Evaluation of RGB and Multispectral Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping and Yield Prediction in Barley Breeding
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material, Environments, and Growing Conditions
2.2. Ground Phenotyping Data
2.3. UAV Data Platforms, Camera Systems and UAV Campaigns
2.4. UAV Data Processing
2.4.1. Initial Pre-Processing of Multispectral Imagery
2.4.2. Photogrammetric Processing
2.4.3. Crop Height Model (CHM) and Vegetation Index (VI) Calculation
2.4.4. Soil Masking
2.4.5. Vegetation Cover (VCOV) Derivation
2.4.6. Plotwise Feature Extraction
2.5. Statistical Analysis
2.5.1. Ground Truth Validation
2.5.2. Growth Rate Modelling
2.5.3. Yield Prediction
- Approach 1: Applying a single linear regression model for each trait at a single time point
- Approach 2: Including all measured and derived traits of the same time point as predictors in a multiple regression
- Approach 3: Extending the multiple regression model across multiple time points, resulting in a multi-temporal stacked prediction
2.5.4. Genotype Association Study
3. Results
3.1. Canopy Height Determination
3.2. Vegetation Cover
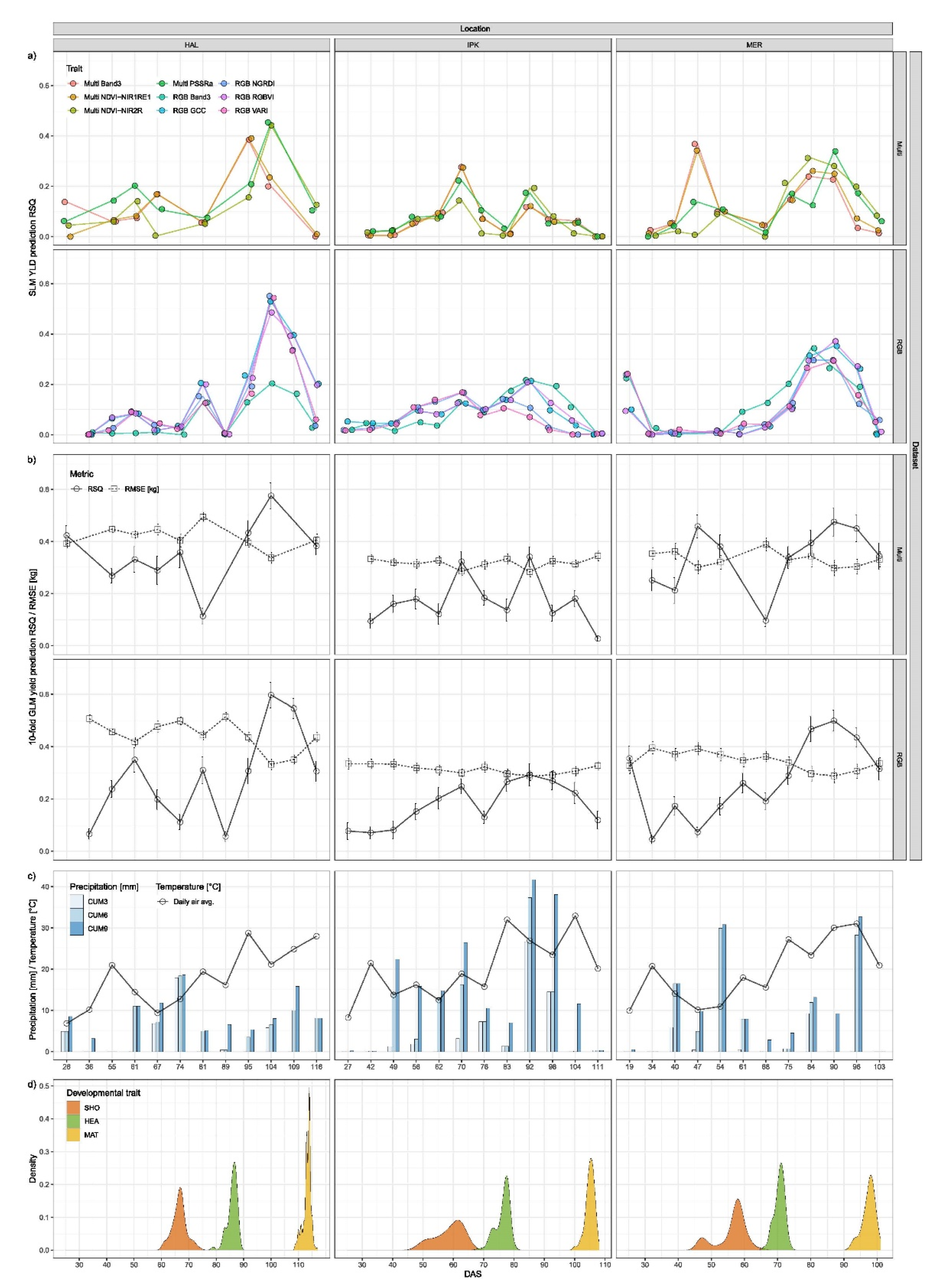
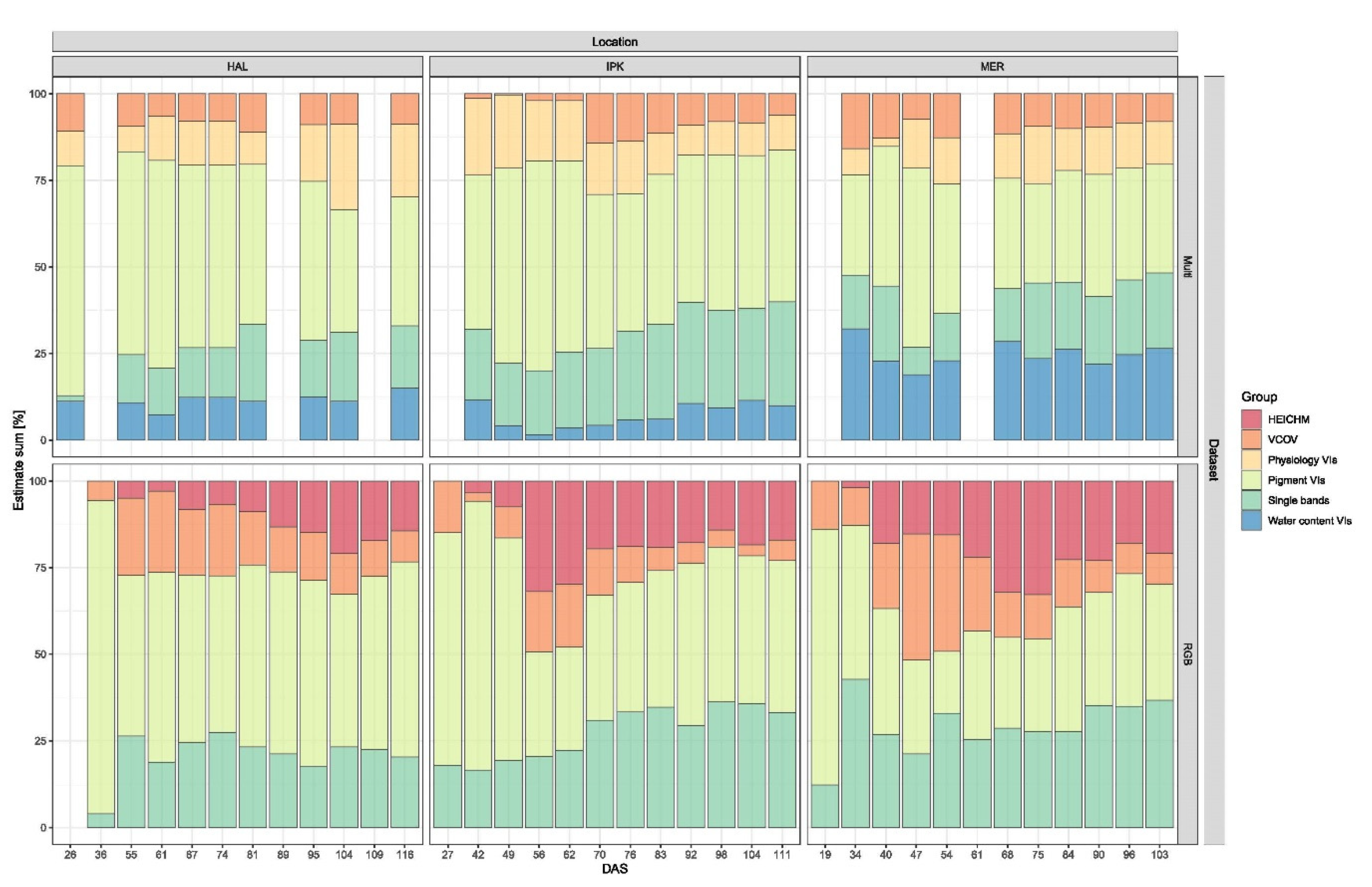
3.3. Yield Prediction
4. Discussion
4.1. Canopy Height Determination
4.2. Vegetation Cover
4.3. Yield Prediction
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Godwin, I.D.; Rutkoski, J.; Varshney, R.K.; Hickey, L.T. Technological perspectives for plant breeding. Theor. Appl. Genet. 2019, 132, 555–557. [Google Scholar] [CrossRef]
- Acreche, M.M.; Briceno-Felix, G.; Sanchez, J.A.M.; Slafer, G.A. Physiological bases of genetic gains in Mediterranean bread wheat yield in Spain. Eur. J. Agron. 2008, 28, 162–170. [Google Scholar] [CrossRef]
- Sadras, V.O.; Lawson, C. Genetic gain in yield and associated changes in phenotype, trait plasticity and competitive ability of South Australian wheat varieties released between 1958 and 2007. Crop Pasture Sci. 2011, 62, 533–549. [Google Scholar] [CrossRef]
- Valin, H.; Sands, R.D.; van der Mensbrugghe, D.; Nelson, G.C.; Ahammad, H.; Blanc, E.; Bodirsky, B.; Fujimori, S.; Hasegawa, T.; Havlik, P.; et al. The future of food demand: Understanding differences in global economic models. Agric. Econ. 2014, 45, 51–67. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef]
- Ghanem, M.E.; Marrou, H.; Sinclair, T.R. Physiological phenotyping of plants for crop improvement. Trends Plant Sci. 2015, 20, 139–144. [Google Scholar] [CrossRef]
- Tardieu, F.; Cabrera-Bosquet, L.; Pridmore, T.; Bennett, M. Plant Phenomics, From Sensors to Knowledge. Curr. Biol. 2017, 27, R770–R783. [Google Scholar] [CrossRef] [PubMed]
- Saiz-Rubio, V.; Rovira-Mas, F. From Smart Farming towards Agriculture 5.0: A Review on Crop Data Management. Agronomy 2020, 10, 207. [Google Scholar] [CrossRef]
- Zhang, C.Y.; Marzougui, A.; Sankaran, S. High-resolution satellite imagery applications in crop phenotyping: An overview. Comput. Electron. Agric. 2020, 175. [Google Scholar] [CrossRef]
- Zeng, L.L.; Wardlow, B.D.; Xiang, D.X.; Hu, S.; Li, D.R. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237. [Google Scholar] [CrossRef]
- Bégué, A.; Arvor, D.; Bellon, B.; Betbeder, J.; de Abelleyra, D.; Ferraz, R.P.D.; Lebourgeois, V.; Lelong, C.; Simões, M.; Verón, S.R. Remote Sensing and Cropping Practices: A Review. Remote Sens. 2018, 10, 99. [Google Scholar] [CrossRef]
- Fieuzal, R.; Sicre, C.M.; Baup, F. Estimation of corn yield using multi-temporal optical and radar satellite data and artificial neural networks. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 14–23. [Google Scholar] [CrossRef]
