Minimax Bridgeness-Based Clustering for Hyperspectral Data
Abstract
:1. Introduction
2. Related Work
3. The Minimax Distance
3.1. Definition
3.2. Minimax Silhouette
3.3. Minimax Distance-Based kNN Clustering
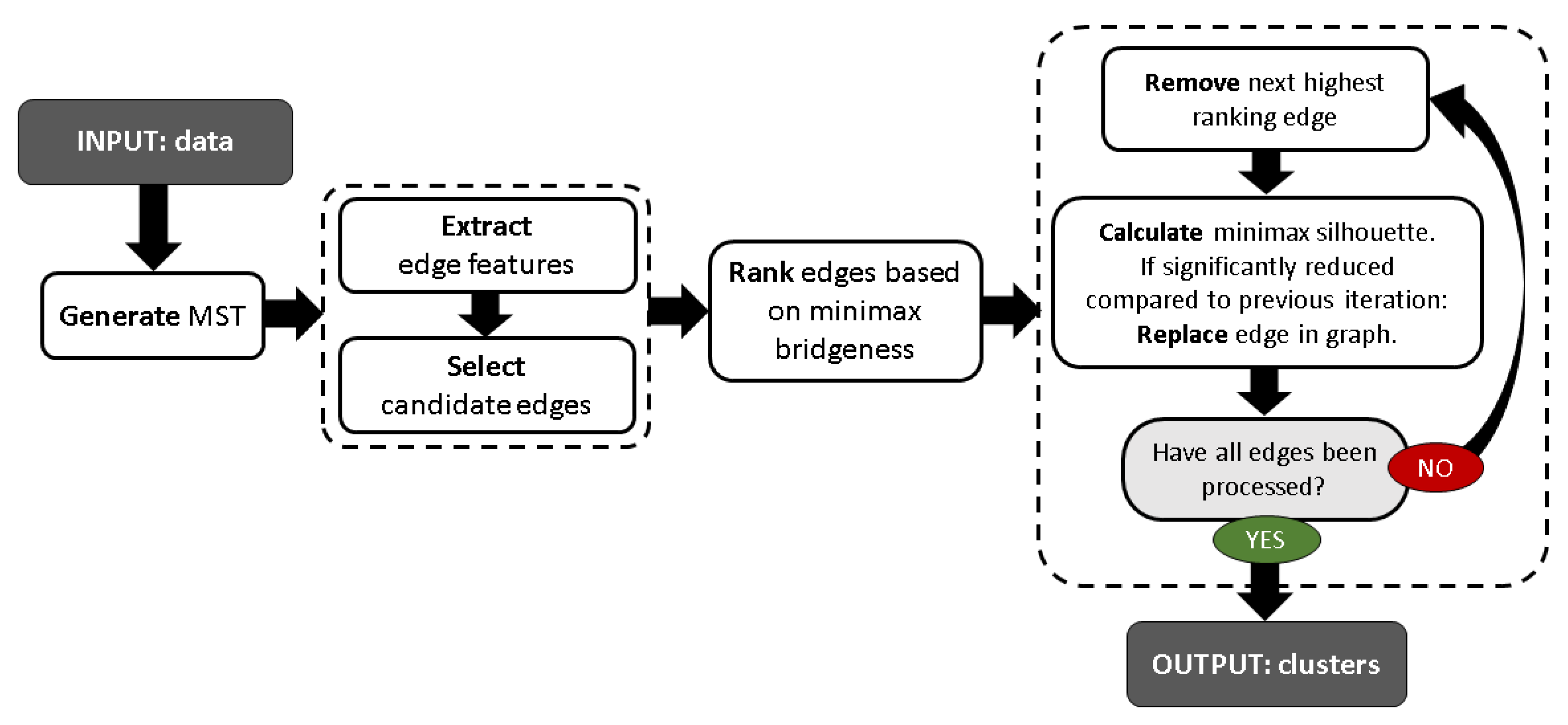
4. Minimax Bridgeness-Based Clustering
- Step 1: Compute minimum spanning tree (MST).
- Step 2: Discard edges that are unlikely to separate clusters.
- Step 3: Rank the remaining edges based on minimax bridgeness.
- Step 4: Remove next highest ranking edge that does not significantly decrease the minimax silhouette. Repeat until all edges have been assessed.
