Comparison between Three Registration Methods in the Case of Non-Georeferenced Close Range of Multispectral Images
Abstract
:1. Introduction
2. Materials and Methods
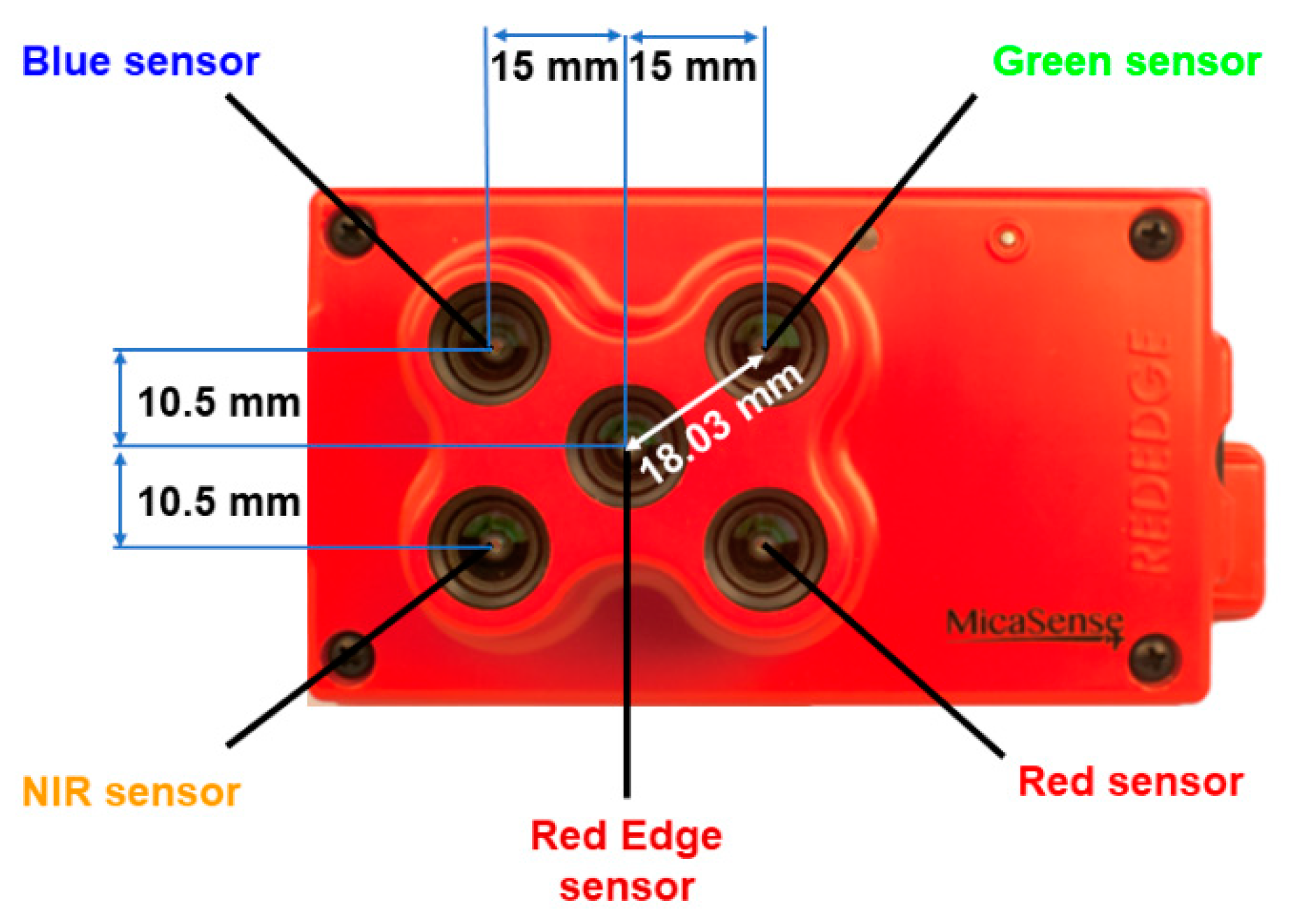
2.1. Image Acquisition
- D = camera displacement to image a new section of the plants row(m);
- P = overlap between images (10%);
- H = camera position height (m);
- vFOV = vertical field of view of the MicaSense® RedEdge camera (0.62).
