Unified Low-Rank Subspace Clustering with Dynamic Hypergraph for Hyperspectral Image
Abstract
:1. Introduction
- (1)
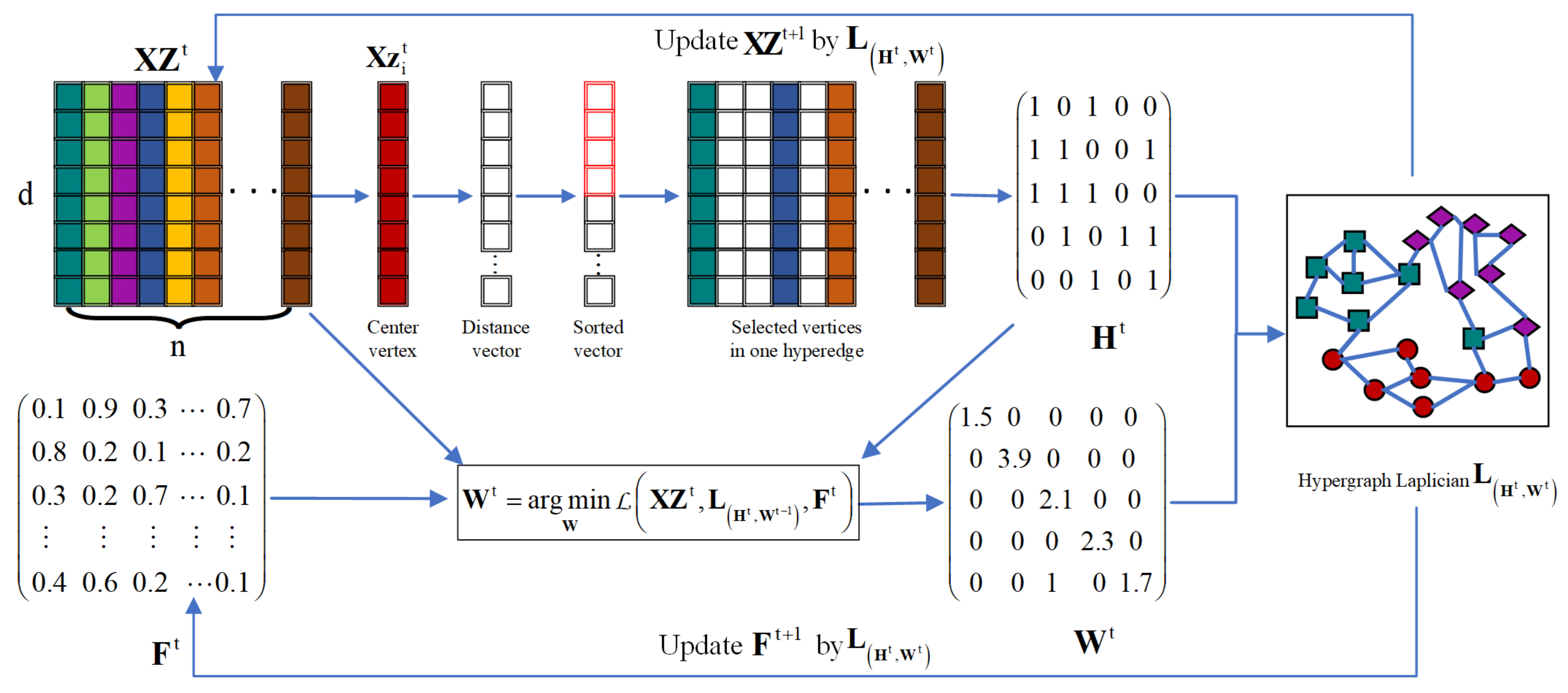
- Instead of pre-constructing a fixed hypergraph incidence and weight matrices, the hypergraph is adaptively learned from the low-rank subspace feature. The dynamically constructed hypergraph is well structured and theoretically suitable for clustering.
- (2)
- The proposed method simultaneously optimizes continuous labels, and discrete cluster labels by a rotation matrix without any relaxing information loss.
- (3)
- It jointly learns the similarity hypergraph from the learned low-rank subspace data and the discrete clustering labels by solving a unified optimization problem, in which the low-rank subspace feature and hypergraph are adaptively learned by considering the clustering performance and the continuous clustering labels just serve as intermediate products.
2. Related Work
2.1. Low-Rank Representation
2.2. Hypergraph
3. Materials and Methods
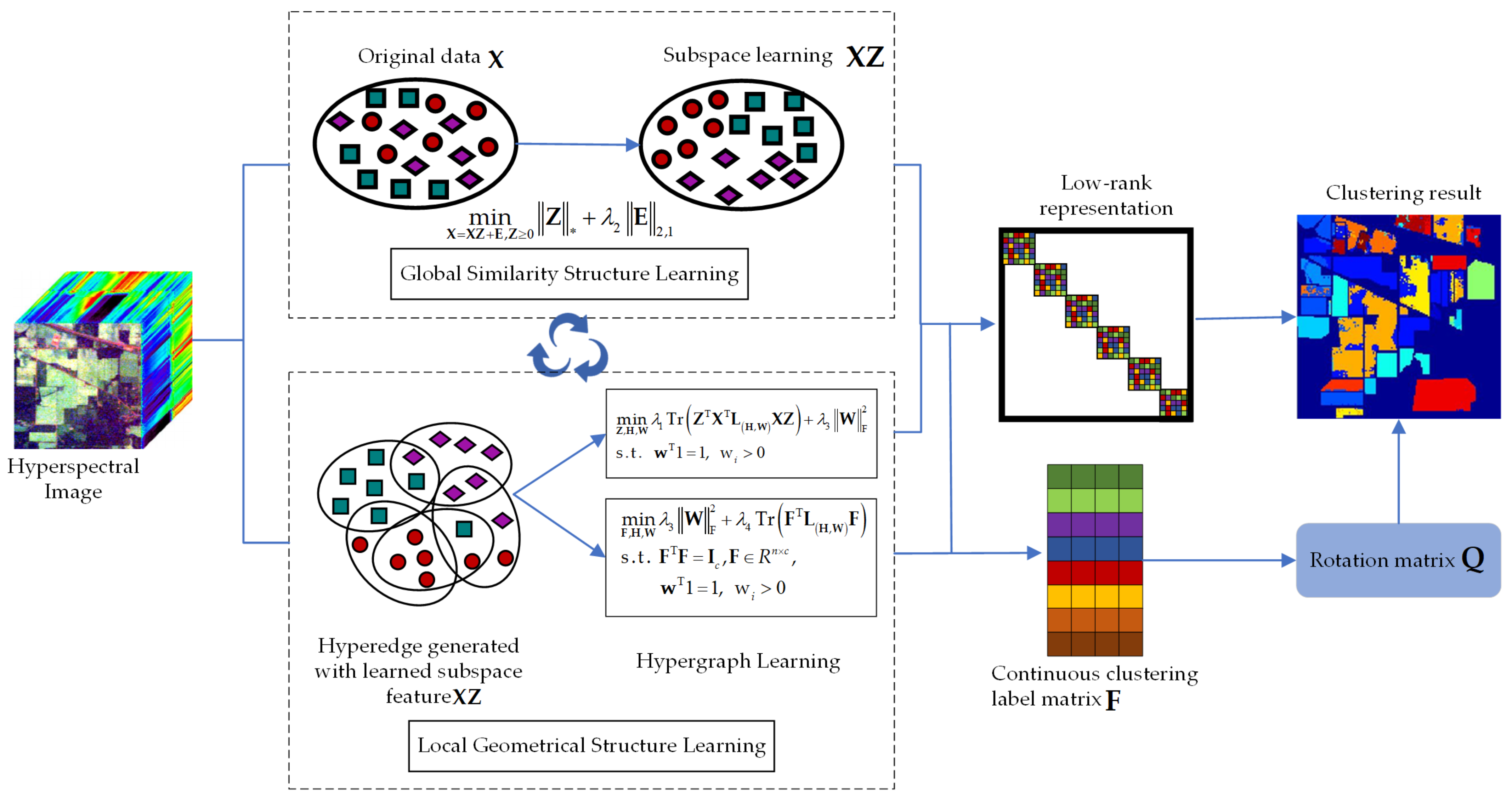
3.1. Dynamic Hypergraph-Based Low-Rank Subspace Clustering
3.2. Optimization Algorithm for Solving Problem (7)
| Algorithm 1 the DHLR algorithm for HSI clustering |
|
3.3. Unified Dynamic Hypergraph-Based Low-Rank Subspace Clustering
3.4. Optimization Algorithm for Solving Problem (25)
| Algorithm 2 the UDHLR algorithm for HSI clustering |
|
4. Results
4.1. Experimental Datasets
4.1.1. Indian pines
4.1.2. Selinas-A
4.1.3. Jasper Ridge
4.2. Experimental Setup
4.2.1. Evaluation Metrics
4.2.2. Compared Methods
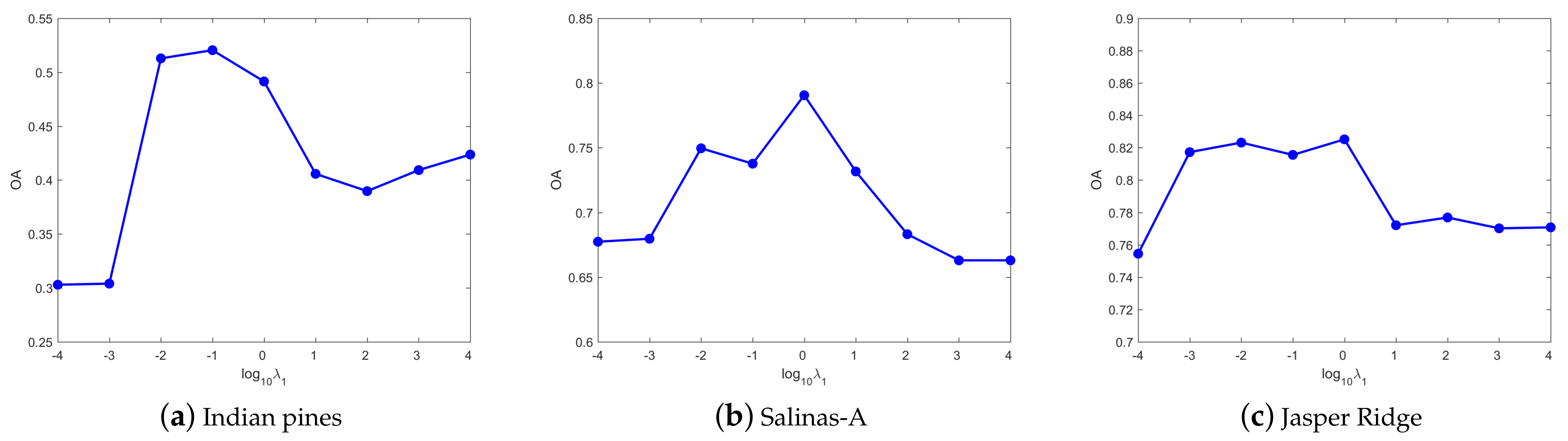
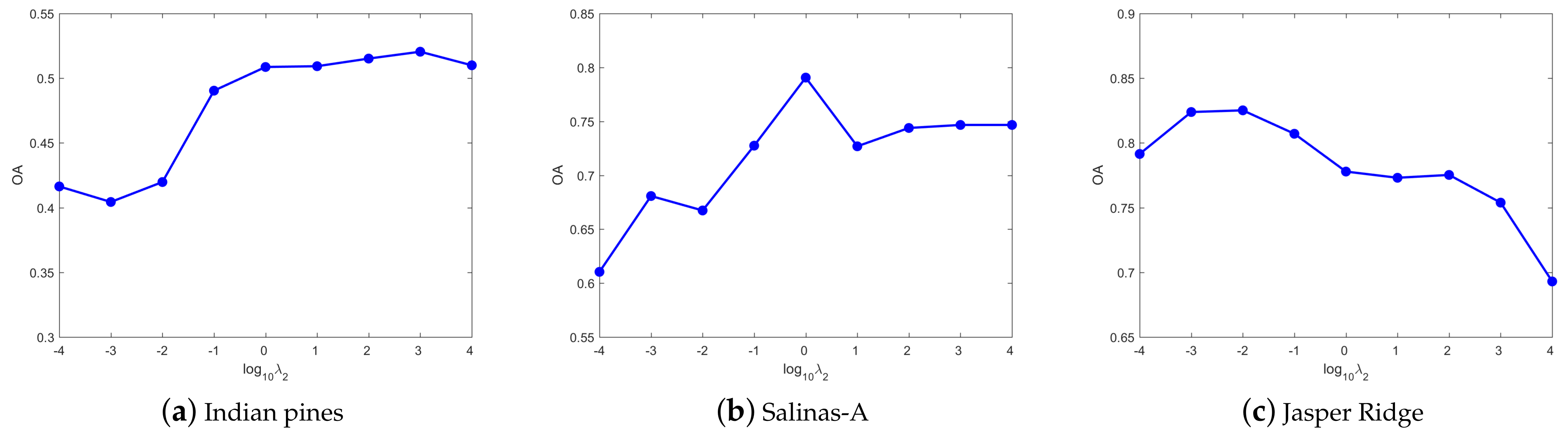
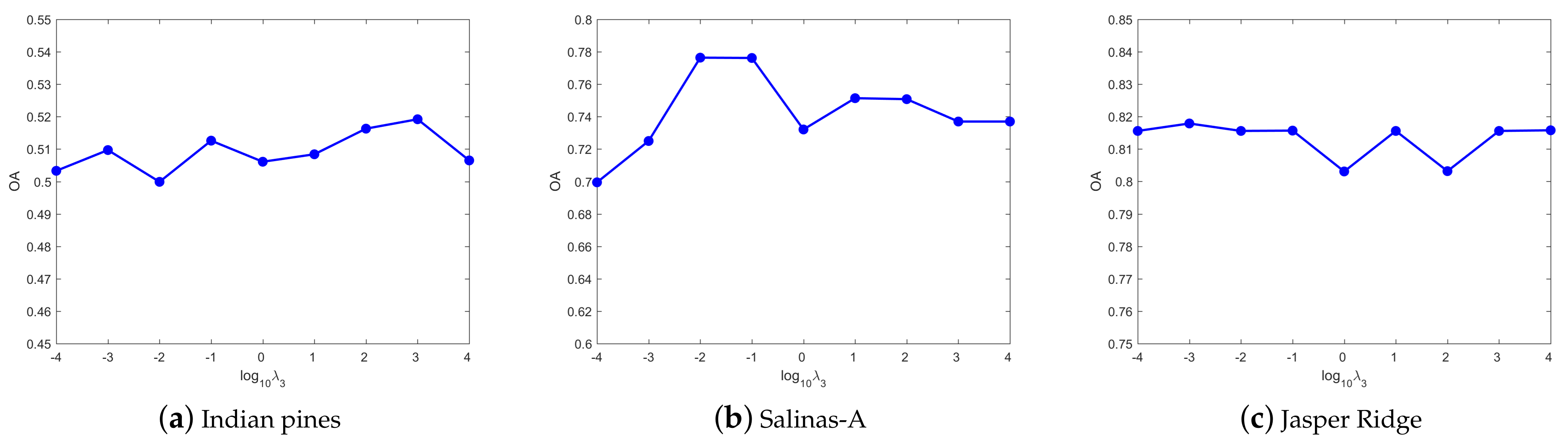
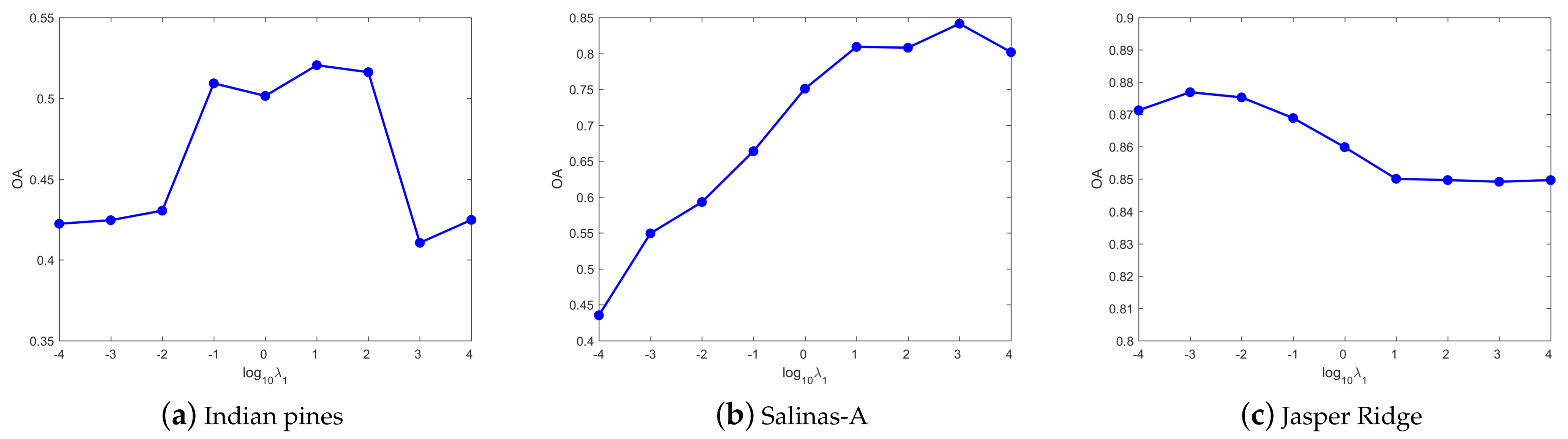
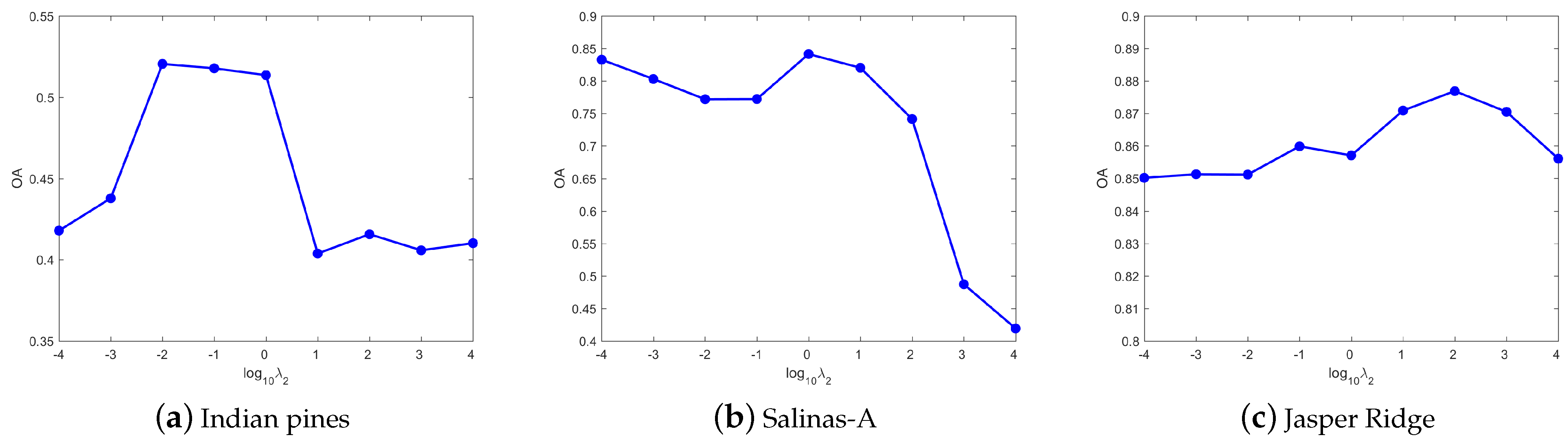
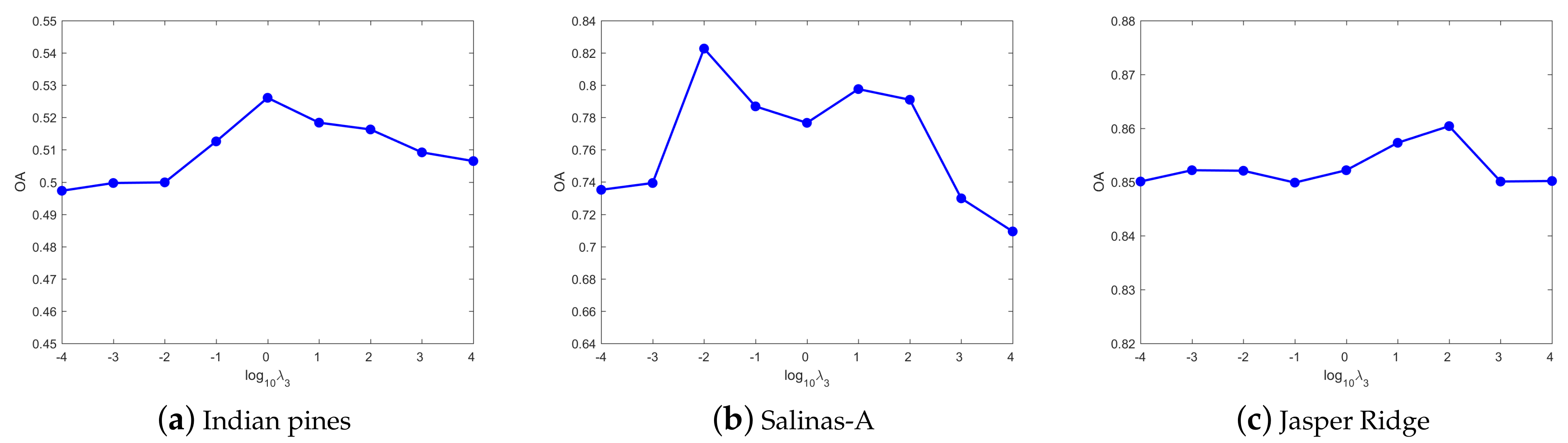
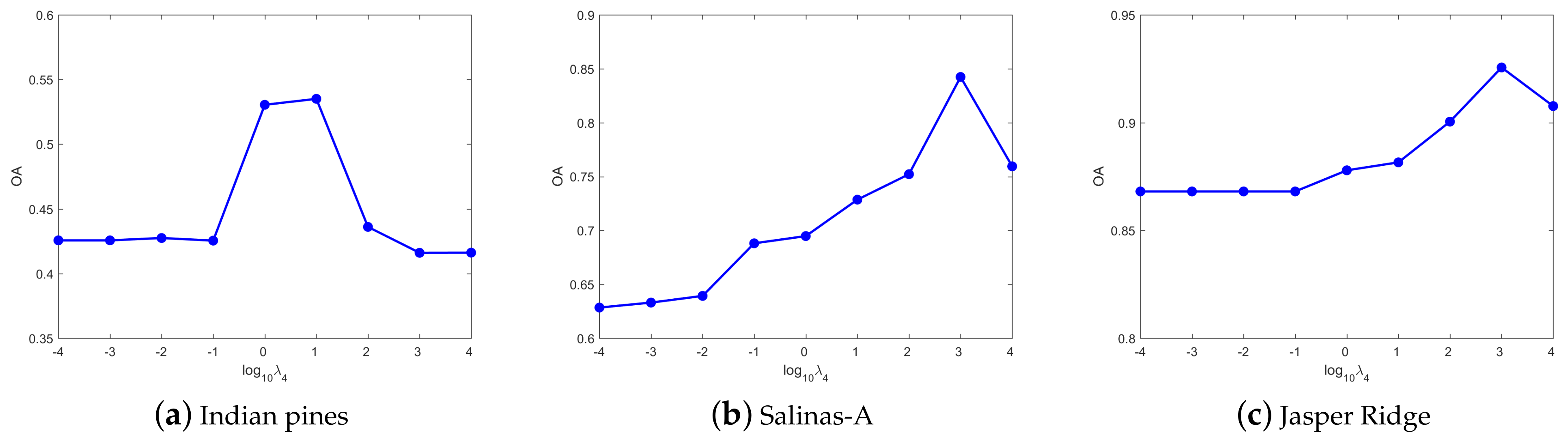
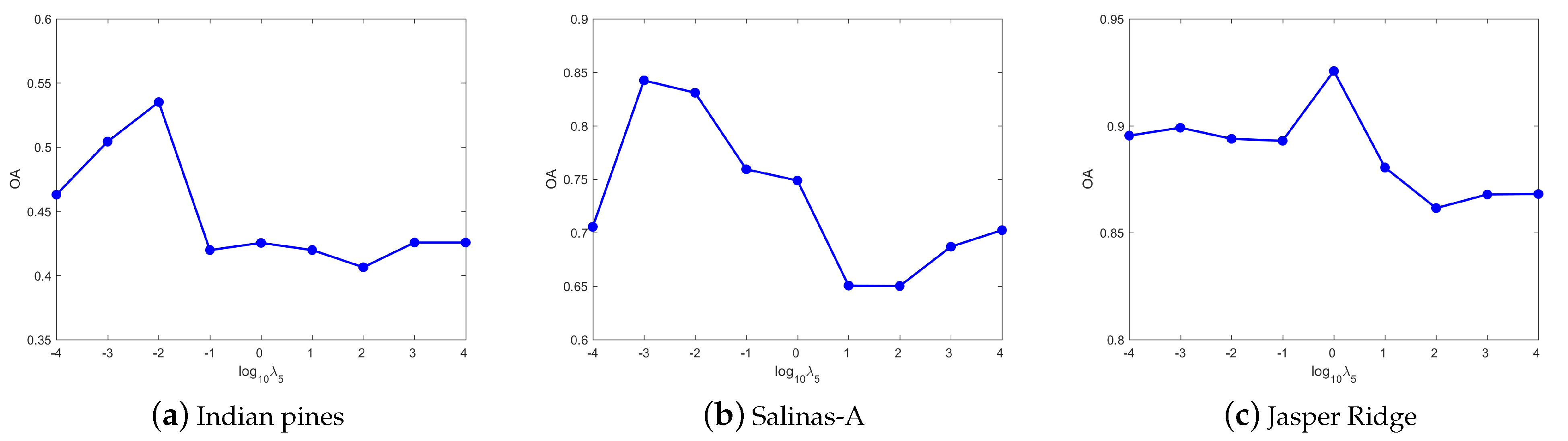
4.3. Parameters Tuning
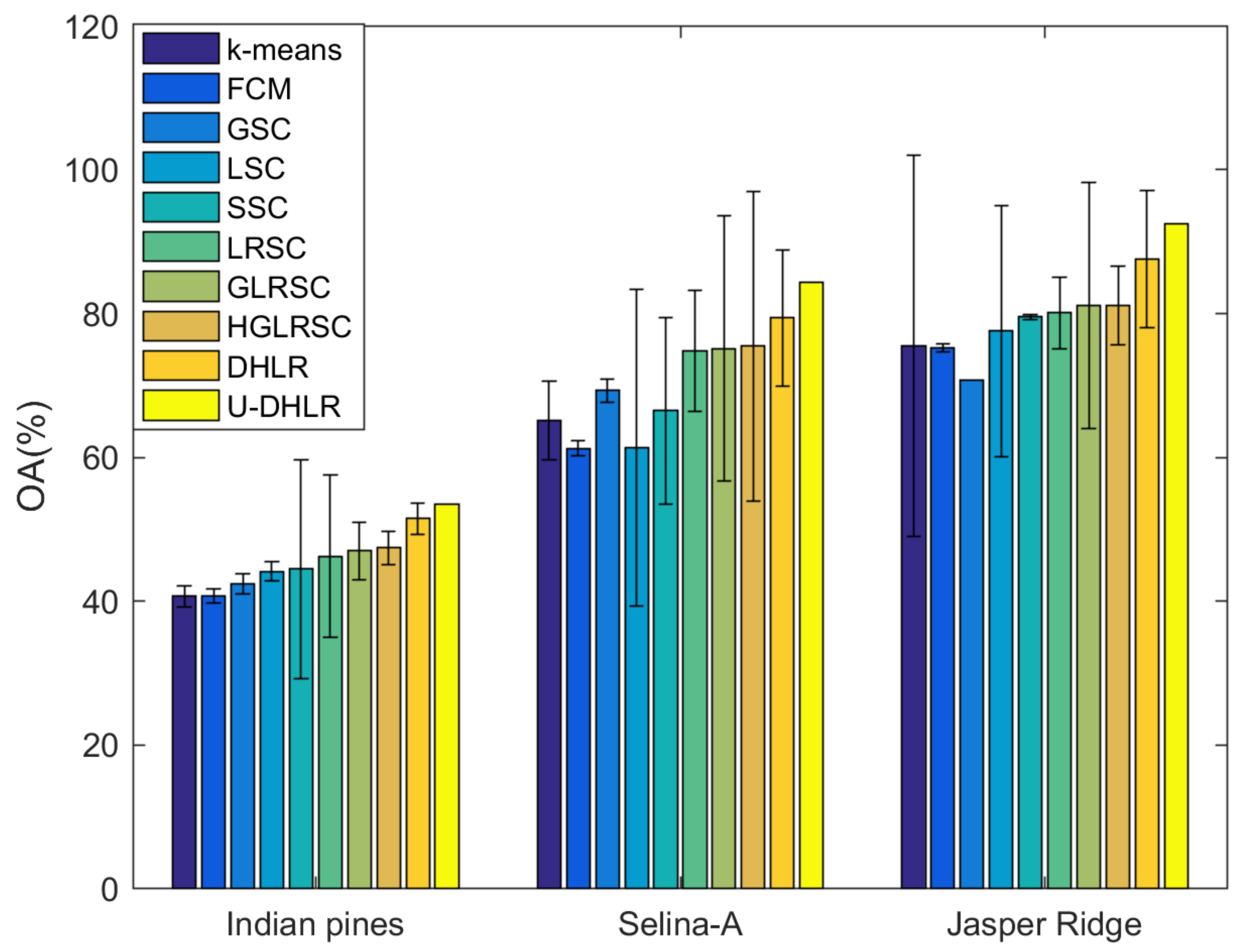
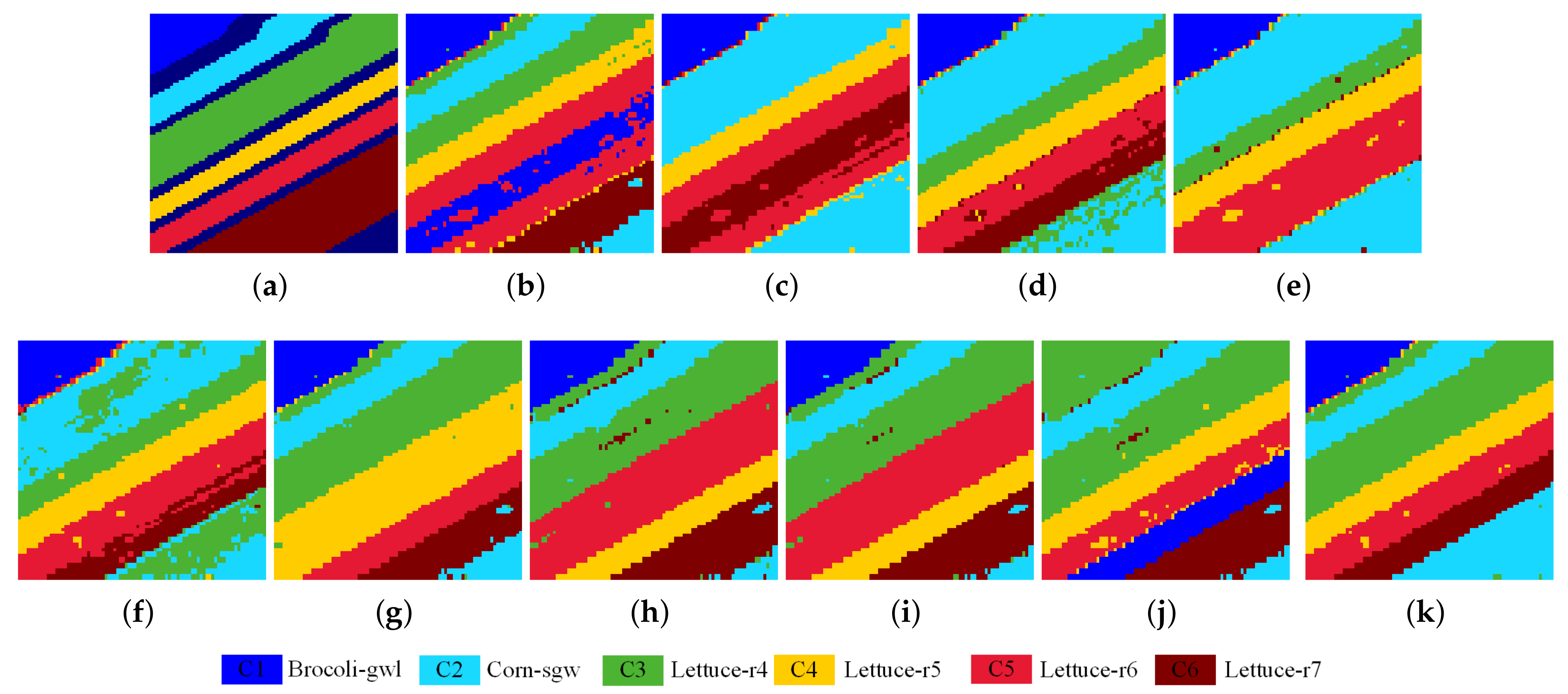
4.4. Investigate of Clustering Performance
5. Discussion
6. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Wang, Q.; He, X.; Li, X. Locality and structure regularized low rank representation for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2018, 57, 911–923. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Xiao, Z.; Chen, Y.; Yang, J. Spatial-spectral graph regularized kernel sparse representation for hyperspectral image classification. ISPRS Int. J. Geo-Inform. 2017, 6, 258. [Google Scholar] [CrossRef]
- Liu, J.; Wu, Z.; Xiao, Z.; Yang, J. Classification of hyperspectral images using kernel fully constrained least squares. ISPRS Int. J. Geo-Inform. 2017, 6, 344. [Google Scholar] [CrossRef] [Green Version]
- Shen, Y.; Xiao, L.; Chen, J.; Pan, D. A Spectral-Spatial Domain-Specific Convolutional Deep Extreme Learning Machine for Supervised Hyperspectral Image Classification. IEEE Access 2019, 7, 132240–132252. [Google Scholar] [CrossRef]
- Zhang, H.; Zhai, H.; Zhang, L.; Li, P. Spectral—spatial sparse subspace clustering for hyperspectral remote sensing images. IEEE Trans. Geosci. Remote Sens. 2016, 54, 3672–3684. [Google Scholar] [CrossRef]
- Lloyd, S. Least squares quantization in PCM. IEEE Trans. Inform. Theory 1982, 28, 129–137. [Google Scholar] [CrossRef]
- Peizhuang, W. Pattern recognition with fuzzy objective function algorithms (James C. Bezdek). SIAM Rev. 1983, 25, 442. [Google Scholar] [CrossRef]
- Ng, A.Y.; Jordan, M.I.; Weiss, Y. On spectral clustering: Analysis and an algorithm. In Advances in Neural Information Processing Systems; MIT Press: Vancouver, BC, Canada, 2002; pp. 849–856. [Google Scholar]
- Belkin, M.; Niyogi, P. Laplacian eigenmaps for dimensionality reduction and data representation. Neur. Comput. 2003, 15, 1373–1396. [Google Scholar] [CrossRef] [Green Version]
- Elhamifar, E.; Vidal, R. Sparse subspace clustering. In Proceedings of the 2009 IEEE Conference on Computer Vision and Pattern Recognition, Los Alamitos, CA, USA, 20–25 June 2009; pp. 2790–2797. [Google Scholar]
- Elhamifar, E.; Vidal, R. Sparse subspace clustering: Algorithm, theory, and applications. IEEE Trans. Pattern Anal. Mach. Intell. 2013, 35, 2765–2781. [Google Scholar] [CrossRef] [Green Version]