- Virlet, N.; Sabermanesh, K.; Sadeghi-Tehran, P.; Hawkesford, M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct. Plant Biol. 2016, 44, 143–153. [Google Scholar] [CrossRef]
- Kicherer, A.; Herzog, K.; Bendel, N.; Kluck, H.C.; Backhaus, A.; Wieland, M.; Rose, J.C.; Klingbeil, L.; Labe, T.; Hohl, C.; et al. Phenoliner: A New Field Phenotyping Platform for Grapevine Research. Sensors 2017, 17, 1625. [Google Scholar] [CrossRef]
- Zhang, J.C.; Huang, Y.B.; Pu, R.L.; Gonzalez-Moreno, P.; Yuan, L.; Wu, K.H.; Huang, W.J. Monitoring plant diseases and pests through remote sensing technology: A review. Comput. Electron. Agric. 2019, 165. [Google Scholar] [CrossRef]
- Busemeyer, L.; Ruckelshausen, A.; Moller, K.; Melchinger, A.E.; Alheit, K.V.; Maurer, H.P.; Hahn, V.; Weissmann, E.A.; Reif, J.C.; Wurschum, T. Precision phenotyping of biomass accumulation in triticale reveals temporal genetic patterns of regulation. Sci. Rep. 2013, 3, 2442. [Google Scholar] [CrossRef]
- Gruber, S.; Kwon, H.; York, G.; Pack, D. Payload Design of Small UAVs. In Handbook of Unmanned Aerial Vehicles; Springer: Dordrecht, The Netherlands, 2018; pp. 1–25. [Google Scholar]
- Aasen, H.; Honkavaara, E.; Lucieer, A.; Zarco-Tejada, P.J. Quantitative Remote Sensing at Ultra-High Resolution with UAV Spectroscopy: A Review of Sensor Technology, Measurement Procedures, and Data Correction Workflows. Remote Sens. 2018, 10, 1091. [Google Scholar] [CrossRef]
- Han, X.; Thomasson, J.A.; Bagnall, G.C.; Pugh, N.A.; Horne, D.W.; Rooney, W.L.; Jung, J.; Chang, A.; Malambo, L.; Popescu, S.C.; et al. Measurement and Calibration of Plant-Height from Fixed-Wing UAV Images. Sensors 2018, 18, 4092. [Google Scholar] [CrossRef] [PubMed]
- Aasen, H.; Kirchgessner, N.; Walter, A.; Liebisch, F. PhenoCams for Field Phenotyping: Using Very High Temporal Resolution Digital Repeated Photography to Investigate Interactions of Growth, Phenology, and Harvest Traits. Front. Plant Sci. 2020, 11, 593. [Google Scholar] [CrossRef]
- Anderegg, J.; Yu, K.; Aasen, H.; Walter, A.; Liebisch, F.; Hund, A. Spectral Vegetation Indices to Track Senescence Dynamics in Diverse Wheat Germplasm. Front. Plant Sci. 2019, 10, 1749. [Google Scholar] [CrossRef]
- Brien, C.; Jewell, N.; Watts-Williams, S.J.; Garnett, T.; Berger, B. Smoothing and extraction of traits in the growth analysis of noninvasive phenotypic data. Plant Methods 2020, 16, 36. [Google Scholar] [CrossRef]
- Shipley, B.; Hunt, R. Regression smoothers for estimating parameters of growth analyses. Ann. Bot. 1996, 78, 569–576. [Google Scholar] [CrossRef][Green Version]
- Richards, R.A. Defining selection criteria to improve yield under drought. Plant Growth Regul. 1996, 20, 157–166. [Google Scholar] [CrossRef]
- Trethowan, R.M.; van Ginkel, M.; Ammar, K.; Crossa, J.; Payne, T.S.; Cukadar, B.; Rajaram, S.; Hernandez, E. Associations among twenty years of international bread wheat yield evaluation environments. Crop Sci. 2003, 43, 1698–1711. [Google Scholar] [CrossRef]
- Slafer, G.A.; Andrade, F.H. Changes in Physiological Attributes of the Dry-Matter Economy of Bread Wheat (Triticum-Aestivum) through Genetic-Improvement of Grain-Yield Potential at Different Regions of the World—A Review. Euphytica 1991, 58, 37–49. [Google Scholar] [CrossRef]
- Kefauver, S.C.; Vicente, R.; Vergara-Diaz, O.; Fernandez-Gallego, J.A.; Kerfal, S.; Lopez, A.; Melichar, J.P.E.; Serret Molins, M.D.; Araus, J.L. Comparative UAV and Field Phenotyping to Assess Yield and Nitrogen Use Efficiency in Hybrid and Conventional Barley. Front. Plant Sci. 2017, 8, 1733. [Google Scholar] [CrossRef] [PubMed]