2.2. Image Processing
2.2.1. SURF Features
Point Detection
Feature Extraction
- = the sum of horizontal the wavelet responses values for all of the 4*4 sub-regions;
- = the sum of absolute horizontal wavelet responses values for all of the 4*4 sub-regions;
- = the sum of the vertical wavelet responses values for all of the 4*4 sub-regions;
- = the sum of the absolute vertical wavelet responses for all of the 4*4 sub-regions.
Matching Features
2.2.2. Geometric Transformations
2.2.3. RMSE Computation
3. Results
3.1. SURF Features
3.2. Geometric Transformation
3.3. RMSE
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Statistics Canada. 2017a. Available online: https://www.agr.gc.ca/eng/horticulture/horticulture-sector-reports/statistical-overview-of-the-canadian-greenhouse-vegetable-industry-2017/?id=1549048060760 (accessed on 26 November 2020).
- Khater, M.; De La Escosura-Muñiz, A.; Merkoçi, A. Biosensors for plant pathogen detection. Biosens. Bioelectron. 2017, 93, 72–86. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hafez, Y.M.; Attia, K.A.; Kamel, S.M.; Alamery, S.F.; El-Gendy, S.; Al-Doss, A.A.; Mehiar, F.; Ghazy, A.I.; Ibrahim, E.I.; Abdelaal, K.A.A. Bacillus subtilis as a bioagent combined with nano molecules can control powdery mildew disease through histochemical and physiobiochemical changes in cucumber plants. Physiol. Mol. Plant Pathol. 2020, 111, 101489. [Google Scholar] [CrossRef]
- Spanu, P.D.; Panstruga, R. Editorial: Biotrophic Plant-Microbe Interactions. Front. Plant Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nishizawa, Y.; Mochizuki, S.; Yokotani, N.; Nishimura, T.; Minami, E. Molecular and cellular analysis of the biotrophic interaction between rice and Magnaporthe oryzae–Exploring situations in which the blast fungus controls the infection. Physiol. Mol. Plant Pathol. 2016, 95, 70–76. [Google Scholar] [CrossRef]
- Eskandari, S.; Sharifnabi, B. The modifications of cell wall composition and water status of cucumber leaves induced by powdery mildew and manganese nutrition. Plant Physiol. Biochem. 2019, 145, 132–141. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Atole, R.R.; Park, D. A multiclass deep convolutional neural network classifier for detection of common rice plants anomalies. Int. J. Adv. Comput. Sci. Appl. 2018, 9, 67–70. [Google Scholar]
- Özgüven, M.M.; Adem, K. Automatic detection and classification of leaf spot disease in sugar beet using deep learning algorithms. Phys. A Stat. Mech. Appl. 2019, 535, 122537. [Google Scholar] [CrossRef]
- Yang, C. Remote Sensing and Precision Agriculture Technologies for Crop Disease Detection and Management with a Practical Application Example. Engineering 2020, 6, 528–532. [Google Scholar] [CrossRef]
- Jung, J.; Maeda, M.; Chang, A.; Bhandari, M.; Ashapure, A.; Landivar, J. The potential of remote sensing and artificial intelligence as tools to improve the resilience of agriculture production systems. Curr. Opin. Biotechnol. 2021, 70, 15–22. [Google Scholar] [CrossRef]
- Bai, X.; Li, X.; Fu, Z.; Lv, X.; Zhang, L. A fuzzy clustering segmentation method based on neighborhood grayscale information for defining cucumber leaf spot disease images. Comput. Electron. Agric. 2017, 136, 157–165. [Google Scholar] [CrossRef]
