1. Introduction
Integrated Administration Control System (IACS) was created in the European Union to control direct payment to agriculture. Under common agricultural policy (CAP), direct payments, without going into different payment schemes, apply to crops, which the farmer declares each year, specifying the type of crop (plant) and its area. Some declarations (ca.
) are controlled using on-the-spot check under which the area of an agricultural parcel is measured and the plant is identified in the field. In order to simplify and automate this procedure, in 2018, the European Commission adopted new rules to control all declared parcels based on Copernicus satellite data: Sentinel-1 (S-1), Sentinel-2 (S-2). In purpose of control farmers’ declarations, the EC research center JRC (Joint Research Center) recommends analysis of time series vegetation indices of any agricultural parcel during the vegetation time [
1,
2,
3].
The most popular index calculated form optical images is NDVI (normalized differential vegetation index) [
4] and in microwave spectral range SIGMA (radar backscattering coefficient) [
5]. Analysis of the variability plots of these parameters over time allows for reliable assessment of the condition of vegetation and type of crop but it is also work, time consuming and nearly not feasible for all declared plots. There are many initiatives and projects dedicated to agriculture or the CAP (e.g., Sen2Agri, Sen4Cap) among others aimed at the control of direct payments based on the time series of images S-1, S-2.
Automatic image classification for declarations’ inspection seems to be very promising but there are no applicable recommendations for automatic image classification to support audits. The key issue is the credibility of the method as control may result in financial penalties for farmers. Moreover, the verification of all declared parcels is a huge undertaking because it affects areas of the whole country, e.g., in Poland concerns ca. 30% of the country’s area, i.e., 140,000 square kilometers and 10 million agricultural plots.
There are many publications on crops identification in various places around the world, for different purposes and with varying levels of credibility. Analyzing practiced by most researchers methods of image classification for crops recognition, machine learning (ML) undoubtedly dominates: random forest, support vector machine (SVM), convolutional neural network (CNN). The literature reports high accuracies for these methods: e.g.,
[
6],
[
7],
[
8],
[
9],
[
10],
[
11]. A common practice in crops identification is applying the time series consisting of several or several dozen images. Acquiring multiple cloudless S-2 images over a large area is difficult, so many researchers perform analyzes on combinations of a different number of images. In such approaches it is efficient to use cloud computing using ML methods, e.g., Google Earth Engine (some accuracy values reported by the authors:
[
12],
[
13],
[
14]).
The level of accuracy achieved with single images is lower, several results can be cited (e.g., [
6,
7,
8]). The accuracy of the classification, using a single S-2 image, of three crops (wheat, sugarcane, fodder), which was performed for the test area located in India, was:
and
[
6]. In turn [
7], for the test area in Australia, the accuracy of classification of a single S-2 image using the
method was 77.4% for annual crops (cotton, rice, maize, canola, wheat, barley) and perennial crops (citrus, almond, cherry, etc.). Especially interesting is [
8], where the authors examined the accuracy of the classification of Sentinel-2 images by various classifications methods (RF, SVM, Decision Tree, k-nearest neighbors). They analyzed the results of the S-2 time series for crop recognition in South Africa: canola, lucerne, pasture, wheat and fallow recognition. The highest accuracy was obtained with the use of the support vector machine (SVM) approx. 80%. The most important conclusion is that it was possible to obtain high accuracy of crop classification (77.2%) using single Sentinel-2 image recorded approx. 8 weeks before the harvest. Moreover, they found that adding more than 5 multi-time images does not increase accuracy, and that “good” images do not compensate for “bad” images.
When comparing the research results, the type of crop must be taken into account. The species of cultivated crops depend on the climatic zone in which the research area is located. In this context it is worth citing publications relating to a similar test area to ours. In [
15], the authors present the results of research conducted in Northrhinewestfalia (Germany) [
15]. They performed a random forest classification of 70 Sentinel-1 mulitemporal images with topographical and cadastral data and reference agricultural parcels obtained from Amtliches Liegenschaftskataster-Informationssystem (ALKIS). With recognition of 11 crops: maize, sugar beet, barley, wheat, rye, spring barley, pasture, rapeseed, potato, pea and carrot very high overall accuracy of
was obtained (comparing to optic data
).
