1 | 2 | 3 | 4 | 5 | 6 | 7 |
|---|---|---|---|---|---|---|---|
| FLM | |||||||
| RLM | |||||||
| OA | 80.37 | 81.94 | 79.50 | 75.43 | 77.83 | 78.37 | 26.55 |
| Class | SVM | LapSVM | TSVM | Res-3D-CNN | SS-CNN | DCGAN+SEMI | GCN | RN-FSC |
|---|---|---|---|---|---|---|---|---|
| Asphalt | 94.08 | 98.12 | 96.55 | 71.67 | 89.89 | 92.18 | 96.00 | 87.28 |
| Meadows | 79.03 | 81.57 | 80.47 | 88.96 | 84.40 | 90.32 | 93.39 | 84.33 |
| Gravel | 27.67 | 30.97 | 11.11 | 23.30 | 59.94 | 41.80 | 50.71 | 90.42 |
| Trees | 57.71 | 62.47 | 48.71 | 88.86 | 57.94 | 86.39 | 95.85 | 78.09 |
| Metal Sheets | 91.67 | 91.39 | 94.92 | 89.39 | 97.11 | 83.30 | 99.19 | 99.56 |
| Bare Soil | 21.10 | 37.78 | 37.91 | 37.88 | 53.01 | 43.63 | 37.54 | 63.25 |
| Bitumen | 35.33 | 37.67 | 20.50 | 38.62 | 36.15 | 44.54 | 57.26 | 52.09 |
| Bricks | 57.31 | 60.47 | 55.36 | 42.59 | 72.70 | 62.11 | 73.31 | 84.81 |
| Shadow | 99.79 | 99.89 | 99.89 | 63.13 | 48.65 | 66.33 | 98.13 | 95.94 |
| OA | 55.79 | 67.06 | 61.92 | 65.44 | 71.73 | 73.52 | 73.40 | 81.94 |
| AA | 62.63 | 66.70 | 60.60 | 60.49 | 66.64 | 67.84 | 77.93 | 81.75 |
| 46.60 | 57.90 | 51.44 | 55.63 | 63.37 | 66.07 | 66.96 | 75.84 |
| Class | SVM | LapSVM | TSVM | Res-3D-CNN | SS-CNN | DCGAN+SEMI | GCN | RN-FSC |
|---|---|---|---|---|---|---|---|---|
| Water | 99.95 | 99.99 | 95.12 | 99.99 | 99.17 | 98.13 | 99.74 | 100.00 |
| Trees | 94.68 | 94.75 | 92.22 | 74.17 | 93.34 | 98.15 | 99.36 | 99.53 |
| Meadows | 40.86 | 60.84 | 40.12 | 80.24 | 75.17 | 65.81 | 61.53 | 67.60 |
| Bricks | 56.47 | 14.57 | 8.12 | 27.11 | 68.85 | 55.64 | 68.22 | 72.43 |
| Bare Soil | 19.51 | 65.47 | 27.15 | 23.08 | 38.25 | 53.42 | 42.77 | 96.91 |
| Asphalt | 63.66 | 61.85 | 46.87 | 67.69 | 81.42 | 84.21 | 81.62 | 85.86 |
| Bitumen | 78.21 | 92.83 | 1.38 | 77.38 | 75.82 | 99.37 | 91.61 | 85.55 |
| Tile | 88.66 | 94.55 | 97.14 | 98.88 | 99.57 | 99.02 | 99.06 | 99.94 |
| Shadow | 99.76 | 99.86 | 93.17 | 87.61 | 95.60 | 77.46 | 98.00 | 91.87 |
| OA | 83.11 | 86.43 | 67.60 | 80.03 | 89.27 | 91.85 | 90.65 | 96.36 |
| AA | 71.31 | 76.08 | 55.70 | 70.69 | 80.80 | 81.24 | 82.43 | 88.86 |
| 76.62 | 81.22 | 56.60 | 73.16 | 88.30 | 91.02 | 89.79 | 95.98 |
| Class | SVM | LapSVM | TSVM | Res-3D-CNN | SS-CNN | DCGAN+SEMI | GCN | RN-FSC |
|---|---|---|---|---|---|---|---|---|
| Brocoli_green_weeds_1 | 85.60 | 78.59 | 80.50 | 39.47 | 93.02 | 56.94 | 100.00 | 99.26 |
| Brocoli_green_weeds_2 | 98.54 | 98.99 | 98.08 | 74.02 | 92.51 | 71.53 | 81.95 | 100.00 |
| Fallow | 65.38 | 82.96 | 65.19 | 49.33 | 84.31 | 87.44 | 83.50 | 97.87 |
| Fallow_rough_plow | 95.82 | 96.64 | 95.46 | 88.71 | 86.43 | 76.45 | 96.99 | 99.50 |
| Fallow_smooth | 95.83 | 88.09 | 64.25 | 77.50 | 90.91 | 94.95 | 96.96 | 97.81 |
| Stubble | 99.92 | 100.00 | 99.95 | 97.52 | 99.55 | 99.47 | 99.82 | 99.35 |
| Celery | 95.29 | 89.61 | 85.10 | 61.53 | 97.54 | 89.63 | 94.66 | 100.00 |
| Grapes_untrained | 57.00 | 63.87 | 44.29 | 68.93 | 73.52 | 70.93 | 86.00 | 66.24 |
| Soil_vinyard_develop | 90.64 | 79.49 | 74.06 | 92.83 | 93.81 | 92.89 | 95.65 | 97.34 |
| Corn_senesced_green_weeds | 85.87 | 56.55 | 64.71 | 69.33 | 77.21 | 63.58 | 81.31 | 93.66 |