- They are longer than most edges.
- They have a higher minimax bridgeness than most edges.
- They have a lower density point at their centre than most edges.
- Neither of the vertices they connect is the other’s first neighbour, nor do they have the same nearest neighbour (see [41]).
5. Experimental Validation
5.1. Alternative Clustering Methods
5.2. Data
5.3. Criteria
5.4. Results
- K-MBC significantly outperforms FCM, FDPC and GWENN on all data-sets except Botswana in terms of OA and NMI. It also yields the best pixel purity on all data-sets except Massey University, where it is surpassed only by GWENN by 0.08.
- The clustering maps in Figure 5 confirm that our approach performs particularly well at creating clusters of high purity.
- Although it is expected that MBC would give the best overall results when applied to the core set , or when the number of clusters is known, this does not appear as obvious from our results. However, we note that MBC tends to find too few clusters in the data (especially on the Botswana data-set, where it misses six classes of pixels).
- FINCH and LPC generally perform poorly overall and especially in terms of number of clusters. They each tend to detect too many clusters on all data-sets. Interestingly, they also yield poor pixel purity values. Usually, high purity is expected when the number of clusters is high. The fact that we observe the contrary indicates that these two methods are really not well suited to deal with raw hyperspectral data.
- On the core-set , MBC performs very well and even finds the right number of clusters in the Pavia University and Salinas scenes. It over-estimates this number by one on the Massey University scene and under-estimate by two on the KSC scene.
- The Botswana scene seems to the most challenging for the proposed methods. The only case where the MBC clustering surpasses the benchmark with this scene is in terms of pixel purity. We noted that this particular scene contains classes of pixels that are among the least pure in the benchmark, with strong variations in reflectance spectra within classes. We hypothesise that this is the main reason for our method under-performing on this scene. On the other hand, it is well known that clustering is generally an ill-posed problem and that different applications may require different types of clustering approaches. Clearly, in this case, FDPC performs better, but it should be noted that it was tuned manually, unlike MBC.
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cilia, C.; Panigada, C.; Rossini, M.; Meroni, M.; Busetto, L.; Amaducci, S.; Boschetti, M.; Picchi, V.; Colombo, R. Nitrogen status assessment for variable rate fertilization in maize through hyperspectral imagery. Remote Sens. 2014, 6, 6549–6565. [Google Scholar] [CrossRef] [Green Version]
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J. Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef] [Green Version]
- Groom, S.B.; Sathyendranath, S.; Ban, Y.; Bernard, S.; Brewin, B.; Brotas, V.; Brockmann, C.; Chauhan, P.; Choi, J.K.; Chuprin, A.; et al. Satellite ocean colour: Current status and future perspective. Front. Mar. Sci. 2019, 6, 485. [Google Scholar] [CrossRef] [Green Version]
- Chehdi, K.; Cariou, C. Learning or assessment of classification algorithms relying on biased ground truth data: What interest? J. Appl. Remote Sens. 2019, 13, 1–26. [Google Scholar] [CrossRef]