The other example are the research concerning the IACS system was conducted on the whole area of Belgium [
16]. Authors performed an experiment to identify 8 crops: wheat, barley, rapeseed, maize, potatoes, beets, flax and grassland. Various combinations of Sentinel-1 and Sentinel-2 time series were tested in the study. The images were classified using the random forest (RF) method. The maximum accuracy, equal to
for the combination of twelve images: six Sentinel-1 and six Sentinel-2 was reported.
The presented two approaches provided high accuracy of crop recognition for control purposes, but required many unclouded images of large areas. This is rather difficult in the case of Sentinel-2, especially considering that the area of Belgium and Northrhinewestfalia is 10 times smaller than the area of Poland.
The similar research were also conducted by our team [
17,
18]. Ten Sentinel-2 images from September 2016 to August 2017, and nine Sentinel-1 images from March to September 2017 were analyzed. A Spectral Angle Mapper (SAM) classifier was used to classify the time series of NDVI images. Accuracy of
= 68.27% was achieved, which is consistent with the accuracy (69%) of other independent studies of similar nature conducted in Poland [
19]. Therefore, instead of NDVI and SIGMA time series, it was decided to check the possibility of using the classification of single S-2 images and a combination of several multitemporal S-2 images. The aim of the research was to develop a simple, fast but reliable screening method for farmers’ declarations control.
However, while reviewing the literature on the currently used image classification methods for the purpose of crop recognition, we encountered the problem of comparing the accuracy of the classification result. In a traditional remote sensing approach, accuracy is calculated from test data independent of the training data. In machine learning, a lot of attention is paid to the selection of hyperparameters, which is carried out iteratively using only a part of the training set. At the same time, the validation accuracy is determined on the basis of the samples from training set not used for learning.
Some authors report validation accuracy and accuracy calculated on independent test data [
7,
12,
20]. Others only provide information on accuracy based on external reference data not included in the training set [
8,
15,
16,
21]. In some publications, there is not enough information on this issue [
13,
22,
23].
There are plenty examples of using only one reference set divided randomly or stratified on training and validation sample, while the distinction between training, validation and independent test data is extremely rare [
24].
It turns out that the problem was also noticed by other researchers, e.g., a key work on rigorous accuracy assessment of land cover products [
25]. Good practice in accuracy assessment, sampling design for training, validation and accuracy analysis was discussed. The key, from the point of view of our research is the statement: “Using the same data for training and validation can lead to optimistically biased accuracy assessments. Consequently, the training sample and the validation sample need to be independent of each other which can be achieved by appropriately dividing a single sample of reference data or, perhaps more commonly, by acquiring separate samples for training and testing”. Similar conclusions can be found in [
26].
Reliability of the classification also depends on the reported accuracy metrics and the unambiguous way of their calculation. Like the previous topic, it is not a trivial issue, although it seems that. Despite many years of research on various accuracy metrics [
27,
28,
29,
30,
31,
32,
33] in the 2019 paper, mentioned above, it was stated that overall accuracy (
), producer accuracy (PA) and user accuracy (UA) are still considered the basic ones [
25].
Unfortunately, the situation in this area has become more complicated due to the common use of ML methods. In recent years, additional accuracy metrics have been developed that are not used in traditional approaches, i.e., specificity and precision. Other metrics calculated automatically in ML tools: sensitivity and precision, correspond, respectively, to producer accuracy (PA) and user accuracy (UA) in traditional image classification.
In extensive reviews of the literature from the past and the latest, comparative analyzes of plenty various metrics can be found [
26,
27,
28,
29,
30,
31,
32,
33], but they do not respect sensitivity and accuracy, despite they are commonly used in ML. It is worth noting that sensitivity and accuracy are appropriate for the classification of one class. A problem arises when they are used in assessing the classification of multiple classes, especially if the average value of the accuracy [
34] or global precision [
35] is reported as
, creating an illusion of higher accuracy [
36].
Summarizing the issue of the reliability of the classification result, especially using ignorantly ML tools, there is a double risk of overestimation accuracy: related to the lack of independent test set and the incorrect calculation of the most frequently compared metric in the research: . Therefore, the accuracy of the classification, which has a very significant impact on the reliability and validity of the remote sensing method for verifying the accuracy of the crops declared by farmers, should be demonstrated with deep attention and carefulness. In the article, we focused on three issues:
- 1.