- García-Martínez, H.; Flores-Magdaleno, H.; Ascencio-Hernández, R.; Khalil-Gardezi, A.; Tijerina-Chávez, L.; Mancilla-Villa, O.R.; Vázquez-Peña, M.A. Corn Grain Yield Estimation from Vegetation Indices, Canopy Cover, Plant Density, and a Neural Network Using Multispectral and RGB Images Acquired with Unmanned Aerial Vehicles. Agriculture 2020, 10, 277. [Google Scholar] [CrossRef]
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Yang, G.; Yang, X.; Fan, L. Estimation of the Yield and Plant Height of Winter Wheat Using UAV-Based Hyperspectral Images. Sensors 2020, 20, 1231. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.G.; Tian, Y.C.; Yao, X.; Zhu, Y.; Cao, W.X. Predicting grain yield and protein content in wheat by fusing multi-sensor and multi-temporal remote-sensing images. Field Crop. Res. 2014, 164, 178–188. [Google Scholar] [CrossRef]
- Yue, J.B.; Feng, H.K.; Yang, G.J.; Li, Z.H. A Comparison of Regression Techniques for Estimation of Above-Ground Winter Wheat Biomass Using Near-Surface Spectroscopy. Remote Sens. 2018, 10, 66. [Google Scholar] [CrossRef]
- Mourtzinis, S.; Arriaga, F.J.; Balkcom, K.S.; Ortiz, B.V. Corn Grain and Stover Yield Prediction at R1 Growth Stage. Agron. J. 2013, 105, 1045–1050. [Google Scholar] [CrossRef]
- Reynolds, M.P.; Rajaram, S.; Sayre, K.D. Physiological and genetic changes of irrigated wheat in the post-green revolution period and approaches for meeting projected global demand. Crop Sci. 1999, 39, 1611–1621. [Google Scholar] [CrossRef]
- Silva-Perez, V.; Molero, G.; Serbin, S.P.; Condon, A.G.; Reynolds, M.P.; Furbank, R.T.; Evans, J.R. Hyperspectral reflectance as a tool to measure biochemical and physiological traits in wheat. J. Exp. Bot. 2018, 69, 483–496. [Google Scholar] [CrossRef] [PubMed]
- Babar, M.A.; Reynolds, M.P.; Van Ginkel, M.; Klatt, A.R.; Raun, W.R.; Stone, M.L. Spectral reflectance to estimate genetic variation for in-season biomass, leaf chlorophyll, and canopy temperature in wheat. Crop Sci. 2006, 46, 1046–1057. [Google Scholar] [CrossRef]
- Xue, J.R.; Su, B.F. Significant Remote Sensing Vegetation Indices: A Review of Developments and Applications. J. Sens. 2017, 2017, 1–17. [Google Scholar] [CrossRef]
- Nebiker, S.; Lack, N.; Abächerli, M.; Läderach, S. Light-Weight Multispectral Uav Sensors and Their Capabilities for Predicting Grain Yield and Detecting Plant Diseases. ISPRS Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, XLI-B1, 963–970. [Google Scholar] [CrossRef]
- Haghighattalab, A.; Gonzalez Perez, L.; Mondal, S.; Singh, D.; Schinstock, D.; Rutkoski, J.; Ortiz-Monasterio, I.; Singh, R.P.; Goodin, D.; Poland, J. Application of unmanned aerial systems for high throughput phenotyping of large wheat breeding nurseries. Plant Methods 2016, 12, 35. [Google Scholar] [CrossRef] [PubMed]
- Gutierrez, M.; Reynolds, M.P.; Raun, W.R.; Stone, M.L.; Klatt, A.R. Spectral Water Indices for Assessing Yield in Elite Bread Wheat Genotypes under Well-Irrigated, Water-Stressed, and High-Temperature Conditions. Crop Sci. 2010, 50, 197–214. [Google Scholar] [CrossRef]
- Veys, C.; Chatziavgerinos, F.; AlSuwaidi, A.; Hibbert, J.; Hansen, M.; Bernotas, G.; Smith, M.; Yin, H.; Rolfe, S.; Grieve, B. Multispectral imaging for presymptomatic analysis of light leaf spot in oilseed rape. Plant Methods 2019, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Raper, T.B.; Varco, J.J. Canopy-scale wavelength and vegetative index sensitivities to cotton growth parameters and nitrogen status. Precis. Agric. 2014, 16, 62–76. [Google Scholar] [CrossRef]
- Filella, I.; Penuelas, J. The Red Edge Postion and Shape as Indicators of Plant Chlorophyll Content, Biomass and Hydric Status. 1994. Available online: https://www.tandfonline.com/doi/abs/10.1080/01431169408954177 (accessed on 7 May 2021).
