1. Introduction
Paddy rice is one of the world’s major food crops and it feeds about half of the global population. The spatial distribution of rice paddy is not only the basis for decision-making in agricultural production, such as crop management, but also the basic data for agricultural research and application, such as paddy rice yield estimation and planting structure adjustment and optimization. Therefore, accurate and effective mapping of rice paddy distribution is helpful to timely and effective monitoring of paddy rice agricultural information, which can improve food crop production and ensure food security. Recently, remote sensing technology has been widely used to map rice paddy distribution, and thus has become an important research issue in agricultural monitoring of paddy rice.
The methods using remote sensing to map rice paddy distribution can be generally divided into two main categories: methods based on knowledge of paddy rice phenology (phenological methods) and those only based on spectral learning. Phenological methods are based on the cyclic and seasonal growing patterns that crops show along their respective life cycle [
1]. Paddy rice has its unique phenological characteristics. For example, during the period of flooding and transplanting, the land surface of rice paddy is covered with the mixture of water, paddy rice, and soil, while other crops and dry land are usually not covered with water. Phenological methods use these characteristics to differentiate rice paddy from other crops. The phenological characteristics can usually be reflected through time series of remote sensing indices calculated from multi-temporal remote sensing images [
2,
3,
4,
5,
6]. There are two representative methods that have been widely used for this purpose.
The first is based on the reflectance nature of the flooding and transplanting dates [
2]. The rice paddy is mainly covered with water during this time compared with non-rice paddy areas (areas whose land cover types are not rice paddy). This method uses the Normalized Difference Water Index (NDWI) and the Normalized Difference Vegetation Index (NDVI) from remote sensing images to depict the differences between rice paddies and non-rice paddy areas. It is believed that during the flooding and rice transplanting period, the NDWI value will come close to or even slightly bigger than the value of NDVI, compared with non-rice paddy areas. In other periods, the NDWI will be smaller. The method uses a time-series of remote sensing images to analyze the phenological changes of NDWI and NDVI. These changes will be used to map rice paddy distribution. However, the success of this method can be impacted by precipitation. The areas flooded after precipitation are likely to be falsely recognized as rice paddy. Thus, the resultant distribution map can be exaggerated if the remote sensing images are acquired right after precipitation during flooding and transplanting seasons.
The second method is based on the comprehensive consideration of vegetation phenology and surface water change [
3]. This method takes the characteristics of development periods aside from the rice transplanting period into consideration. They believe that from the tillering to heading period, the change of Land Surface Water Index (LSWI) is smaller while the change of two-band Enhanced Vegetation Index (EVI2) is larger, compared to other crops. Therefore, this method uses the ratio between the changing amplitude of LSWI and the difference between the EVI2 on the tillering date and the heading date to identify the rice paddy through a threshold. This method is not dependent on the short transplanting period, rather over the major periods of paddy rice life cycle. Therefore, it is more robust under the disturbance of precipitation. However, the values used for paddy rice identification constructed with phenological characteristics are simple ratios, which would not be applicable over complicated areas because it would change with different vegetation environments.
Spectral learning methods are purely based on the spectral signatures of rice paddy. The spectral learning methods can be grouped into three general types. The first type uses classic statistical methods to map rice paddy distribution [
7,
8,
9,
10,
11]. Classic statistical methods usually extract distinguishable spectral signature values of the rice paddies first, and then divide the feature space according to the principle of statistical decision-making to identify paddy rice. The classic statistical classification relies heavily on the statistical characteristics (signature) of spectral data from remote sensing images. When the spectral signature of rice paddies is not distinct (unique), this type of methods is less effective.
The second type utilizes the machine learning methods [
12,
13,
14,
15,
16,
17]. Currently, the machine learning methods commonly used in mapping rice paddy distribution include random forest, support vector machine, and neural networks. These methods first train classification models to learn the hidden features in the spectral data from a large amount of training samples and then the trained models (trained trees, trained forests, trained vector machines, or trained networks) are applied to map rice paddy distribution from remote sensing images. For machine learning methods, theoretically, models with more parameters are more capable of learning complex patterns. The key requirement of machine learning methods is the large set of training samples, which is often obtained through field sampling.