Image classification in the aspect of screening method of controlling farmers’ declaration based on a limited number of images (verification in Polish conditions of the hypothesis from the publication [
8]).
- 2.
Analysis of the classification’ results made using extremely different sampling design (we did not focus on the description of the model fitting).
- 3.
Comparing the traditional accuracy metrics with those used in ML (also discussing incorrect calculations), in order to confirm the hypothesis about artificially overestimating the accuracy of the classification result.
To the best of our knowledge, there are no publications on a quick and reliable screening method to control the declarations submitted by millions of farmers in each EU country each year. In addition, despite there are some publications [
7,
25] containing information on artificially overestimating the accuracy of the classification if the
calculated from the validation set, instead of the test set, but there is no broader discussion of this issue. On the other hand, the issue of incorrect calculation of
is completely ignored in publications.
2. Materials and Methods
The research consists of three parts
Figure 1:
2.1. Materials and Data Preparation
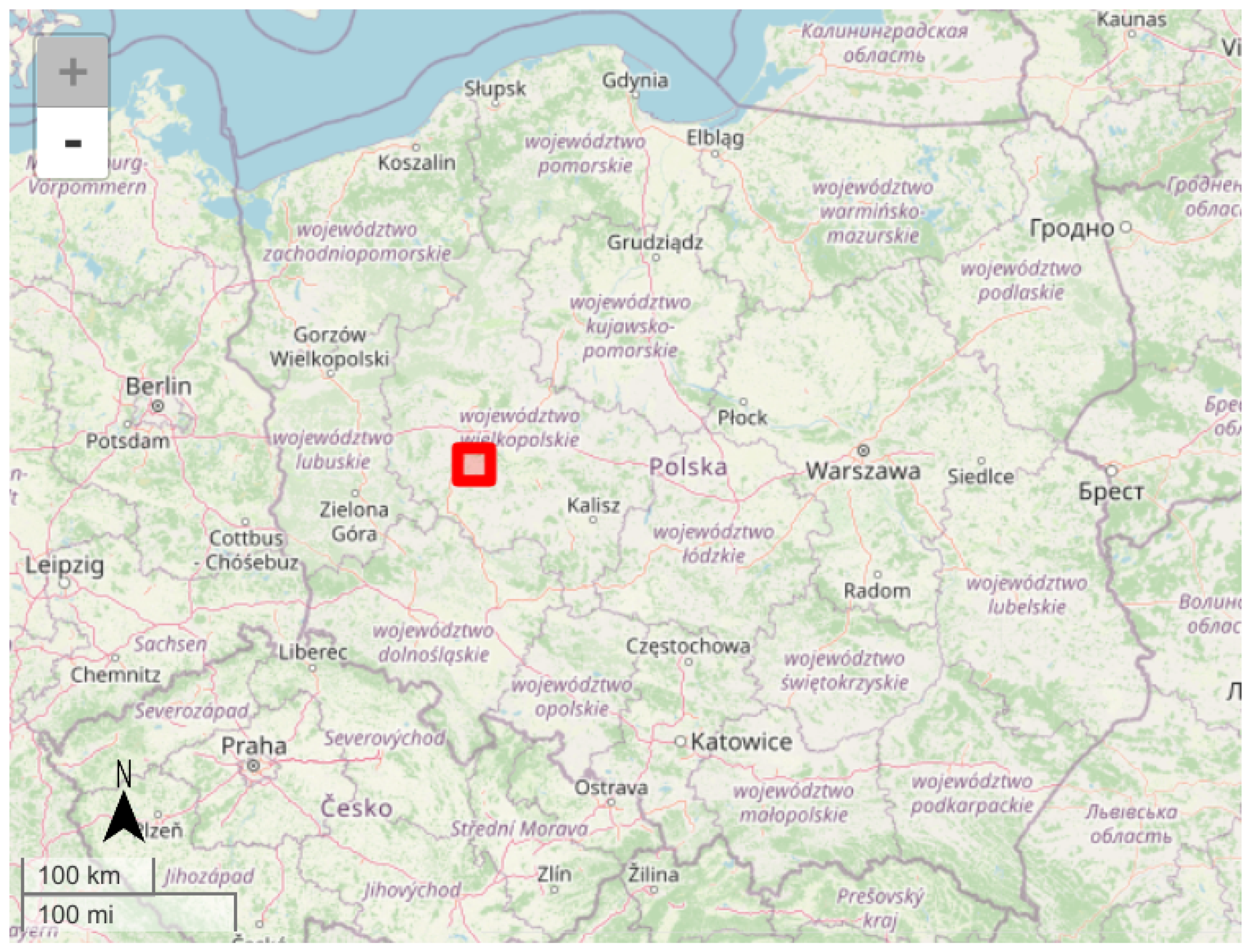
The test area of 625 square kilometers (25 km × 25 km) was located in central Poland, near Poznań (
Figure 2), and included 5500 agriculture parcels declared by farmers for the subsidies. Data on farmers’ declarations were provided by Agency for Restructuring and Modernisation of Agriculture (ARMA) in Poland and included size of the agricultural parcel, type of crop, geometry of the agricultural parcel (polygon). The critical size of the agricultural plot is 0.5 ha (this size should exclude the influence of the shape [
1]). We selected parcels of the area of 1ha or bigger to avoid technical problems with identifying small parcels. In order to reduce the number of plots and eliminate errors, farmers’ declarations were statistically analyzed for:
The parcels’ set was randomly divided into 2 groups: training fields (2190 parcels) and test fields (2386 parcels), which were used for classification and accuracy assessment.