- Aggarwal, C.C.; Hinneburg, A.; Keim, D.A. On the Surprising Behavior of Distance Metrics in High Dimensional Space; Lecture Notes in Computer Science; Springer: Berlin/Heidelberg, Germany, 2001; Volume 1973, pp. 420–434. [Google Scholar]
- Murphy, J.M.; Maggioni, M. Unsupervised Clustering and Active Learning of Hyperspectral Images With Nonlinear Diffusion. IEEE Trans. Geosci. Remote Sens. 2018, 57, 1829–1845. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chang, C. Independent component analysis-based dimensionality reduction with applications in hyperspectral image analysis. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1586–1600. [Google Scholar] [CrossRef]
- Fauvel, M.; Chanussot, J.; Benediktsson, J. Kernel principal component analysis for feature reduction in hyperspectrale images analysis. In Proceedings of the 7th Nordic Signal Processing Symposium—NORSIG 2006, Rejkjavik, Iceland, 7–9 June 2006; IEEE: Piscataway, NJ, USA, 2006; pp. 238–241. [Google Scholar]
- Pandey, P.C.; Tate, N.J.; Balzter, H. Mapping tree species in coastal portugal using statistically segmented principal component analysis and other methods. IEEE Sens. J. 2014, 14, 4434–4441. [Google Scholar] [CrossRef] [Green Version]
- Bachmann, C.M.; Ainsworth, T.L.; Fusina, R.A. Exploiting manifold geometry in hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2005, 43, 441–454. [Google Scholar] [CrossRef]
- Chang, C.; Wang, S. Constrained band selection for hyperspectral imagery. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1575–1585. [Google Scholar] [CrossRef]
- Jarvis, R.A.; Patrick, E.A. Clustering using a similarity measure based on shared near neighbors. IEEE Trans. Comput. 1973, 100, 1025–1034. [Google Scholar] [CrossRef]
- Schnitzer, D.; Flexer, A.; Schedl, M.; Widmer, G. Local and global scaling reduce hubs in space. J. Mach. Learn. Res. 2012, 13, 2871–2902. [Google Scholar]
- Stevens, J.R.; Resmini, R.G.; Messinger, D.W. Spectral-Density-Based Graph Construction Techniques for Hyperspectral Image Analysis. IEEE Trans. Geosci. Remote Sens. 2017, 55, 5966–5983. [Google Scholar] [CrossRef]
- Chehreghani, M.H. Classification with Minimax Distance Measures. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence, San Francisco, CA, USA, 4–10 February 2017; pp. 1784–1790. [Google Scholar]
- Grygorash, O.; Zhou, Y.; Jorgensen, Z. Minimum spanning tree based clustering algorithms. In Proceedings of the 18th IEEE International Conference on Tools with Artificial Intelligence, ICTAI’06, Arlington, VA, USA, 13–15 November 2006; IEEE: Piscataway, NJ, USA, 2006; pp. 73–81. [Google Scholar]
- Little, A.; Maggioni, M.; Murphy, J.M. Path-based spectral clustering: Guarantees, robustness to outliers, and fast algorithms. arXiv 2017, arXiv:1712.06206. [Google Scholar]
- Saxena, A.; Prasad, M.; Gupta, A.; Bharill, N.; Patel, O.P.; Tiwari, A.; Er, M.J.; Ding, W.; Lin, C.T. A review of clustering techniques and developments. Neurocomputing 2017, 267, 664–681. [Google Scholar] [CrossRef] [Green Version]
- Xie, J.; Girshick, R.; Farhadi, A. Unsupervised deep embedding for clustering analysis. In Proceedings of the International Conference on Machine Learning, New York, NY, USA, 19–24 June 2016; pp. 478–487. [Google Scholar]
- Yang, J.; Parikh, D.; Batra, D. Joint unsupervised learning of deep representations and image clusters. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 5147–5156. [Google Scholar]
- Ng, A.Y.; Jordan, M.I.; Weiss, Y. On spectral clustering: Analysis and an algorithm. In Advances in Neural Information Processing Systems; MIT Press: Cambridge, MA, USA, 2002; pp. 849–856. [Google Scholar]