The third type is deep learning. Deep learning is a further development of the neural networks. It contains more hidden layers than regular neural networks, capable of representing the complex relationships in a progressive and layered manner. It has more nonlinear transformations and a better generalization capability, thus can extract deeper hidden features in spectral data. It has received wide attention in the field of mapping rice paddy with remote sensing and has achieved better performance than the machine learning techniques [
18,
19,
20]. However, deep learning, like other machine learning methods, requires a large number of training samples which are expensive and difficult to acquire.
As stated above, the phenological methods are simple but accuracy is low, the spectral learning methods, especially the deep learning methods, have strong feature learning ability and can improve the model performance. However, these methods rely heavily on a large number of training samples. It might be possible to combine the simplicity of the phenological methods with the learning ability of the deep learning methods to improve the efficiency and accuracy of rice paddy distribution mapping with remote sensing without field sampling.
Contributions
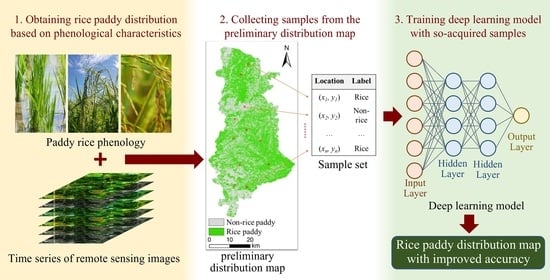
This paper aims to propose a method for mapping rice paddy distribution by coupling a phenological method with a deep learning method (referred to as pheno-deep method hereafter). The basic idea is to use the phenological method to extract initial samples from remote sensing images and then use these samples to train the deep learning model. Finally apply the trained model to map rice paddy distribution. The pheno-deep method is expected to achieve high mapping accuracy without field sampling efforts. This method also explores the feasibility of combining knowledge-based method and data-driven method for the spatial prediction of geographic variables in general.
The remainder of the article is divided into five sections.
Section 2 presents the pheno-deep method. The experiment design (including case study and evaluation) is illustrated in
Section 3.
Section 4 contains the results, and the discussions is presented in
Section 5.
Section 6 draws the conclusions.
2. The Pheno-Deep Method
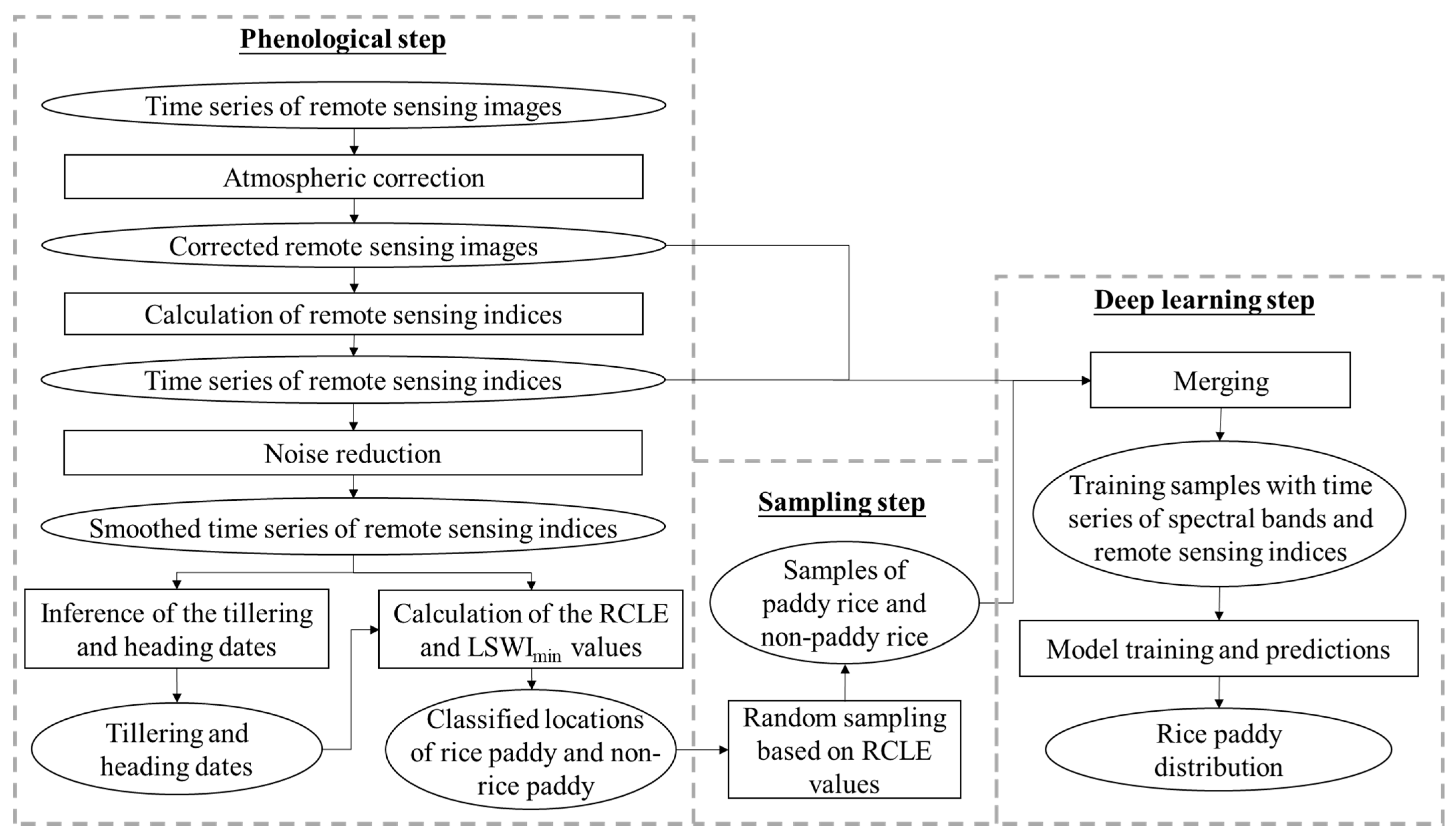
The pheno-deep method consists of three major steps and the overall workflow is illustrated in