| Lettuce_romaine_4wk | 38.32 | 38.02 | 47.56 | 59.07 | 42.37 | 83.81 | 60.05 | 73.96 |
| Lettuce_romaine_5wk | 87.56 | 92.71 | 92.56 | 70.59 | 95.85 | 97.33 | 95.65 | 99.84 |
| Lettuce_romaine_6wk | 88.66 | 46.88 | 47.87 | 75.38 | 99.23 | 97.53 | 89.39 | 100.00 |
| Lettuce_romaine_7wk | 87.87 | 93.26 | 86.81 | 89.12 | 92.98 | 87.09 | 86.41 | 96.39 |
| Vinyard_untrained | 33.18 | 49.84 | 32.31 | 47.62 | 50.37 | 74.78 | 51.00 | 68.85 |
| Vinyard_vertical_trellis | 81.64 | 91.00 | 54.24 | 88.90 | 80.54 | 77.17 | 95.07 | 99.89 |
| OA | 73.64 | 74.99 | 64.63 | 67.60 | 79.23 | 80.11 | 80.90 | 86.99 |
| AA | 80.45 | 77.91 | 70.81 | 71.87 | 84.38 | 82.60 | 97.15 | 93.12 |
| 70.70 | 72.05 | 60.95 | 64.28 | 77.04 | 77.86 | 78.95 | 85.44 |
| University of Pavia | Pavia Center | Salinas |
|---|---|---|
| t/significant? | t/significant? | t/significant? |
| RN-FSC vs. SVM | ||
| 75.53/yes | 26.34/yes | 34.40/yes |
| RN-FSC vs. LapSVM | ||
| 24.36/yes | 34.79/yes | 33.75/yes |
| RN-FSC vs. TSVM | ||
| 47.44/yes | 71.68/yes | 37.29/yes |
| RN-FSC vs. Res-3D-CNN | ||
| 35.62/yes | 30.08/yes | 29.19/yes |
| RN-FSC vs. SS-CNN | ||
| 27.17/yes | 22.60/yes | 19.33/yes |
| RN-FSC vs. DCGAN+SEMI | ||
| 23.56/yes | 19.88/yes | 16.86/yes |
| RN-FSC vs. GCN | ||
| 21.05/yes | 20.05/yes | 15.98/yes |
| The Target Data Set | EPF-B-g | EPF-B-c | EPF-G-g | EPF-G-c | IEPF-G-g | RN-FSC | |
|---|---|---|---|---|---|---|---|
| Salinas | OA AA | 96.46 98.17 | 96.23 98.08 | 96.61 98.24 | 97.11 98.56 | 98.36 99.19 | 98.86 99.26 |
| Indian Pines | OA AA | 94.82 95.96 | 95.30 95.44 | 93.94 96.64 | 94.87 96.52 | 98.19 98.67 | 97.46 96.68 |
| Target Data Set | Meta-Learning | |||||
|---|---|---|---|---|---|---|
| UP | yes | 81.94 | 82.71 | 85.70 | 89.20 | 91.05 |
| not | 61.74 | 69.56 | 74.12 | 78.93 | 80.22 | |
| PC | yes | 96.36 | 97.21 | 97.70 | 98.03 | 98.72 |
| not | 85.63 | 86.46 | 87.41 | 90.53 | 92.10 | |
| Salinas | yes | 86.99 | 87.79 | 88.68 | 90.49 | 92.15 |
| not | 71.08 | 74.08 | 75.96 | 77.34 | 80.45 |
| Target Data Set | DCGAN+SEMI | GCN | RN-FSC |
|---|---|---|---|
| UP | 1355.86 + 2.57 | 1915.29 + 0.98 | 217.27 + 72.57 |
| PC | 1401.31 + 8.13 | 3042.18 + 1.44 | 214.98 + 198.09 |
| Salinas | 2386.74 + 3.03 | 1224.03 + 1.10 | 632.98 + 81.23 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, K.; Liu, B.; Yu, X.; Qin, J.; Zhang, P.; Tan, X. Deep Relation Network for Hyperspectral Image Few-Shot Classification. Remote Sens. 2020, 12, 923. https://doi.org/10.3390/rs12060923
Gao K, Liu B, Yu X, Qin J, Zhang P, Tan X. Deep Relation Network for Hyperspectral Image Few-Shot Classification. Remote Sensing. 2020; 12(6):923. https://doi.org/10.3390/rs12060923
Chicago/Turabian StyleGao, Kuiliang, Bing Liu, Xuchu Yu, Jinchun Qin, Pengqiang Zhang, and Xiong Tan. 2020. "Deep Relation Network for Hyperspectral Image Few-Shot Classification" Remote Sensing 12, no. 6: 923. https://doi.org/10.3390/rs12060923
APA StyleGao, K., Liu, B., Yu, X., Qin, J., Zhang, P., & Tan, X. (2020). Deep Relation Network for Hyperspectral Image Few-Shot Classification. Remote Sensing, 12(6), 923. https://doi.org/10.3390/rs12060923