Images from Sentinel-2 and Sentinel-1 satellites of the European Copernicus program (ESA, 2020) were used for the analysis.
Table 2 and
Table 3 contain a list and description of satellite images used in the study. The images were downloaded from the Copernicus Services Data Hub (CSDH) (
https://cophub.copernicus.eu/ (accessed on 1 September 2018)).
The data were collected as granules with a size of 100 per 100 km. Three Sentinel-2 images registered in September 2017, May 2018 and July 2018 were selected for analysis. Images of Level-2A were acquired, which means after geometric, radiometric, and atmospheric correction.
Additionally, one Sentinel-1 image was included in the tests, which had been pre-processed for the sigma coefficient, according to the following workflow for two polarization modes (VV and HV):
radiometric transformation of pixel value to backscatter coefficient (sigma0),
geometric transformation by Range Doppler orthorectification method with SRTM 3 sek as DEM and bilinear interpolation,
removing the salt pepper effect called speckle effect using refined Lee filter,
logarithmic transformation of backscatter coefficient to dB.
In the next step, the classifications were performed on the basis of the following image sets:
3 single Sentinel-2,
1 combination of 3 Sentinel-2,
2 combinations of 4 images: 3 Sentinel-2 and one Sentinel-1 (VV), 3 Sentinel-2 and one Sentinel-1 (HV).
In the single classification, all 10 channels were used, while the channels with a resolution of 20 m were previously resampled to a spatial resolution of 10 m. The classification of the combination of 3 images consists of the classification of 30 channels, 10 from each Sentinel-2 image. The simultaneous classification of optical and radar images was based on the classification of a stack of 31 channels: 30 optical, Sentinel-2 and 1 Sentinel-1 (VV or HV).
2.2. Images’ Classifications
The idea was to use SNAP (ESA) software, because it is open-source commonly used for image processing Sentinel-1, Sentinel-2 and is likely to be used in IACS control. However, it has limitations in the size of the training set. Therefore we prepare our own Python scripts to complete the research.