- Wang, X.F.; Xu, Y. Fast clustering using adaptive density peak detection. Stat. Methods Med. Res. 2017, 26, 2800–2811. [Google Scholar] [CrossRef] [PubMed]
- Fukunaga, K.; Hostetler, L.D. The estimation of the gradient of a density function, with applications in pattern recognition. IEEE Trans. Inf. Theory 1975, 21, 32–40. [Google Scholar] [CrossRef] [Green Version]
- Comaniciu, D.; Meer, P. Mean Shift: A Robust Approach Toward Feature Space Analysis. IEEE Trans. Pattern Anal. Mach. Intell. 2002, 24, 603–619. [Google Scholar] [CrossRef] [Green Version]
- Rodriguez, A.; Laio, A. Clustering by fast search and find of density peaks. Science 2014, 344, 1492–1496. [Google Scholar] [CrossRef] [Green Version]
- Lu, L.; Jiang, H.; Wong, W.H. Multivariate density estimation by bayesian sequential partitioning. J. Am. Stat. Assoc. 2013, 108, 1402–1410. [Google Scholar] [CrossRef]
- Mantero, P.; Moser, G.; Serpico, S.B. Partially supervised classification of remote sensing images through SVM-based probability density estimation. IEEE Trans. Geosci. Remote Sens. 2005, 43, 559–570. [Google Scholar] [CrossRef]
- Ester, M.; Kriegel, H.P.; Sander, J.; Xu, X. A density-based algorithm for discovering clusters in large spatial databases with noise. Kdd 1996, 96, 226–231. [Google Scholar]
- Loftsgaarden, D.O.; Quesenberry, C.P. A nonparametric estimate of a multivariate density function. Ann. Math. Stat. 1965, 36, 1049–1051. [Google Scholar] [CrossRef]
- Cariou, C.; Chehdi, K. Nearest neighbor-density-based clustering methods for large hyperspectral images. In Image and Signal Processing for Remote Sensing XXIII; International Society for Optics and Photonics: Bellingham WA, USA, 2017; Volume 10427, p. 104270I. [Google Scholar]
- Geng, Y.A.; Li, Q.; Zheng, R.; Zhuang, F.; He, R.; Xiong, N. RECOME: A new density-based clustering algorithm using relative KNN kernel density. Inf. Sci. 2018, 436, 13–30. [Google Scholar] [CrossRef] [Green Version]
- Thorndike, R.L. Who belongs in the family. In Psychometrika; Citeseer: Princeton, NJ, USA, 1953. [Google Scholar]
- Goutte, C.; Toft, P.; Rostrup, E.; Nielsen, F.A.; Hansen, L.K. On clustering fMRI time series. NeuroImage 1999, 9, 298–310. [Google Scholar] [CrossRef] [Green Version]
- Botev, Z.I.; Grotowski, J.F.; Kroese, D.P. Kernel density estimation via diffusion. Ann. Stat. 2010, 38, 2916–2957. [Google Scholar] [CrossRef] [Green Version]
- Ketchen, D.J.; Shook, C.L. The application of cluster analysis in strategic management research: An analysis and critique. Strateg. Manag. J. 1996, 17, 441–458. [Google Scholar] [CrossRef]
- Keogh, E.; Lonardi, S.; Ratanamahatana, C.A. Towards parameter-free data mining. In Proceedings of the Tenth ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, Seattle, WA, USA, 22–25 August 2004; ACM: New York, NY, USA, 2004; pp. 206–215. [Google Scholar]
- Koonsanit, K.; Jaruskulchai, C.; Eiumnoh, A. Parameter-free K-means clustering algorithm for satellite imagery application. In Proceedings of the International Conf. on Information Science and Applications, Suwon, Korea, 23–25 May 2012; IEEE: Piscataway, NJ, USA, 2012; pp. 1–6. [Google Scholar]
- Yang, X.H.; Zhu, Q.P.; Huang, Y.J.; Xiao, J.; Wang, L.; Tong, F.C. Parameter-free Laplacian centrality peaks clustering. Pattern Recognit. Lett. 2017, 100, 167–173. [Google Scholar] [CrossRef]
- Cesario, E.; Manco, G.; Ortale, R. Top-down parameter-free clustering of high-dimensional categorical data. IEEE Trans. Knowl. Data Eng. 2007, 19, 1607–1624. [Google Scholar] [CrossRef]