- Ma, J.; Du, K.; Zhang, L.; Zheng, F.; Chu, J.; Sun, Z. A segmentation method for greenhouse vegetable foliar disease spots images using color information and region growing. Comput. Electron. Agric. 2017, 142, 110–117. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, X.; You, Z.; Zhang, L. Leaf image based cucumber disease recognition using sparse representation classification. Comput. Electron. Agric. 2017, 134, 135–141. [Google Scholar] [CrossRef]
- Zhang, S.; Zhang, S.; Zhang, C.; Wang, X.; Shi, Y. Cucumber leaf disease identification with global pooling dilated convolutional neural network. Comput. Electron. Agric. 2019, 162, 422–430. [Google Scholar] [CrossRef]
- Kuppala, K.; Sandhya, B.; Barige, T.R. An overview of deep learning methods for image registration with focus on feature-based approaches. Int. J. Image Data Fusion 2020, 11, 113–135. [Google Scholar] [CrossRef]
- Dawn, S.; Saxena, V.; Sharma, B. Remote sensing image registration techniques: A survey. In Proceedings of the Image and Signal Processing, 4th International Conference, ICISP, Trois-Rivières, QC, Canada, 30 June–2 July 2010; pp. 103–112. [Google Scholar]
- Goshtasby, A.A. Image Registration Principles Tools and Methods, 1st ed.; Springer: London, UK, 2012; pp. 7–31. [Google Scholar]
- Bay, H.; Tuytelaars, T.; Van Gool, L. SURF: Speeded Up Robust Features. In Proceedings of the 9th European Conference on Computer Vision (ECCV 2006), Graz, Austria, 7–13 May 2006; pp. 404–417. [Google Scholar]
- Bay, H.; Ess, A.; Tuytelaars, T.; Van Gool, L. Speeded-Up Robust Features (SURF). Comput. Vis. Image Underst. 2008, 110, 346–359. [Google Scholar] [CrossRef]
- Tareen, S.A.K.; Saleem, Z. A comparative analysis of SIFT, SURF, KAZE, AKAZE, ORB, and BRISK. In Proceedings of the Inter-national Conference on Computing, Mathematics and Engineering Technologies (iCoMET 2018), Sukkur, Pakistan, 3–4 March 2018; pp. 1–10. [Google Scholar]
- Lowe, D.G. Distinctive Image Features from Scale-Invariant Keypoints. Int. J. Comput. Vis. 2004, 60, 91–110. [Google Scholar] [CrossRef]
- Hassaballah, M.; Alshazly, H.; Ali, A.A. Analysis and Evaluation of Keypoint Descriptors for Image Matching. In Recent Advances in Computer Vision, Studies in Computational Intelligence; Hassaballah, M., Hosny, K., Eds.; Springer: Cham, Switzerland, 2019; Volume 854, pp. 113–140. [Google Scholar]
- Torr, P.H.S.; Zisserman, A. MLESAC: A new robust estimator with application to estimating image geometry. Comput. Vis. Image Underst. 2000, 78, 138–156. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; White, J.M.; Summer, R.M. Automated detection of blob structures by hessian analysis and object scale. In Proceedings of the 2010 IEEE 17th International Conference on Image Processing, Hong Kong, China, 26–29 September 2010; pp. 841–844. [Google Scholar]
- Neubeck, A.; Van Gool, L. Efficient Non-Maximum Suppression. In Proceedings of the 18th International Conference on Pattern Recognition (ICPR’06), Hong Kong, China, 20–24 August 2006; pp. 850–855. [Google Scholar]
- Brown, M.; Lowe, D. Invariant Features from Interest Point Groups. In Proceedings of the 13th British Machine Vision Confer-ence (BMVC 2002), Cardiff, UK, 2–5 September 2002; pp. 25.1–26.10. [Google Scholar]
- Hassaballah, M.; Ali, A.A.; Alshazly, H. Image features detection, description, and matching. In Image Feature Detectors and Descriptors: Foundations and Applications; Awad, A., Hassaballah, M., Eds.; Springer: Cham, Switzerland, 2016; Volume 630, pp. 11–45. [Google Scholar]