- Zarco-Tejada, P.J.; Berni, J.A.J.; Suarez, L.; Sepulcre-Canto, G.; Morales, F.; Miller, J.R. Imaging chlorophyll fluorescence with an airborne narrow-band multispectral camera for vegetation stress detection. Remote Sens. Environ. 2009, 113, 1262–1275. [Google Scholar] [CrossRef]
- Nijland, W.; de Jong, R.; de Jong, S.M.; Wulder, M.A.; Bater, C.W.; Coops, N.C. Monitoring plant condition and phenology using infrared sensitive consumer grade digital cameras. Agric. For. Meteorol. 2014, 184, 98–106. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Vonbargen, K.; Mortensen, D.A. Color Indexes for Weed Identification under Various Soil, Residue, and Lighting Conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef]
- Ullman, S. The interpretation of structure from motion. Proc. R Soc. Lond. B Biol. Sci. 1979, 203, 405–426. [Google Scholar] [CrossRef] [PubMed]
- Bendig, J.; Bolten, A.; Bennertz, S.; Broscheit, J.; Eichfuss, S.; Bareth, G. Estimating Biomass of Barley Using Crop Surface Models (CSMs) Derived from UAV-Based RGB Imaging. Remote Sens. 2014, 6, 10395–10412. [Google Scholar] [CrossRef]
- Di Gennaro, S.F.; Rizza, F.; Badeck, F.W.; Berton, A.; Delbono, S.; Gioli, B.; Toscano, P.; Zaldei, A.; Matese, A. UAV-based high-throughput phenotyping to discriminate barley vigour with visible and near-infrared vegetation indices. Int. J. Remote Sens. 2017, 39, 5330–5344. [Google Scholar] [CrossRef]
- Igawa, M.; Mano, M. A Nondestructive Method to Estimate Plant Height, Stem Diameter and Biomass of Rice under Field Conditions Using Digital Image Analysis. J. Environ. Sci. Nat. Resour. 2017, 10, 1–7. [Google Scholar] [CrossRef][Green Version]
- Travlos, I.; Mikroulis, A.; Anastasiou, E.; Fountas, S.; Bilalis, D.; Tsiropoulos, Z.; Balafoutis, A. The use of RGB cameras in defining crop development in legumes. Adv. Anim. Biosci. 2017, 8, 224–228. [Google Scholar] [CrossRef]
- Marcial-Pablo, M.d.J.; Gonzalez-Sanchez, A.; Jimenez-Jimenez, S.I.; Ontiveros-Capurata, R.E.; Ojeda-Bustamante, W. Estimation of vegetation fraction using RGB and multispectral images from UAV. Int. J. Remote Sens. 2018, 40, 420–438. [Google Scholar] [CrossRef]
- Yousfi, S.; Kellas, N.; Saidi, L.; Benlakehal, Z.; Chaou, L.; Siad, D.; Herda, F.; Karrou, M.; Vergara, O.; Gracia, A.; et al. Comparative performance of remote sensing methods in assessing wheat performance under Mediterranean conditions. Agric. Water Manag. 2016, 164, 137–147. [Google Scholar] [CrossRef]
- Mutka, A.M.; Fentress, S.J.; Sher, J.W.; Berry, J.C.; Pretz, C.; Nusinow, D.A.; Bart, R. Quantitative, Image-Based Phenotyping Methods Provide Insight into Spatial and Temporal Dimensions of Plant Disease. Plant Physiol. 2016, 172, 650–660. [Google Scholar] [CrossRef]
- Zhou, B.; Elazab, A.; Bort, J.; Vergara, O.; Serret, M.D.; Araus, J.L. Low-cost assessment of wheat resistance to yellow rust through conventional RGB images. Comput. Electron. Agric. 2015, 116, 20–29. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Ustin, S.L.; Verdebout, J.; Schmuck, G.; Andreoli, G.; Hosgood, B. Estimating leaf biochemistry using the PROSPECT leaf optical properties model. Remote Sens. Environ. 1996, 56, 194–202. [Google Scholar] [CrossRef]
- Hunt, E.R.; Cavigelli, M.; Daughtry, C.S.T.; McMurtrey, J.E.; Walthall, C.L. Evaluation of Digital Photography from Model Aircraft for Remote Sensing of Crop Biomass and Nitrogen Status. Precis. Agric. 2005, 6, 359–378. [Google Scholar] [CrossRef]
- Torres-Sanchez, J.; Lopez-Granados, F.; De Castro, A.I.; Pena-Barragan, J.M. Configuration and specifications of an Unmanned Aerial Vehicle (UAV) for early site specific weed management. PLoS ONE 2013, 8, e58210. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multispectral and digital imagery. Isprs J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Lu, B.; He, Y.; Liu, H.H.T. Mapping vegetation biophysical and biochemical properties using unmanned aerial vehicles-acquired imagery. Int. J. Remote Sens. 2017, 39, 5265–5287. [Google Scholar] [CrossRef]
- Liakos, K.G.; Busato, P.; Moshou, D.; Pearson, S.; Bochtis, D. Machine Learning in Agriculture: A Review. Sensors 2018, 18, 2674. [Google Scholar] [CrossRef]
- Han, L.; Yang, G.; Dai, H.; Xu, B.; Yang, H.; Feng, H.; Li, Z.; Yang, X. Modeling maize above-ground biomass based on machine learning approaches using UAV remote-sensing data. Plant Methods 2019, 15, 10. [Google Scholar] [CrossRef]
- Fernandez-Gallego, J.A.; Lootens, P.; Borra-Serrano, I.; Derycke, V.; Haesaert, G.; Roldan-Ruiz, I.; Araus, J.L.; Kefauver, S.C. Automatic wheat ear counting using machine learning based on RGB UAV imagery. Plant J. 2020, 103, 1603–1613. [Google Scholar] [CrossRef]