- Hou, J.; Gao, H.; Li, X. DSets-DBSCAN: A parameter-free clustering algorithm. IEEE Trans. Image Process. 2016, 25, 3182–3193. [Google Scholar] [CrossRef]
- Sarfraz, S.; Sharma, V.; Stiefelhagen, R. Efficient Parameter-free Clustering Using First Neighbor Relations. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Long Beach, CA, USA, 16–20 June 2019; pp. 8934–8943. [Google Scholar]
- Ertöz, L.; Steinbach, M.; Kumar, V. Finding clusters of different sizes, shapes, and densities in noisy, high dimensional data. In Proceedings of the International Conference on Data Mining, San Francisco, CA, USA, 1–3 May 2003; SIAM: Philadelphia, PA, USA, 2003; pp. 47–58. [Google Scholar]
- Andoni, A.; Indyk, P. Nearest neighbors in high-dimensional spaces. In Handbook of Discrete and Computational Geometry; Taylor & Francis: Abingdon, UK, 2017; pp. 1135–1155. [Google Scholar]
- Li, Q.; Kecman, V.; Salman, R. A chunking method for euclidean distance matrix calculation on large dataset using multi-gpu. In Proceedings of the 2010 Ninth International Conference on Machine Learning and Applications, Washington, DC, USA, 12–14 December 2010; IEEE: Piscataway, NJ, USA, 2010; pp. 208–213. [Google Scholar]
- Chehreghani, M.H. Efficient Computation of Pairwise Minimax Distance Measures. In Proceedings of the 2017 IEEE International Conference on Data Mining (ICDM), New Orleans, LA, USA, 18–21 November 2017; IEEE: Piscataway, NJ, USA, 2017; pp. 799–804. [Google Scholar]
- Pettie, S.; Ramachandran, V. An optimal minimum spanning tree algorithm. J. ACM (JACM) 2002, 49, 16–34. [Google Scholar] [CrossRef]
- Huang, J.; Xu, R.; Cheng, D.; Zhang, S.; Shang, K. A Novel Hybrid Clustering Algorithm Based on Minimum Spanning Tree of Natural Core Points. IEEE Access 2019, 7, 43707–43720. [Google Scholar] [CrossRef]
- Le Moan, S.; Cariou, C. Parameter-Free Density Estimation for Hyperspectral Image Clustering. In Proceedings of the 2018 International Conference on Image and Vision Computing New Zealand (IVCNZ), Auckland, New Zealand, 19–21 November 2018; IEEE: Piscataway, NJ, USA, 2018; pp. 1–6. [Google Scholar]
- Tran, T.N.; Wehrens, R.; Buydens, L.M. KNN-kernel density-based clustering for high-dimensional multivariate data. Comput. Stat. Data Anal. 2006, 51, 513–525. [Google Scholar] [CrossRef]
- Cariou, C.; Chehdi, K. A new k-nearest neighbor density-based clustering method and its application to hyperspectral images. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; IEEE: Piscataway, NJ, USA, 2016; pp. 6161–6164. [Google Scholar]
- Manning, C.D.; Raghavan, P.; Schütze, H. Introduction to Information Retrieval; Cambridge University Press: Cambridge, UK, 2008. [Google Scholar]
- Bezdek, J.C. Pattern Recognition with Fuzzy Objective Function Algorithms; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Pullanagari, R.R.; Kereszturi, G.; Yule, I.J.; Ghamisi, P. Assessing the performance of multiple spectral-spatial features of a hyperspectral image for classification of urban land cover classes using support vector machines and artificial neural network. J. Appl. Remote Sens. 2017, 11, 026009. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, H. The Hungarian method for the assignment problem. Nav. Res. Logist. Q. 1955, 2, 83–97. [Google Scholar] [CrossRef] [Green Version]




| Pavia U. | KSC | Salinas | Botswana | Massey | ||
|---|---|---|---|---|---|---|
| Regular | SE | −0.744 | −0.693 | −0.461 | −0.604 | −0.742 |