Eventually, images’ classifications have been carried out with the random forest algorithm using:
As mentioned above, classification in SNAP has some limitations. For example, it is not possible to load a relatively large number of training fields, as in our case (2386 parcels). In addition, the choice of classification parameters, such as, e.g., the number of sample pixels is also limited. The training fields must allow the selection of the required number of training pixels. The total number of assigned sample pixels is divided into the number of classes and from each class the algorithm tries to select this number of pixels if possible. It may be problematic to put the number of sample pixels exceeding the total number of pixels in the class. Therefore, the default settings in SNAP are 5000 sample pixels (due to the sampling method of the training set) and 10 trees (due to the computation time). As part of the research, many different variants of classification with different settings were carried out, especially since the default values were insufficient. The commonly used a grid of parameter method was used to select the best hyper parameters of RF. The GridSearchCV class implemented in scikit-learn was applied. The investigated parameter grid included:
number of trees in the forest (n_estimators), the range of values: 30, 50, 100, 150, 500,
the maximum depth of tree (max_depth), the range of values: None, 10, 30, 60, 100,
the minimum number of samples required to split a node, (min_samples_split), the range of values: 2, 4, 6, 8, 10,
the minimum number of samples that must be in a leaf node, (min_samples_leaf), the range of values: 1, 2, 4, 6, 8,
the number of features to consider when split a node, (max_features), the range of values: None, auto,
bootstrap, the range of values: True, False.
Five k-fold (CV = 5) cross validation was aplied. Three metrics were used to assess the quality of the model: accuracy, mean value of recall (balanced_accuracy), a weighted average of the precision and recall (f1_weighted). In all hyper parameter estimation simulations, all 3 metrics assessed the parameters at the same level. Usually, the set of hyper parameters is selected that best suits the computational capabilities. By increasing the number of trees, you can achieve better results, but it is very limited by the size of the available RAM. Moreover, by increasing the number of trees above 100, the differences in accuracy for the considered problems are negligible, in the order of tenths of a percent (mean_accuracy: 0.8150, 0.8164 i 0.8177, respectively, 50, 100 i 500 trees).
Eventually, a possible large number of sample pixels and trees were assumed:
There are no such limitations generally in Python, and the whole training set (2190 parcels, 1,412,092 pixels) was possible to use for training. We tested different settings with the k-fold cross-validations and decided to apply the following settings:
2.3. Accuracy Analysis
Based on a literature review, main metrics were selected for the analysis:
, additionally
and two metrics from ML, usually not used in remote sensing: accuracy and specificity (
Table 4, please notice difference between
and accuracy). The meaning of these metrics can be illustrated on any full cross matrix, for example (
Table 5) taken from the publication from 2021 [
37] (it is
Table 4—transposed for our purposes).
Accuracy of the classification results is estimated on the basis of the confusion matrix: a full confusion matrix, typically implemented in remote sensing or from a binary confusion matrix used in machine learning.
Full confusion matrix represents the complete error matrix (
Table 5), i.e., the combination of all classes with each other using the peer-to-peer method, which includes all commission and omission errors for each class.
Binary confusion matrix contains only cumulative information: number of samples correctly classified as a given class (TP true positives), correctly not classified as this class (TN true negatives), falsely classified as this class (FP false positives) and falsely not classified as this class (FN false negatives). One binary confusion matrix is assigned to one class (e.g., for C1
Table 6). In our case we therefore have 6 binary confusion matrices (
Table 7) which are flattened and each matrix written on one row.
From the complete confusion matrix, the binary confusion matrices can be computed, but the reverse operation is impossible.
From both matrices it is possible to compute all metrics and their values are of course the same. However, it should be also noted that there is more information in the full confusion matrix than in the binary confusion matrix. In the case of more than 4 classes, the size of full confusion matrix is larger then size of binary confusion matrix, because the binary confusion matrix for one class is always 2 × 2 (
Table 6), after flattering one row in (
Table 7). The main advantage of the full confusion matrix is the possibility of exhausting analysis of testing samples and errors (so-called omission and commission errors).
More important, however, is the distinction between and the mean value of accuracy (). The sum of the number of correctly classified samples is used in the numerator to calculate . In the classification of many classes it is the sum of TP. For one class, we are dealing with samples correctly classified as a given class and correctly not classified to it, i.e., on the diagonal of the binary confusion matrix there is the sum of TP and TN. So, for class C1, and are equal to (87 + 220)/(87 + 3 + 10 + 220) = 0.9594.
Analyzing individual classes separately, the values correspond to . While the metrics for all classes is 0.9000 and is not the mean of the classes, which is 0.9667. In this case, the difference is ca. 7% but one should also take into account the relatively small number of TN, because as can be seen from the formula , the more TN the greater the accuracy ().