- Li, B.; Xu, X.; Zhang, L.; Han, J.; Bian, C.; Li, G.; Liu, J.; Jin, L. Above-ground biomass estimation and yield prediction in potato by using UAV-based RGB and hyperspectral imaging. ISPRS J. Photogramm. Remote Sens. 2020, 162, 161–172. [Google Scholar] [CrossRef]
- Yuan, H.H.; Yang, G.J.; Li, C.C.; Wang, Y.J.; Liu, J.G.; Yu, H.Y.; Feng, H.K.; Xu, B.; Zhao, X.Q.; Yang, X.D. Retrieving Soybean Leaf Area Index from Unmanned Aerial Vehicle Hyperspectral Remote Sensing: Analysis of RF, ANN, and SVM Regression Models. Remote Sens. 2017, 9, 309. [Google Scholar] [CrossRef]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization Paths for Generalized Linear Models via Coordinate Descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef]
- Breimann, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Reynolds, M.; Chapman, S.; Crespo-Herrera, L.; Molero, G.; Mondal, S.; Pequeno, D.N.L.; Pinto, F.; Pinera-Chavez, F.J.; Poland, J.; Rivera-Amado, C.; et al. Breeder friendly phenotyping. Plant Sci. 2020, 295, 110396. [Google Scholar] [CrossRef] [PubMed]
- Schmalenbach, I.; March, T.J.; Bringezu, T.; Waugh, R.; Pillen, K. High-Resolution Genotyping of Wild Barley Introgression Lines and Fine-Mapping of the Threshability Locus thresh-1 Using the Illumina GoldenGate Assay. G3 (Bethesda) 2011, 1, 187–196. [Google Scholar] [CrossRef] [PubMed]
- Honsdorf, N.; March, T.J.; Pillen, K. QTL controlling grain filling under terminal drought stress in a set of wild barley introgression lines. PLoS ONE 2017, 12, e0185983. [Google Scholar] [CrossRef]
- von Korff, M.; Wang, H.; Leon, J.; Pillen, K. Development of candidate introgression lines using an exotic barley accession (Hordeum vulgare ssp. spontaneum) as donor. Theor. Appl. Genet. 2004, 109, 1736–1745. [Google Scholar] [CrossRef] [PubMed]
- Schmalenbach, I.; Korber, N.; Pillen, K. Selecting a set of wild barley introgression lines and verification of QTL effects for resistance to powdery mildew and leaf rust. Theor. Appl. Genet. 2008, 117, 1093–1106. [Google Scholar] [CrossRef]
- Wang, G.; Schmalenbach, I.; von Korff, M.; Leon, J.; Kilian, B.; Rode, J.; Pillen, K. Association of barley photoperiod and vernalization genes with QTLs for flowering time and agronomic traits in a BC2DH population and a set of wild barley introgression lines. Theor. Appl. Genet. 2010, 120, 1559–1574. [Google Scholar] [CrossRef]
- Schmalenbach, I.; Leon, J.; Pillen, K. Identification and verification of QTLs for agronomic traits using wild barley introgression lines. Theor. Appl. Genet. 2009, 118, 483–497. [Google Scholar] [CrossRef][Green Version]
- Hoffmann, A.; Maurer, A.; Pillen, K. Detection of nitrogen deficiency QTL in juvenile wild barley introgression lines growing in a hydroponic system. BMC Genet. 2012, 13, 88. [Google Scholar] [CrossRef]
- Soleimani, B.; Sammler, R.; Backhaus, A.; Beschow, H.; Schumann, E.; Mock, H.P.; von Wiren, N.; Seiffert, U.; Pillen, K. Genetic regulation of growth and nutrient content under phosphorus deficiency in the wild barley introgression library S42IL. Plant Breed. 2017, 136, 892–907. [Google Scholar] [CrossRef]
- Honsdorf, N.; March, T.J.; Hecht, A.; Eglinton, J.; Pillen, K. Evaluation of juvenile drought stress tolerance and genotyping by sequencing with wild barley introgression lines. Mol. Breed. 2014, 34, 1475–1495. [Google Scholar] [CrossRef]
- Honsdorf, N.; March, T.J.; Berger, B.; Tester, M.; Pillen, K. High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS ONE 2014, 9, e97047. [Google Scholar] [CrossRef]
- Muzammil, S.; Shrestha, A.; Dadshani, S.; Pillen, K.; Siddique, S.; Leon, J.; Naz, A.A. An Ancestral Allele of Pyrroline-5-carboxylate synthase1 Promotes Proline Accumulation and Drought Adaptation in Cultivated Barley. Plant Physiol. 2018, 178, 771–782. [Google Scholar] [CrossRef] [PubMed]
- Zahn, S.; Koblenz, B.; Christen, O.; Pillen, K.; Maurer, A. Evaluation of wild barley introgression lines for agronomic traits related to nitrogen fertilization. Euphytica 2020, 216. [Google Scholar] [CrossRef]
- Lancashire, P.D.; Bleiholder, H.; Vandenboom, T.; Langeluddeke, P.; Stauss, R.; Weber, E.; Witzenberger, A. A Uniform Decimal Code for Growth-Stages of Crops and Weeds. Ann. Appl. Biol. 1991, 119, 561–601. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical computing, Foundation for Statistical Computing; European Environment Agency: Vienna, Austria, 2020. [Google Scholar]
- Hijmans, R.J. ‘Raster’—Geographic Data Analysis and Modeling. 2020. Available online: https://CRAN.R-project.org/package=raster (accessed on 1 May 2020).