Figure 1. First, the phenological method is used to determine phenological characteristics and classify the cell locations into rice paddies and non-rice paddy areas based these characteristics. Second, training samples are collected from the results of phenological method for deep learning model. Third, a deep learning model is constructed and trained using the samples so acquired for mapping rice paddy.
2.1. Classification of Rice Paddy Based on Phenological Method
The growing process of paddy rice can be divided into two major stages: the growth stage and the reproductive stage. The growth stage includes seedling, transplanting, and rice tillering. At this stage, the paddy rice grows rapidly, so is its vegetation coverage. Over this stage the surface is covered with mixture of water, paddy rice, and soil. The reproductive stage consists of heading and ripening. During this stage, the leaves age and turn yellow, its vegetation coverage begins to decline. These phenological characteristics can usually be reflected in time series of remote sensing images.
The phenological method proposed by Qiu et al. [
3] is used to capture these phenological characteristics and map the preliminary distribution of rice paddy areas. In this method, the two-band (the red and the infrared bands) enhanced vegetation index [
21] (EVI2) and the land surface water index [
22] (LSWI) to characterize the phenological characteristics of paddy rice. EVI2 is used for the detection for vegetation growth status and coverage. It is calculated with the reflectance of near-infrared (NIR) and red (R) bands of remote sensing images:
LSWI is sensitive to the surface moisture changes and is commonly used to detect changes in soil moisture and vegetation moisture content. It is calculated with near-infrared (NIR) and shortwave-infrared (SWIR) bands of the remote sensing images:
During the paddy rice growing process, the changes of water and vegetation in the fields are two key factors reflecting the changes of paddy rice. EVI2 can be used to effectively detect the changes of vegetation while LSWI can effectively detect the changes of water content. Therefore, the time series of these two remote sensing indices can capture well the changes of paddy rice in the whole growing process. Due to the noise resulting from varying atmospheric conditions and other factors, a noise reduction process is needed to smooth the time series of LSWI and EVI2 for further computation.