In our research, the accuracy analysis was performed adequately to the classification design. In the case of learning on a selected number of samples, an accuracy analysis was performed 2 times, on the basis of the validation set and of the test set. In the case of using the entire training set in learning, the accuracy analysis was performed only on the test set.
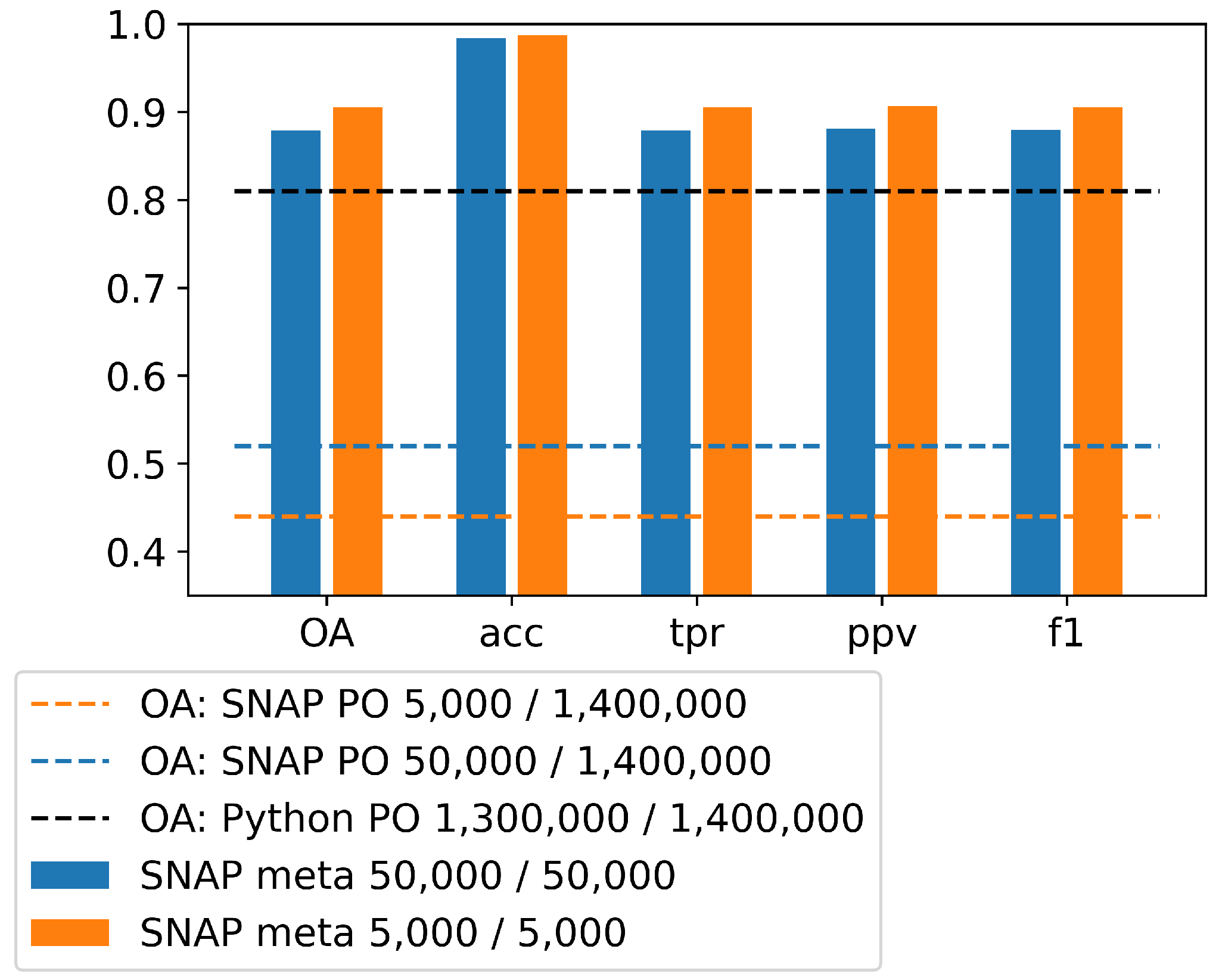
Binary confusion matrices have been calculated for validation simultaneously with the classification in SNAP on the basis of randomly selected pixels from the training set (results are available in the text file, default name: “classifier.txt”, SNAP_META). SNAP does not provide accuracy analysis on the independent test set, therefore we made the analysis externally in our own Python scripts.