- Jiang, Z.Y.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Rouse, J.W., Jr.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring the Vernal Advancement and Retrogradation (Green Wave Effect) of Natural Vegetation; Prog. Rep. RSC 1978-1, NTIS No. E73-106393; Remote Sensing Center, Texas A&M Univ.: College Station, TX, USA, 1973; 93p. [Google Scholar]
- Penuelas, J.; Pinol, J.; Ogaya, R.; Filella, I. Estimation of plant water concentration by the reflectance water index WI (R900/R970). Int. J. Remote Sens. 1997, 18, 2869–2875. [Google Scholar] [CrossRef]
- Blackburn, G.A. Spectral indices for estimating photosynthetic pigment concentrations: A test using senescent tree leaves. Int. J. Remote Sens. 2010, 19, 657–675. [Google Scholar] [CrossRef]
- Roujean, J.L.; Breon, F.M. Estimating Par Absorbed by Vegetation from Bidirectional Reflectance Measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Gitelson, A.; Merzlyak, M.N. Quantitative Estimation of Chlorophyll-a Using Reflectance Spectra—Experiments with Autumn Chestnut and Maple Leaves. J. Photoch Photobiol. B 1994, 22, 247–252. [Google Scholar] [CrossRef]
- Guyot, G.; Baret, F.; Major, D.J. High spectral resolution: Determination of spectral shifts between the red and the near infrared. In Proceedings of the ISPRS Congress, Kyoto, Japan, 1–10 July 1988. [Google Scholar]
- Horler, D.N.H.; Dockray, M.; Barber, J.; Barringer, A.R. Red edge measurements for remotely sensing plant chlorophyll content. Int. J. Remote. Sens. 1983, 4, 273–288. [Google Scholar] [CrossRef]
- Vogelmann, J.E.; Rock, B.N.; Moss, D.M. Red edge spectral measurements from sugar maple leaves. Int. J. Remote Sens. 2007, 14, 1563–1575. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Miller, J.R.; Noland, T.L.; Mohammed, G.H.; Sampson, P.H. Scaling-up and model inversion methods with narrowband optical indices for chlorophyll content estimation in closed forest canopies with hyperspectral data. IEEE Trans. Geosci. Remote 2001, 39, 1491–1507. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Hunt, E.R.; Doraiswamy, P.C.; McMurtrey, J.E.; Daughtry, C.S.T.; Perry, E.M.; Akhmedov, B. A visible band index for remote sensing leaf chlorophyll content at the canopy scale. Int. J. Appl. Earth Obs. Geoinf. 2013, 21, 103–112. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Zur, Y.; Stark, R.; Gritz, U. Non-destructive and remote sensing techniques for estimation of vegetation status. In Proceedings of the 3rd European Conference on Precision Agriculture, Montpelier, France, 18–20 June 2001. [Google Scholar]
- Otsu, N. A Threshold Selection Method from Gray-Level Histograms. IEEE Trans. Syst. ManCybern. 1979, 9, 62–66. [Google Scholar] [CrossRef]
- Landini, G.; Randell, D.A.; Fouad, S.; Galton, A. Automatic thresholding from the gradients of region boundaries. J. Microsc. 2017, 265, 185–195. [Google Scholar] [CrossRef]
- Brenning, A. Statistical Geocomputing Combining R and SAGA: The Example of Landslide Susceptibility Analysis with Generalized Additive Models. Hamburger Beiträge zur Physischen Geographie und Landschaftsökologie 2008, 19, 23–32. [Google Scholar]
- Kuhn, M.; Wickham, H. Tidymodels: Easily Install and Load the ’Tidymodels’ Packages. R package version 0.1.2. Available online: https://CRAN.R-project.or/ackage=tidymodels (accessed on 1 May 2020).
- Bates, D.; Kliegl, R.; Vasishth, S.; Baayen, H. Parsimonious Mixed Models. arXiv 2018, arXiv:1506.04967. [Google Scholar]
- Dunnett, C.W. A Multiple Comparison Procedure for Comparing Several Treatments with a Control. J. Am. Stat. Assoc. 1955, 50, 1096–1121. [Google Scholar] [CrossRef]
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous inference in general parametric models. Biom J. 2008, 50, 346–363. [Google Scholar] [CrossRef] [PubMed]
- Bretz, F.; Hothorn, T.; Westfall, P. Multiple Comparisons Using R; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar]
- Bareth, G.; Bendig, J.; Tilly, N.; Hoffmeister, D.; Aasen, H.; Bolten, A. A Comparison of UAV- and TLS-derived Plant Height for Crop Monitoring: Using Polygon Grids for the Analysis of Crop Surface Models (CSMs). Photogramm Fernerkun 2016. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Michalski, A.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. High Throughput Field Phenotyping of Wheat Plant Height and Growth Rate in Field Plot Trials Using UAV Based Remote Sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Aasen, H.; Burkart, A.; Bolten, A.; Bareth, G. Generating 3D hyperspectral information with lightweight UAV snapshot cameras for vegetation monitoring: From camera calibration to quality assurance. Isprs J. Photogramm. Remote Sens. 2015, 108, 245–259. [Google Scholar] [CrossRef]
- Pask, A.; Pietragalla, J.; Mullan, D.M.; Reynolds, M.P. Physiological Breeding II: A Field Guide to Wheat Phenotyping; CIMMYT: Distrito Federal, Mexico, 2012. [Google Scholar]
- Grenzdörffer, G.J. Crop height determination with UAS point clouds. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2014, XL-1, 135–140. [Google Scholar] [CrossRef]
- Wang, J.M.; Yang, J.M.; McNeil, D.L.; Zhou, M.X. Identification and molecular mapping of a dwarfing gene in barley (Hordeum vulgare L.) and its correlation with other agronomic traits. Euphytica 2010, 175, 331–342. [Google Scholar] [CrossRef]
- Resop, J.P.; Lehmann, L.; Hession, W.C. Drone Laser Scanning for Modeling Riverscape Topography and Vegetation: Comparison with Traditional Aerial Lidar. Drones 2019, 3, 35. [Google Scholar] [CrossRef]
- Lin, Y. LiDAR: An important tool for next-generation phenotyping technology of high potential for plant phenomics? Comput. Electron. Agric. 2015, 119, 61–73. [Google Scholar] [CrossRef]
- Sankey, T.; Donager, J.; McVay, J.; Sankey, J.B. UAV lidar and hyperspectral fusion for forest monitoring in the southwestern USA. Remote Sens. Environ. 2017, 195, 30–43. [Google Scholar] [CrossRef]