- Dai, X.; Khorram, S. A feature-based image registration algorithm using improved chain-code representation combined with Invariant Moments. IEEE Trans. Geosci. Remote Sens. 1999, 37, 2351–2362. [Google Scholar] [CrossRef] [Green Version]
- Fischler, M.A.; Bolles, R.C. Random sample consensus: A paradigm for model fitting with applications to image analysis and automated cartography. Commun. ACM 1981, 24, 381–395. [Google Scholar] [CrossRef]
- Szeliski, R. Image alignment and stitching: A tutorial. Found. Trends Comput. Graph. Vis. 2006, 2, 1–104. [Google Scholar] [CrossRef]
- Zitová, B.; Flusser, J. Image registration methods: A survey. Image Vision. Comput. 2003, 21, 977–1000. [Google Scholar] [CrossRef] [Green Version]
- Goshtasby, A.A. Piecewise linear mapping functions for image registration. Pattern Recognit. 1986, 19, 459–466. [Google Scholar] [CrossRef]
- Chai, T.; Draxler, R. Root mean square error (RMSE) or mean absolute error (MAE)?—Arguments against avoiding RMSE in the literature. Geosci. Model Dev. 2014, 7, 1247–1250. [Google Scholar] [CrossRef] [Green Version]
- Jin, H.; Fan, C.; Li, Y.; Xu, L. A novel coarse-to-fine method for registration of multispectral images. Infrared Phys. Technol. 2016, 77, 219–225. [Google Scholar] [CrossRef]
- Hong, G.; Zhang, Y. Wavelet-based range image registration technique for high-resolution remote sensing images. Comput. Geosci. 2008, 34, 1708–1720. [Google Scholar] [CrossRef]
- Zheng, J.; Xu, Q.; Zhai, B.; Wang, Y. Accurate hyperspectral and infrared satellite image registration method using structured topological constraints. Infrared Phys. Technol. 2020, 104, 103122. [Google Scholar] [CrossRef]
- Yasir, R.; Eramian, M.; Stavness, I.; Shirtliffe, S.; Duddu, H. Data-driven multispectral image registration. In Proceedings of the 2018 15th Conference on Computer and Robot Vision (CRV), Toronto, ON, Canada, 8–10 May 2018; pp. 230–237. [Google Scholar]
- Hassapour, M.; Javan, F.D.; Azizi, A. Band to band registration of multi-spectral aerial imagery-relief displacement and miss-registration error. In Proceedings of the International Archives Photogrammetry, Remote Sensing, and Spatial Information Sciences, GeoSpatial Conference, Karaj, Iran, 12–14 October 2019; Volume 42, pp. 467–474. [Google Scholar]
- Kerkech, M.; Hafiane, A.; Canals, R. Vine disease detection in UAV multispectral images using optimized image registration and deep learning segmentation approach. Comput. Electron. Agr. 2020, 174, 105446. [Google Scholar] [CrossRef]
- Haddadi, A.; Leblon, B. Developing a UAV-Based Camera for Precision Agriculture, Final Report; Mitacs # IT07423 Grant; University of New Brunswick: Fredericton, NB, Canada, 2018; 106p. [Google Scholar]
- Harris, C.; Stephens, M. A combined corner and edge detector. In Proceedings of the 4th Alvey Vision Conference, Manchester, UK, 31 August–2 September 1988; pp. 147–151. [Google Scholar]
- Schowengerdt, R.A. Remote Sensing: Models and Methods for Image Processing, 3rd ed.; Academic Press: Burlington, MA, USA, 2007; pp. 285–354. [Google Scholar]
- Wang, X.; Yang, W.; Wheaton, A.; Cooley, N.; Moran, B. Efficient registration of optical and IR images for automatic plant water stress assessment. Comput. Electron. Agric. 2010, 74, 230–237. [Google Scholar] [CrossRef]