Accuracy analysis was performed on the test set in the pixel and object-oriented approach, using our own scripts, Python (PP) and Python (PO), respectively. In the object-oriented approach, 2386 samples equal to the number of all parcels in test set were analyzed, corresponding to 1,412,092 pixels (10 m pixel size), which is the number of samples analyzed in the pixel approach.
In Python (PP), test polygons were converted to raster form and cross with classification result for the computation of confusion matrix. In Python (PO), using a zonal statistics algorithm, the modal value of the classification score located within each polygon was calculated. This provided the basis for calculating the confusion matrix. The full confusion matrix, binary confusion matrices and accuracy metrics were calculated for each classification results.
4. Discussion
In the discussion, we refer to the aim of the research, i.e., the analysis of the results of image classification for an effective and reliable screening method to control farmers’ declarations. In temperate climates, an efficient method that is applicable to a large area must be based on as few images as possible, preferably one. The method implemented for the inspection of farmers’ declarations must be reliable as it may result in financial penalties for the farmer. The reliability of the method can be determined on the basis of a properly performed accuracy analysis. In this case, we are not interested in the accuracy of fitting the hyperparameters of the classification method. Accuracy of validation does not determine the actual accuracy of the product, which is the classification result. The phenomenon of accuracy overestimation using only validation data set, emphasized in the literature review [
25], is confirmed in other literature, e.g., [
7,
20], and also in our research.
The accuracy analysis should be performed on the training set (if possible, e.g., in the SAM method), on the validation set and on the test set. In most methods, it is not possible to obtain accuracy on the training set, but only on the validation and test set. In many publications the accuracy of validation (
) is reported, which in almost all cases is above 80% (e.g.,
[
7],
[
20],
[
13],
[
22],
[
23],
[
39], Dynamic Time Warping algorithm, NDVI time series classification = 72–89%, multi-band classification = 76–88% [
40]). In some cases, the accuracy for test data is also delivered:
[
7],
[
20], which means in the case of 13.5% [
7] less value than the accuracy of the validation and in the case of 10.02% [
20] lower.
In our research, the accuracy of the validation was also over 90%, and the accuracy of the test data was approx. 45%. We used training set composed of 2190 parcels/1,412,092 pixels, test set of 2386 parcels/1,412,092 pixels and the number of samples for learning was 5000 and 50,000. Since the classification accuracy based on selected sample delivered not satisfactory results, the entire training set was used for training and the accuracy on the test data increased to 80% (all accuracy metrics: ).
It is difficult to compare our experiment with the research design mentioned above. Note the number of training and test samples: 2005/341 points [
7] and 2281/1239 pixels [
20].
When analyzing the credibility of the method, the issue of selecting accuracy metrics cannot be ignored. The most frequently reported accuracy metric is , regardless of whether it is traditional approach or ML. Confusion may arise when the mean accuracy () value is given in the ML instead of the correct value. In our research, we obtained an overestimation of up to 45%. It is impossible to refer to the literature on this topic because to our knowledge this problem has not been discussed so far.
Many studies exist regarding the application of remote sensing for crop recognition. They are typically based on time series of optical images, radar images, or both simultaneously. The authors do not always provide sufficient information about the accuracy analysis and they use different metrics. Nevertheless, several examples can be given in this area.
Integration of multi-temporal S-1 and S-2 images resulted in higher classification accuracy compared to classification of S-2 and S-1 data alone [
41] (max. kappa for two crops: wheat-0.82 and rapeseed-0.92). Using only S-2 data images it was obtained max. kappa = 0.75 and 0.86 for wheat and rapeseed, respectively. Using only S-1 data images obtained max. kappa = 0.61 and 0.64 for wheat and rapeseed, respectively.
The kappa coefficient was also used in the evaluation of in-season mapping of irrigated crops using Landsat 8, Sentinel-1 time series and Shuttle Radar Topography Mission (SRTM) [
42]. Reported classification accuracy using the RF method for integrated data was: kappa = 0.89 compared to kappa = 0.84 for each type of data separately.