- Zhou, L.F.; Gu, X.H.; Cheng, S.; Yang, G.J.; Shu, M.Y.; Sun, Q. Analysis of Plant Height Changes of Lodged Maize Using UAV-LiDAR Data. Agriculture 2020, 10, 146. [Google Scholar] [CrossRef]
- ten Harkel, J.; Bartholomeus, H.; Kooistra, L. Biomass and Crop Height Estimation of Different Crops Using UAV-Based Lidar. Remote Sens. 2019, 12, 17. [Google Scholar] [CrossRef]
- Maesano, M.; Khoury, S.; Nakhle, F.; Firrincieli, A.; Gay, A.; Tauro, F.; Harfouche, A. UAV-Based LiDAR for High-Throughput Determination of Plant Height and Above-Ground Biomass of the Bioenergy Grass Arundo donax. Remote Sens. 2020, 12, 3464. [Google Scholar] [CrossRef]
- Lóopez-Castañeda, C.; Richards, R.A.; Farquhar, G.D. Variation in Early Vigor between Wheat and Barley. Crop Sci. 1995, 35. [Google Scholar] [CrossRef]
- Mullan, D.J.; Reynolds, M.P. Quantifying genetic effects of ground cover on soil water evaporation using digital imaging. Funct. Plant Biol. 2010, 37, 703–712. [Google Scholar] [CrossRef]
- Grieder, C.; Hund, A.; Walter, A. Image based phenotyping during winter: A powerful tool to assess wheat genetic variation in growth response to temperature. Funct. Plant Biol. 2015, 42, 387–396. [Google Scholar] [CrossRef]
- Ballesteros, R.; Ortega, J.F.; Hernandez, D.; Moreno, M.A. Onion biomass monitoring using UAV-based RGB imaging. Precis. Agric. 2018, 19, 840–857. [Google Scholar] [CrossRef]
- Kim, S.L.; Chung, Y.S.; Ji, H.; Lee, H.; Choi, I.; Kim, N.; Lee, E.; Oh, J.; Kang, D.Y.; Baek, J.; et al. New Parameters for Seedling Vigor Developed via Phenomics. Appl. Sci. 2019, 9, 1752. [Google Scholar] [CrossRef]
- Liu, J.G.; Pattey, E. Retrieval of leaf area index from top-of-canopy digital photography over agricultural crops. Agric. For. Meteorol. 2010, 150, 1485–1490. [Google Scholar] [CrossRef]
- Liebisch, F.; Kirchgessner, N.; Schneider, D.; Walter, A.; Hund, A. Remote, aerial phenotyping of maize traits with a mobile multi-sensor approach. Plant Methods 2015, 11, 9. [Google Scholar] [CrossRef]
- Yu, K.; Kirchgessner, N.; Grieder, C.; Walter, A.; Hund, A. An image analysis pipeline for automated classification of imaging light conditions and for quantification of wheat canopy cover time series in field phenotyping. Plant Methods 2017, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.J.; Li, L.Y.; Coy, A.; Mu, X.H.; Chen, S.B.; Xie, D.H.; Zhang, W.M.; Shen, Q.F.; Zhou, H.M. Improving the estimation of fractional vegetation cover from UAV RGB imagery by colour unmixing. Isprs J. Photogramm. Remote Sens. 2019, 158, 23–34. [Google Scholar] [CrossRef]
- Torres-Sánchez, J.; Peña, J.M.; de Castro, A.I.; López-Granados, F. Multi-temporal mapping of the vegetation fraction in early-season wheat fields using images from UAV. Comput. Electron. Agric. 2014, 103, 104–113. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X.; et al. Unmanned Aerial Vehicle Remote Sensing for Field-Based Crop Phenotyping: Current Status and Perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef]
- Oehlschlager, J.; Schmidhalter, U.; Noack, P.O. UAV-Based Hyperspectral Sensing for Yield Prediction in Winter Barley. In Proceedings of the 2018 9th Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Amsterdam, The Netherlands, 23–26 September 2018. [Google Scholar] [CrossRef]
- Geipel, J.; Link, J.; Claupein, W. Combined Spectral and Spatial Modeling of Corn Yield Based on Aerial Images and Crop Surface Models Acquired with an Unmanned Aircraft System. Remote Sens. 2014, 6, 10335–10355. [Google Scholar] [CrossRef]
- Rischbeck, P.; Elsayed, S.; Mistele, B.; Barmeier, G.; Heil, K.; Schmidhalter, U. Data fusion of spectral, thermal and canopy height parameters for improved yield prediction of drought stressed spring barley. Eur. J. Agron. 2016, 78, 44–59. [Google Scholar] [CrossRef]
- Yue, J.B.; Yang, G.J.; Li, C.C.; Li, Z.H.; Wang, Y.J.; Feng, H.K.; Xu, B. Estimation of Winter Wheat Above-Ground Biomass Using Unmanned Aerial Vehicle-Based Snapshot Hyperspectral Sensor and Crop Height Improved Models. Remote Sens. 2017, 9, 708. [Google Scholar] [CrossRef]
- Yue, J.B.; Feng, H.K.; Jin, X.L.; Yuan, H.H.; Li, Z.H.; Zhou, C.Q.; Yang, G.J.; Tian, Q.J. A Comparison of Crop Parameters Estimation Using Images from UAV-Mounted Snapshot Hyperspectral Sensor and High-Definition Digital Camera. Remote Sens. 2018, 10, 1138. [Google Scholar] [CrossRef]
- Herzig, P.; Backhaus, A.; Seiffert, U.; von Wiren, N.; Pillen, K.; Maurer, A. Genetic dissection of grain elements predicted by hyperspectral imaging associated with yield-related traits in a wild barley NAM population. Plant Sci. 2019, 285, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Yang, G.; Reynolds, M.; Xia, X.; Xiao, Y.; He, Z. A rapid monitoring of NDVI across the wheat growth cycle for grain yield prediction using a multi-spectral UAV platform. Plant Sci. 2019, 282, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Roth, L.; Streit, B. Predicting cover crop biomass by lightweight UAS-based RGB and NIR photography: An applied photogrammetric approach. Precis. Agric. 2017, 19, 93–114. [Google Scholar] [CrossRef]
- Babar, M.A.; Reynolds, M.P.; van Ginkel, M.; Klatt, A.R.; Raun, W.R.; Stone, M.L. Spectral reflectance indices as a potential indirect selection criteria for wheat yield under irrigation. Crop Sci. 2006, 46, 578–588. [Google Scholar] [CrossRef]
- Main, R.; Cho, M.A.; Mathieu, R.; O’Kennedy, M.M.; Ramoelo, A.; Koch, S. An investigation into robust spectral indices for leaf chlorophyll estimation. Isprs J. Photogramm. Remote Sens. 2011, 66, 751–761. [Google Scholar] [CrossRef]
- Bowman, B.C.; Chen, J.; Zhang, J.; Wheeler, J.; Wang, Y.; Zhao, W.; Nayak, S.; Heslot, N.; Bockelman, H.; Bonman, J.M. Evaluating Grain Yield in Spring Wheat with Canopy Spectral Reflectance. Crop Sci. 2015, 55, 1881–1890. [Google Scholar] [CrossRef]