- Vidal, R.; Favaro, P. Low rank subspace clustering (LRSC). Pattern Recognit. Lett. 2014, 43, 47–61. [Google Scholar] [CrossRef] [Green Version]
- Liu, G.; Lin, Z.; Yan, S.; Sun, J.; Yu, Y.; Ma, Y. Robust recovery of subspace structures by low-rank representation. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35, 171–184. [Google Scholar] [CrossRef] [Green Version]
- Zheng, M.; Bu, J.; Chen, C.; Wang, C.; Zhang, L.; Qiu, G.; Cai, D. Graph regularized sparse coding for image representation. IEEE Trans. Image Process. 2010, 20, 1327–1336. [Google Scholar] [CrossRef] [PubMed]
- Lu, X.; Wang, Y.; Yuan, Y. Graph-regularized low-rank representation for destriping of hyperspectral images. IEEE Trans. Geosci. Remote Sens. 2013, 51, 4009–4018. [Google Scholar] [CrossRef]
- Berge, C. Hypergraphs; North-Holland: Amsterdam, The Netherlands, 1989. [Google Scholar]
- Zhou, D.; Huang, J.; Schölkopf, B. Learning with hypergraphs: Clustering, classification, and embedding. Adv. Neural Inf. Process. Syst. 2006, 19, 1601–1608. [Google Scholar]
- Yuan, H.; Tang, Y.Y. Learning with hypergraph for hyperspectral image feature extraction. IEEE Geosci. Remote Sens. Lett. 2015, 12, 1695–1699. [Google Scholar] [CrossRef]
- Bai, X.; Guo, Z.; Wang, Y.; Zhang, Z.; Zhou, J. Semisupervised hyperspectral band selection via spectral—Spatial hypergraph model. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2774–2783. [Google Scholar] [CrossRef] [Green Version]
- Du, W.; Qiang, W.; Lv, M.; Hou, Q.; Zhen, L.; Jing, L. Semi-supervised dimension reduction based on hypergraph embedding for hyperspectral images. Int. J. Remote Sens. 2018, 39, 1696–1712. [Google Scholar] [CrossRef]
- Chang, Y.; Yan, L.; Zhong, S. Hyper-laplacian regularized unidirectional low-rank tensor recovery for multispectral image denoising. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 21–26 July 2017; pp. 4260–4268. [Google Scholar]