In other studies, simultaneous classification of S-1, S-2, Landsat-8 data was applied to crops: wheat, rapeseed, and corn recognition [
43]. Classification accuracy performed with the Classification and Regression Trees (CART) algorithm in Google Earth Engine (GEE), estimated in this case by metric: overall accuracy, was
.
The issue of the effect of different time intervals on early season crop mapping (rice, corn and soyabean) has been the subject of other studies [
44]. Based on the analysis of time profiles of different features computed from satellite images, optimal classification sets were selected. The study resulted in maximum accuracy of
and slightly lower 91–92% in specific periods of plant phenology.
Wheat area mapping and phenology detection using S-1 and S-2 data has been the subject of other studies by [
45]. Classifications were performed using the RF method in GEE obtaining accuracy for integrated data 88.31% (accuracy drops to 87.19% and 79.16% while using only NDVI or VV-VH, respectively).
Time series of various features from S-2 were analysed in the context of three crops recognition rice, corn and soyabean [
46]. The research included 126 features from Sentinel-2A images: spectral reflectance of 12 bands, 96 texture parameters, 7 vegetation indices, and 11 phenological parameters. The results of the study indicated 13 features as optimal. Overall accuracies obtained by different methods were, respectively, SVM 98.98%, RF 98.84%, maximum likelihood classifier (MCL) 88.96%.
In conclusion of this brief review, it is important to note the dissimilarity of the metrics when comparing validation accuracy with accuracy based on a test set. In the following discussion, the accuracy data cited from the literature and from our study applies only to metrics computed on the training-independent test data set.
Ultimately, in the context of the literature, we would like to discuss the results of our research on the accuracy of single image classification for crop recognition. Recently, in 2020 reported [
8] that it was possible to obtain high accuracy of crop classification using one Sentinel-2 image registered in the appropriate plant phenological phase. Ref. [
8] presented the results of the Sentinel-2 time series classification performed on the test area in South Africa, Western Cape Province, for 5 crops: canola, lucerne, pasture (grass), wheat, fallow. The most important conclusion from the research is that it is possible to obtain high accuracy of crop classification (
by SVM Supported Vector Machine method) using one Sentinel-2 image recorded approx. 8 weeks before harvest (comparing max. of
).
Some other researchers compare the results of classification of various combinations of time series with the results of classification of single images. This is particularly important in temperate climatic zones, where acquiring many cloudless images over large areas is problematic.
One example can be noticed in [
7] where the authors examined perennial crops using various combinations of multispectral Sentinel-1 and Sentinel-2 images. They obtained the maximum accuracy for the combination of ten images Sentinel-2 and ten Sentinel-1
, for comparison, the classification accuracy of the combination of ten images Sentinel-2 was
.
The influence of the classification of all Sentinel-2 channels was also tested in comparison to the classification of channels with a resolution of 10 m. A single optical image with four 10 m channels resulted in an accuracy of
, while the use of 10 channels improved the accuracy of
. In addition, these studies show one more conclusion that the NDVI time series classification gives worse results than the classification of the original images (which was also observed during our research [
17,
18].
Results of the research most similar to ours can be found in the paper [
16] (cited also in Introduction). The maximum accuracy of
was achieved for the combination: 6 × Sentinel-1 + 6 × Sentinel-2, much bigger then for the single Sentinel-2 image for which it was 39%.
In our case, the highest accuracy (81%) was obtained in RF classification using entire training set in object-oriented approach with accuracy estimation for Combination_3x, Combination_4VV, Combination_4VH (for comparison to pixel approach - 78%). It was also astonishing that there was no large decrease in accuracy for a single image S2_20180526 (79% in Python PO and 73% in Python PP). We obtained better accuracy for one image then [
16] (one Sentinel-1: 47%, one Sentinel-2: 39%), but comparable with [
7,
8]. The highest accuracy of 79% was for a single image registered on 26 May 2018, while the classification of the image of 20 July—just before the harvest was slightly less accurate.
The high accuracies obtained in crop recognition using time series only radar images (
[
47],
[
48],
[
15]) provide valuable inspiration for future research in our test area.