| Band # | Spectral Region | Central Wavelength (nm) | Bandwidth (nm) |
|---|---|---|---|
| 1 | Blue | 475 | 20 |
| 2 | Green | 560 | 20 |
| 3 | Red | 668 | 10 |
| 4 | NIR | 840 | 40 |
| 5 | Red-edge | 717 | 10 |
| Parameter | Default Value | Applied Value |
|---|---|---|
| MetricThreshold | 1000 | 1 |
| NumOctaves | 3 | 4 |
| NumScaleLevels | 4 | 6 |
| Parameter | Band | Default Value | Applied Value |
|---|---|---|---|
| MatchThreshold | All bands | 10 | 50 |
| MaxRatio | Blue–Red–NIR | 0.60 | 0.90 |
| Green | 0.60 | 0.75 | |
| Unique | All bands | false | true |
| Parameter | Projection | Default Value | Applied Value |
|---|---|---|---|
| MaxNumTrials | Affine, similarity, and projective | 1000 | 10,000 |
| MaxDistance | Affine and similarity | 1.5 | 1.5 |
| Projective | 1.5 | 3.0 |
| Band | Minimum | Maximum | Mean | Standard Deviation | Standard Error |
|---|---|---|---|---|---|
| Blue | 1250 | 1599 | 1415.80 | 72.35 | 10.78 |
| Green | 1125 | 2089 | 1708.20 | 216.96 | 32.34 |
| Red | 1314 | 1670 | 1457.40 | 79.13 | 11.79 |
| NIR | 1383 | 1770 | 1579.80 | 96.38 | 14.36 |
| Geometric Transformation | Band | Min | Max | Mean | Standard Deviation | Standard Error | Mean Percentage (%) (1) |
|---|---|---|---|---|---|---|---|
| Similarity | Blue | 78 | 282 | 162.40 | 45.27 | 6.74 | 11.39 |
| Green | 318 | 1295 | 777.02 | 219 | 32.64 | 44.74 | |
| Red | 88 | 280 | 169.95 | 43.17 | 6.43 | 11.59 | |
| NIR | 124 | 413 | 210.26 | 54.95 | 8.19 | 13.22 | |
| Affine | Blue | 73 | 274 | 171.06 | 46.75 | 6.96 | 11.99 |
| Green | 347 | 1242 | 808.57 | 224.22 | 33.42 | 46.58 | |
| Red | 89 | 291 | 183.62 | 46.46 | 6.92 | 12.50 | |
| NIR | 130 | 367 | 225.95 | 57.77 | 8.61 | 14.21 | |
| Projective | Blue | 136 | 444 | 292.20 | 64.78 | 9.65 | 20.52 |
| Green | 622 | 1692 | 1273.50 | 245.95 | 36.66 | 73.97 | |
| Red | 165 | 457 | 320.84 | 67.05 | 9.99 | 21.89 | |
| NIR | 249 | 672 | 414.17 | 94.57 | 14.19 | 26.05 |
| Function | Geometric Transformation | Time (s) | Cumulative time (s) | Average Time Per Band (1) | ||
|---|---|---|---|---|---|---|
| detectSURFFeatures | 14.18 | 14.18 | 0.079 | |||
| extractSURFFeatures | 18.62 | 32.80 | 0.182 | |||
| matchFeatures | 114.77 | 147.57 | 0.820 | |||
| estimateGeometricTransform | Affine | 144.55 | 292.13 | 1.623 | ||
| Similarity | 16.81 | 164.38 | 0.913 | |||
| Projective | 224.77 | 372.35 | 2.069 | |||
| imwarp | Affine | 0.83 | 292.95 | 0.005 | ||
| Similarity | 0.75 | 165.13 | 0.004 | |||
| Projective | 1.87 | 374.22 | 0.010 | |||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, C.I.; Haddadi, A.; Leblon, B.; Wang, J.; Wang, K. Comparison between Three Registration Methods in the Case of Non-Georeferenced Close Range of Multispectral Images. Remote Sens. 2021, 13, 396. https://doi.org/10.3390/rs13030396
Fernández CI, Haddadi A, Leblon B, Wang J, Wang K. Comparison between Three Registration Methods in the Case of Non-Georeferenced Close Range of Multispectral Images. Remote Sensing. 2021; 13(3):396. https://doi.org/10.3390/rs13030396
Chicago/Turabian StyleFernández, Claudio Ignacio, Ata Haddadi, Brigitte Leblon, Jinfei Wang, and Keri Wang. 2021. "Comparison between Three Registration Methods in the Case of Non-Georeferenced Close Range of Multispectral Images" Remote Sensing 13, no. 3: 396. https://doi.org/10.3390/rs13030396