| SAM | −0.742 | −0.500 | −0.428 | −0.616 | −0.686 | |
| SNN | SE | −0.160 | −0.245 | −0.214 | −0.216 | −0.094 |
| SAM | −0.087 | −0.209 | −0.189 | −0.236 | −0.184 | |
| Mutual Proximity | SE | −0.123 | −0.207 | −0.035 | −0.028 | −0.182 |
| SAM | −0.156 | −0.071 | −0.087 | −0.084 | −0.075 | |
| Minimax | SE | −0.047 | −0.019 | 0.021 | 0.015 | −0.256 |
| SAM | −0.123 | −0.256 | −0.021 | −0.104 | −0.261 | |
| Mutual Proximity on | SE | −0.038 | −0.108 | 0.038 | 0.037 | −0.115 |
| SAM | −0.164 | −0.042 | 0.062 | −0.194 | 0.026 | |
| Minimax on | SE | 0.128 | 0.260 | 0.212 | 0.174 | −0.115 |
| SAM | 0.196 | −0.087 | 0.194 | −0.046 | −0.349 |
| Parameters | Pros | Cons | |
|---|---|---|---|
| FCM [52] | K, m | fuzzy | fitted to convex clusters |
| FDPC [25] | fast | requires decision graph | |
| DBSCAN [28] | , | fast | not scalable |
| GWENN [50] | k | non iterative | requires NN search |
| FINCH [41] | - | parameter-free | requires NN search |
| Laplacian centrality-based (LPC) [38] | - | parameter-free | not scalable |
| Data-Set | N | D | Classes |
|---|---|---|---|
| Pavia University | 42,776 | 103 | 9 |
| Kennedy Space Centre | 5211 | 176 | 13 |
| Salinas | 54,129 | 204 | 16 |
| Botswana | 3248 | 145 | 14 |
| Massey University | 9564 | 339 | 23 |
| Number of Clusters Is | Pavia U. | KSC | Salinas | Botswana | Massey U. | |
|---|---|---|---|---|---|---|
| Known | FCM | 41.9 | 52.5 | 54.6 | 61.4 | 43.3 |
| FDPC | 44.2 | 47.6 | 62.2 | 62.4 | 54.0 | |
| GWENN | 47.9 | 49.8 | 65.3 | 53.5 | 44.3 | |
| K-MBC | 65.9 | 58.2 | 76.5 | 51.2 | 70.0 | |
| Unknown | FINCH | 37.0 | 39.7 | 55.7 | 56.2 | 38.6 |
| LPC | 35.2 | 42.4 | 59.2 | 54.2 | 41.6 | |
| MBC | 64.2 | 51.2 | 76.2 | 42.3 | 53.3 | |
| MBC on | 70.1 | 59.1 | 80.0 | 60.5 | 65.7 |
| Number of Clusters Is | Pavia U. | KSC | Salinas | Botswana | Massey U. | |
|---|---|---|---|---|---|---|
| Known | FCM | 0.47 | 0.56 | 0.63 | 0.65 | 0.47 |
| FDPC | 0.54 | 0.64 | 0.71 | 0.81 | 0.59 | |
| GWENN | 0.62 | 0.69 | 0.77 | 0.73 | 0.82 | |
| K-MBC | 0.91 | 0.74 | 0.93 | 0.97 | 0.74 | |
| Unknown | FINCH | 0.42 | 0.42 | 0.59 | 0.58 | 0.42 |
| LPC | 0.39 | 0.44 | 0.59 | 0.59 | 0.60 | |
| MBC | 0.97 | 0.70 | 0.91 | 0.96 | 0.60 | |
| MBC on | 0.96 | 0.73 | 0.95 | 0.97 | 0.80 |
| Number of Clusters Is | Pavia U. | KSC | Salinas | Botswana | Massey U. | |
|---|---|---|---|---|---|---|
| Known | FCM | 0.49 | 0.58 | 0.69 | 0.68 | 0.60 |
| FDPC | 0.49 | 0.59 | 0.74 | 0.74 | 0.69 | |
| GWENN | 0.48 | 0.60 | 0.72 | 0.61 | 0.57 | |
| K-MBC | 0.62 | 0.71 | 0.85 | 0.47 | 0.76 | |
| Unknown | FINCH | 0.49 | 0.60 | 0.75 | 0.72 | 0.69 |
| LPC | 0.49 | 0.57 | 0.72 | 0.69 | 0.53 | |
| MBC | 0.62 | 0.62 | 0.84 | 0.44 | 0.77 | |
| MBC on | 0.63 | 0.70 | 0.87 | 0.61 | 0.79 |
| Pavia U. | KSC | Salinas | Botswana | Massey U. | |
|---|---|---|---|---|---|
| FINCH | −13 | −20 | −17 | −12 | −15 |
| LPC | −14 | −20 | −16 | −14 | −17 |
| MBC | 3 | 2 | 1 | 6 | −4 |
| MBC on | 0 | 2 | 0 | 4 | −1 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Le Moan, S.; Cariou, C. Minimax Bridgeness-Based Clustering for Hyperspectral Data. Remote Sens. 2020, 12, 1162. https://doi.org/10.3390/rs12071162
Le Moan S, Cariou C. Minimax Bridgeness-Based Clustering for Hyperspectral Data. Remote Sensing. 2020; 12(7):1162. https://doi.org/10.3390/rs12071162
Chicago/Turabian StyleLe Moan, Steven, and Claude Cariou. 2020. "Minimax Bridgeness-Based Clustering for Hyperspectral Data" Remote Sensing 12, no. 7: 1162. https://doi.org/10.3390/rs12071162