- Huang, H.; Chen, M.; Duan, Y. Dimensionality reduction of hyperspectral image using spatial-spectral regularized sparse hypergraph embedding. Remote Sens. 2019, 11, 1039. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Tsang, I.W.H.; Chia, L.T. Laplacian sparse coding, hypergraph laplacian sparse coding, and applications. IEEE Trans. Pattern Anal. Mach. Intell. 2012, 35, 92–104. [Google Scholar] [CrossRef] [PubMed]
- Zeng, K.; Yu, J.; Li, C.; You, J.; Jin, T. Image clustering by hyper-graph regularized non-negative matrix factorization. Neurocomputing 2014, 138, 209–217. [Google Scholar] [CrossRef]
- Wang, W.; Qian, Y.; Tang, Y.Y. Hypergraph-regularized sparse NMF for hyperspectral unmixing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 681–694. [Google Scholar] [CrossRef]
- Yin, M.; Gao, J.; Lin, Z. Laplacian regularized low-rank representation and its applications. IEEE Trans. Pattern Anal. Mach. Intell. 2015, 38, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Zeng, M.; Ning, B.; Hu, C.; Gu, Q.; Cai, Y.; Li, S. Hyper-Graph Regularized Kernel Subspace Clustering for Band Selection of Hyperspectral Image. IEEE Access 2020, 8, 135920–135932. [Google Scholar] [CrossRef]
- Zhang, Z.; Bai, L.; Liang, Y.; Hancock, E. Joint hypergraph learning and sparse regression for feature selection. Pattern Recognit. 2017, 63, 291–309. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Lin, H.; Gao, Y.; BNRist, K. Dynamic Hypergraph Structure Learning. In Proceedings of the International Joint Conferences on Artificial Intelligence Organization, Stockholm, Sweden, 13–19 June 2018; pp. 3162–3169. [Google Scholar]
- Zhu, X.; Zhu, Y.; Zhang, S.; Hu, R.; He, W. Adaptive Hypergraph Learning for Unsupervised Feature Selection. In Proceedings of the Twenty-Seventh International Joint Conference on Artificial Intelligence IJCAI, Melbourne, VIC, Australia, 19–25 August 2017; pp. 3581–3587. [Google Scholar]
- Zhu, X.; Zhang, S.; Zhu, Y.; Zhu, P.; Gao, Y. Unsupervised Spectral Feature Selection with Dynamic Hyper-graph Learning. IEEE Trans. Knowl. Data Eng. 2020. [Google Scholar] [CrossRef]
- Tang, C.; Liu, X.; Wang, P.; Zhang, C.; Li, M.; Wang, L. Adaptive hypergraph embedded semi-supervised multi-label image annotation. IEEE Trans. Multimed. 2019, 21, 2837–2849. [Google Scholar] [CrossRef]