- Smith, K.L.; Steven, M.D.; Colls, J.J. Use of hyperspectral derivative ratios in the red-edge region to identify plant stress responses to gas leaks. Remote Sens. Environ. 2004, 92, 207–217. [Google Scholar] [CrossRef]
- Lamb, D.W.; Steyn-Ross, M.; Schaare, P.; Hanna, M.M.; Silvester, W.; Steyn-Ross, A. Estimating leaf nitrogen concentration in ryegrass (Lolium spp.) pasture using the chlorophyll red-edge: Theoretical modelling and experimental observations. Int. J. Remote Sens. 2010, 23, 3619–3648. [Google Scholar] [CrossRef]
- Boochs, F.; Kupfer, G.; Dockter, K.; KÜHbauch, W. Shape of the red edge as vitality indicator for plants. Int. J. Remote Sens. 2007, 11, 1741–1753. [Google Scholar] [CrossRef]
- Wiegmann, M.; Backhaus, A.; Seiffert, U.; Thomas, W.T.B.; Flavell, A.J.; Pillen, K.; Maurer, A. Optimizing the procedure of grain nutrient predictions in barley via hyperspectral imaging. PLoS ONE 2019, 14, e0224491. [Google Scholar] [CrossRef] [PubMed]
- Freeman, P.K.; Freeland, R.S. Agricultural UAVs in the U.S.: Potential, policy, and hype. Remote Sens. Appl. Soc. Environ. 2015, 2, 35–43. [Google Scholar] [CrossRef]





| Trait | Abbreviation | Unit | Instrument of Determination | Measurement |
|---|---|---|---|---|
| Time to shooting | SHO | days | visually | Number of days from sowing until first node noticeable 1 cm above soil surface for 50% of all plants of a plot, i.e., BBCH 31 [81] |
| Time to heading | HEA | days | visually | Number of days from sowing until awn emergence for 50% of all plants of a plot, i.e., BBCH 49 [81] |
| Time to maturity | MAT | days | visually | Number of days from sowing until hard dough: grain content firm and fingernail impression held, BBCH 87 [81] |
| Canopy height | HEIGT a/ HEICHM b/ HEICHMred c | cm | visually/ UAV data (RGB) | Average canopy height of all plants of a plot measured once a week. UAV RGB Data were used to construct digital elevation models (DEM), which led to the determination of growth parameters (HEIGRi d, HEIGRd e, HEIMAX f) |
| Vegetation cover | VCOV | % | UAV data (RGB, Multispectral) | Area of a plot covered by plants, which led to the determination of growth parameters (VCOVGRi g, VCOV90 h, VCOVsmoothed i) |
| Plot yield | YLD | kg | Harvester/ UAV data (RGB, Multispectral) | Grain weight harvesting the whole plot (7.5m2)/ Modelling based on UAV data, VCOV and HEI |
| Index | Index Full Name | Platform | Group/Sensitivity | Formula | Reference |
|---|---|---|---|---|---|
| B1-NIR1 | Near infrared band 1 | Multi | Single band | - | more information see Table S4 |
| B2-RED | Red band | Multi | Single band | - | |
| B3-RE1 | Red edge band 1 | Multi | Single band | ||
| B4-RE2 | Red edge band 2 | Multi | Single band | ||
| B5-NIR2 | Near infrared band 2 | Multi | Single band | ||
| B6-WA | Water band | Multi | Single band | ||
| EVI2 | Enhanced vegetation index 2 | Multi | Pigments | Jiang et al. [84] | |
| NDVI | Normalized difference vegetation index | Multi | Pigments | Rouse et al. [85] | |
| ND-NIR1RE1 | Normalized difference NIR1-RE1 | Multi | Pigments | - | |
| ND-NIR1RE2 | Normalized difference NIR1-RE2 | Multi | Pigments | - | |
| ND-NIR2RED | Normalized difference NIR2-RED | Multi | Pigments | - | |
| ND-NIR2RE1 | Normalized difference NIR2-RE1 | Multi | Pigments | - | |
| ND-NIR2RE2 | Normalized difference NIR2-RE2 | Multi | Pigments | - | |
| NDWI | Normalized difference water index | Multi | Water content | Penuelas et al. [86] | |
| PSSRa | Pigment Specific Simple Ratio | Pigments | Blackburn [87] | ||
| SR980_R700 | Simple Ratio | Multi | Water content | ||
| RDVI | Renormalized difference vegetation index | Multi | Pigments | Roujean et al. [88] | |
| RENDVI | Red edge normalized difference vegetation index | Multi | Pigments | Gitelson et al. [89] | |
| REP | Red edge position | Multi | Physiology | Guyot [90] | |
| RVSI | Red edge vegetation stress index | Multi | Physiology | Horler et al. [91] | |
| VOG | Vogelmann ratio | Multi | Pigments | Vogelmann et al. [92] | |
| B1-R | Red | RGB | Single band | - | |
| B2-G | Green | RGB | Single band | - | |
| B3-B | Blue | RGB | Single band | - | |
| EG | Excess greenness | RGB | Pigments | Nijland et al. [44] | |
| GCC | Green chromatic coordinate | RGB | Pigments | Nijland et al. [44] | |
| NGRDI | Normalized green red difference index | RGB | Pigments | Zarco-Tejada et al. [93] | |
| RGBVI | Red green blue vegetation index | RGB | Pigments | Bendig et al. [94] | |
| TGI | Triangular greenness index | RGB | Pigments | Hunt et al. [95] | |
| VARI | Visible atmospheric resistant index | RGB | Pigments | Gitelson et al. [96] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Herzig, P.; Borrmann, P.; Knauer, U.; Klück, H.-C.; Kilias, D.; Seiffert, U.; Pillen, K.; Maurer, A. Evaluation of RGB and Multispectral Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping and Yield Prediction in Barley Breeding. Remote Sens. 2021, 13, 2670. https://doi.org/10.3390/rs13142670
Herzig P, Borrmann P, Knauer U, Klück H-C, Kilias D, Seiffert U, Pillen K, Maurer A. Evaluation of RGB and Multispectral Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping and Yield Prediction in Barley Breeding. Remote Sensing. 2021; 13(14):2670. https://doi.org/10.3390/rs13142670
Chicago/Turabian StyleHerzig, Paul, Peter Borrmann, Uwe Knauer, Hans-Christian Klück, David Kilias, Udo Seiffert, Klaus Pillen, and Andreas Maurer. 2021. "Evaluation of RGB and Multispectral Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping and Yield Prediction in Barley Breeding" Remote Sensing 13, no. 14: 2670. https://doi.org/10.3390/rs13142670
APA StyleHerzig, P., Borrmann, P., Knauer, U., Klück, H.-C., Kilias, D., Seiffert, U., Pillen, K., & Maurer, A. (2021). Evaluation of RGB and Multispectral Unmanned Aerial Vehicle (UAV) Imagery for High-Throughput Phenotyping and Yield Prediction in Barley Breeding. Remote Sensing, 13(14), 2670. https://doi.org/10.3390/rs13142670