The determination of a cell location to be a rice paddy or not depends on the values of two indices. The first is the ratio of change amplitude of LSWI from tillering to heading to the difference between EVI2 at a prescribed tillering date and at a heading date. This index is referred to as RCLE (Ratio of Change amplitude of LSWI to EVI2) [
3], as shown in Equation (3):
The LSWI
max and LSWI
min are the maximum and minimum values of the LSWI between tillering and heading dates, while EVI2
heading and EVI2
tillering are the values of EVI2 at heading date and tillering date. The heading date for a given cell location was recognized as the date when EVI2 value reaches the primary maximum and the tillering date was set to be 40 days ahead of that [
3]. The second is the minimum LSWI value from tillering date to heading date. It is referred to as LSWI
min and is used to state the minimum value to be maintained for rice paddy. For a cell location to be rice paddy, the RCLE must be less than a prescribed threshold while LSWI
min must be greater than another prescribed threshold [
3].
2.2. Collection of Training Samples
After the rice paddy cell locations in the study area are identified using the phenological method discussed above, samples are collected based on the phenological results. The purpose of sampling is not just to obtain samples but also to increase the representativeness of the samples. Previous studies [
23,
24,
25,
26] have indicated that even for locations classified as a particular class, their respective representativeness of that class are different for different locations. Thus, it is expected that the locations classified as rice paddies and non-rice paddy areas based on the phenological method would exhibit the same nature. The different levels of representativeness can be indicated by their respective RCLE values. The smaller the RCLE the more representative to rice paddy while the larger the RCLE the more representative to non-rice paddy.
Therefore, intervals of RCLE values are designated to confine the area over which samples for rice paddy and non-rice paddy will be drawn, respectively, to increase the representativeness of the collected samples. Random sampling method is used to obtain the samples from areas whose RCLE values are within the selected ranges. Each sample consists of the tag of rice paddy or non-rice paddy and the time series of remote sensing indices and spectral bands from remote sensing images.
2.3. Deep Learning Model for Mapping Rice Paddy
The deep learning architecture used in the pheno-deep method is the Long Short-Term Memory [
27] (LSTM). The LSTM is a modification to recursive neural network (RNN). In RNN, the current status is dependent on all previous status, making it suitable for processing time series data. However, as the time series gets longer, the influence of initial status becomes faint as the time series progresses. The LSTM network is designed to solve this problem by dividing stored information into long-term memory and short-term memory. The structure of the basic LSTM unit is illustrated in
Figure 2. As is shown in
Figure 2, an LSTM unit consists of a cell state, a forget gate, an input gate, and an output gate. The cell state keeps track of the long-term memory, including that of the previous unit (
ct−1) and the output long-term memory of this unit (
ct). When the input data (
xt) enters the LSTM unit, it first goes through the forget gate together with short-term memory from the previous unit (
ht−1). The forget gate decides the extent to which the long-term information (
ct−1) remains. Then, the input data goes through the input gate where the extent to which the new information needs to be stored in long-term information is decided. Finally, the input data goes through the output gate which controls the output of new long-term memory (
ct) and new short-term memory (
ht). The
σf in
Figure 2 represents activation function, while the
tanh refers to hyperbolic tangent function. The divided store of long-term and short-term memory enables more delicate computing process in LSTM unit, which makes LSTM network suitable for long time series data analysis. In this paper the input data for deep learning model is the time series of remote sensing indices and spectral bands. Therefore, the LSTM networks serve as an excellent basis to construct the deep learning model for mapping rice paddy distribution.
The deep learning model implementing the LSTM networks consists of four main components: the input layer, the stacked LSTM layers, the dense layer, and the output layer. The samples consisting of labels and time series of remote sensing indices and spectral bands are used as the input data. The input layer transfers the time series data of the samples to the hidden LSTM layers. The stacked LSTM layers then stack multiple LSTM units into a deep architecture. This architecture improves the network ability of representing time series data. The final output of the stacked LSTM units goes into the dense layer. In this layer, the number of computational nodes is the same as the number of predicted classes (rice paddy and non-rice paddy area in this case). It bases on activation function to convert the output from the stacked LSTM layers into a probability distribution of the predicted classes. Finally, the output layer assigns the cell to a class which has the higher probability for this cell.