- Ding, D.; Yang, X.; Xia, F.; Ma, T.; Liu, H.; Tang, C. Unsupervised feature selection via adaptive hypergraph regularized latent representation learning. Neurocomputing 2020, 378, 79–97. [Google Scholar] [CrossRef]
- Kang, Z.; Peng, C.; Cheng, Q.; Xu, Z. Unified Spectral Clustering with Optimal Graph. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI-17), San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Han, Y.; Zhu, L.; Cheng, Z.; Li, L.; Liu, X. Discrete Optimal Graph Clustering. IEEE Trans. Cybernet. 2020, 50, 1697–1710. [Google Scholar] [CrossRef] [Green Version]
- Yang, Y.; Shen, F.; Huang, Z.; Shen, H.T. A Unified Framework for Discrete Spectral Clustering. In Proceedings of the Twenty-Fifth International Joint Conference on Artificial Intelligence, Phoenix, AZ, USA, 12–17 February 2016; pp. 2273–2279. [Google Scholar]
- Lin, Z.; Chen, M.; Ma, Y. The Augmented Lagrange Multiplier Method for Exact Recovery of Corrupted Low-Rank Matrices. arXiv 2010, arXiv:1009.5055. [Google Scholar]
- Liu, G.; Yan, S. Latent Low-Rank Representation for subspace segmentation and feature extraction. In Proceedings of the 2011 International Conference on Computer Vision, Barcelona, Spain, 6–13 November 2011; pp. 1615–1622. [Google Scholar] [CrossRef]
- Boley, D.; Chen, Y.; Bi, J.; Wang, J.Z.; Huang, J.; Nie, F.; Huang, H.; Rahimi, A.; Recht, B. Spectral Rotation versus K-Means in Spectral Clustering. In Proceedings of the Twenty-Seventh AAAI Conference on Artificial Intelligence, Bellevue, WA, USA, 14–18 July 2013. [Google Scholar]
- Kang, Z.; Peng, C.; Cheng, Q. Twin Learning for Similarity and Clustering: A Unified Kernel Approach. In Proceedings of the Thirty-First AAAI Conference on Artificial Intelligence (AAAI-17), San Francisco, CA, USA, 4–9 February 2017. [Google Scholar]
- Mohar, B. The Laplacian spectrum of graphs. In Graph Theory, Combinatorics, and Applications; Wiley: Hoboken, NJ, USA, 1991; pp. 871–898. [Google Scholar]
- Fan, K. On a Theorem of Weyl Concerning Eigenvalues of Linear Transformations I. Proc. Natl. Acad. Sci. USA 1949, 35, 652–655. [Google Scholar] [CrossRef] [Green Version]
- Yu, S.X.; Shi, J. Multiclass Spectral Clustering. In Proceedings of the IEEE International Conference on Computer Vision, Nice, France, 13–16 October 2003. [Google Scholar]
- Wen, Z.; Yin, W. A feasible method for optimization with orthogonality constraints. Math. Programm. 2013, 142, 397–434. [Google Scholar] [CrossRef] [Green Version]
- Nie, F.; Zhang, R.; Li, X. A generalized power iteration method for solving quadratic problem on the Stiefel manifold. Sci. China Inf. Sci. 2017, 60, 112101. [Google Scholar] [CrossRef]
- Zhong, Y.; Zhang, L.; Gong, W. Unsupervised remote sensing image classification using an artificial immune network. Int. J. Remote Sens. 2011, 32, 5461–5483. [Google Scholar] [CrossRef]
- Ji, R.; Gao, Y.; Hong, R.; Liu, Q. Spectral-Spatial Constraint Hyperspectral Image Classification. IEEE Trans. Geosci. Remote Sens. 2014, 3, 1811–1824. [Google Scholar] [CrossRef]
- Ul Haq, Q.S.; Tao, L.; Sun, F.; Yang, S. A Fast and Robust Sparse Approach for Hyperspectral Data Classification Using a Few Labeled Samples. Geosci. Remote Sens. IEEE Trans. 2012, 50, 2287–2302. [Google Scholar] [CrossRef]
- Strehl, A.; Ghosh, J. Cluster Ensembles—A Knowledge Reuse Framework for Combining Multiple Partitions. J. Mach. Learn. Res. 2002, 3, 583–617. [Google Scholar]
- Lovasz, L.; Plummer, M.D. Matching Theory; AMS Chelsea Publishing: Amsterdam, North Holland, 1986. [Google Scholar]
- Murphy, J.M.; Maggioni, M. Unsupervised Clustering and Active Learning of Hyperspectral Images with Nonlinear Diffusion. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1829–1845. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Fowler, J.E.; Xiao, L. Hypergraph-Regularized Low-Rank Subspace Clustering Using Superpixels for Unsupervised Spatial-Spectral Hyperspectral Classification. IEEE Geosci. Remote Sens. Lett. 2020, 1–5. [Google Scholar] [CrossRef]


| Notation | Definition |
|---|---|
| d | Number of bands |
| n | Number of pixels |
| c | Number of the classes |
| X | Hyperspectral image |
| Z | Low-rank representation matrix |
| N | Noise matrix |
| G | A hypergraph |
| V | The vertexes of hypergraph |
| E | The hyperedges of hypergraph |
| W | The weight of hyperedges |
| H | The incidence matrix of hypergraph |
| L | Hypergraph Laplacian matrix |
| Q | Rotation matrix |
| F | The continuous label indicator matrix |
| Y | The label matrix |
| t | Number of iterations |
| Datasets | Size(N) | Dim(D) | Classes(C) |
|---|---|---|---|
| Salinas-A | 7138 | 204 | 6 |
| Jasper Ridge | 10,000 | 198 | 4 |
| Indian Pines | 21,025 | 200 | 9 |
| IP | Class | Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | k-Means | FCM | GSC | LSC | SSC | LRSC | GLRSC | HGLRSC | DHLR | UDHLR | |
| User’s accuracy (%) | C1 | 26.50 | 42.61 | 29.15 | 11.16 | 56.97 | 56.97 | 56.90 | 23.84 | 00.55 | 0.62 |
| C2 | 0.00 | 15.83 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 0.35 | 0.00 | 0.00 | |
| C3 | 7.04 | 12.27 | 18.51 | 4.63 | 28.77 | 6.04 | 44.26 | 57.34 | 65.79 | 65.79 | |
| C4 | 40.70 | 35.48 | 31.73 | 30.12 | 88.35 | 95.85 | 90.22 | 76.43 | 83.93 | 87.81 | |
| C5 | 99.39 | 98.98 | 91.41 | 99.59 | 99.80 | 100 | 99.79 | 99.59 | 99.59 | 99.59 | |
| C6 | 17.46 | 31.51 | 0.00 | 4.24 | 0.00 | 0.72 | 0.72 | 0.61 | 50.20 | 0.72 | |
| C7 | 66.05 | 43.56 | 73.95 | 88.86 | 45.87 | 45.87 | 46.23 | 81.28 | 59.92 | 85.61 | |
| C8 | 0.49 | 0.00 | 27.52 | 0.00 | 31.11 | 30.78 | 31.92 | 0.32 | 41.20 | 42.50 | |
| C9 | 61.28 | 67.23 | 59.35 | 76.89 | 56.11 | 72.72 | 65.45 | 56.10 | 88.17 | 88.17 | |
| Producer’s accuracy (%) | C1 | 23.39 | 31.17 | 30.13 | 27.35 | 27.98 | 27.98 | 28.20 | 22.55 | 21.05 | 22.50 |
| C2 | 0.00 | 14.66 | 0.00 | 0.00 | 0.00 | 0.00 | 0.00 | 2.63 | 0.00 | 0.00 | |
| C3 | 6.27 | 17.13 | 26.13 | 17.42 | 50.88 | 11.53 | 31.65 | 35.62 | 93.42 | 93.96 | |
| C4 | 80.21 | 88.92 | 86.49 | 90.00 | 89.30 | 72.91 | 78.92 | 80.19 | 83.15 | 81.38 | |
| C5 | 63.36 | 64.79 | 71.86 | 49.74 | 99.18 | 90.72 | 96.06 | 84.25 | 100 | 100 | |
| C6 | 18.15 | 21.52 | 0.00 | 25.30 | 0.00 | 12.50 | 12.50 | 12.76 | 24.01 | 2.80 | |
| C7 | 43.87 | 53.80 | 44.11 | 39.67 | 43.70 | 43.48 | 43.69 | 42.30 | 41.90 | 40.57 | |
| C8 | 60.00 | 0.00 | 21.91 | 0.00 | 25.67 | 25.36 | 25.42 | 100 | 24.80 | 24.53 | |
| C9 | 72.88 | 73.85 | 73.00 | 75.78 | 69.40 | 75.94 | 88.69 | 87.05 | 100 | 100 | |
| OA (%) | 40.66 | 40.70 | 42.33 | 44.13 | 44.48 | 46.24 | 46.96 | 47.38 | 51.45 | 53.52 | |
| AA (%) | 35.43 | 38.61 | 36.85 | 35.05 | 45.22 | 45.44 | 48.39 | 43.99 | 54.37 | 52.32 | |
| 0.284 | 0.308 | 0.299 | 0.303 | 0.341 | 0.361 | 0.372 | 0.353 | 0.423 | 0.429 | ||
| NMI (%) | 43.08 | 41.17 | 43.55 | 46.68 | 47.28 | 45.31 | 46.12 | 46.58 | 48.68 | 54.26 | |
| IP | Class | Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | k-means | FCM | GSC | LSC | SSC | LRSC | GLRSC | HGLRSC | DHLR | UDHLR | |
| User’s accuracy (%) | C1 | 0.00 | 99.74 | 100 | 99.74 | 99.48 | 99.74 | 99.48 | 99.48 | 0.00 | 99.74 |
| C2 | 92.85 | 0.00 | 100 | 0.00 | 62.50 | 94.48 | 87.98 | 89.28 | 90.58 | 92.04 | |
| C3 | 53.83 | 48.06 | 49.63 | 46.62 | 54.03 | 99.86 | 97.70 | 99.27 | 98.22 | 100 | |
| C4 | 100 | 99.85 | 99.40 | 99.85 | 99.85 | 0.00 | 0.00 | 0.00 | 99.85 | 99.70 | |
| C5 | 87.23 | 95.11 | 92.86 | 95.11 | 98.12 | 99.74 | 99.37 | 99.24 | 90.48 | 97.49 | |
| C6 | 53.53 | 53.46 | 39.38 | 55.69 | 37.30 | 52.71 | 59.86 | 58.74 | 59.04 | 42.88 | |
| Producer’s accuracy (%) | C1 | 0.00 | 100 | 100 | 100 | 100 | 100 | 100 | 100 | 0.00 | 100 |
| C2 | 94.23 | 0.00 | 33.02 | 0.00 | 30.55 | 88.58 | 95.59 | 96.49 | 96.20 | 42.53 | |
| C3 | 94.15 | 89.28 | 70.35 | 97.26 | 50.30 | 97.19 | 96.06 | 95.94 | 77.77 | 96.76 | |
| C4 | 52.41 | 55.66 | 98.82 | 90.57 | 96.69 | 0.00 | 0.00 | 0.00 | 89.85 | 97.39 | |
| C5 | 63.42 | 94.88 | 98.01 | 58.64 | 90.74 | 54.47 | 54.68 | 54.38 | 99.86 | 99.74 | |
| C6 | 100 | 33.75 | 91.04 | 34.45 | 100 | 100 | 93.16 | 97.04 | 95.77 | 99.65 | |
| OA (%) | 65.12 | 61.21 | 69.27 | 61.36 | 66.49 | 74.79 | 75.15 | 75.45 | 79.37 | 84.31 | |
| AA (%) | 64.57 | 66.04 | 80.21 | 66.17 | 75.21 | 74.42 | 74.07 | 74.34 | 73.03 | 88.64 | |
| 0.582 | 0.515 | 0.631 | 0.517 | 0.589 | 0.689 | 0.691 | 0.696 | 0.742 | 0.808 | ||
| NMI (%) | 70.84 | 62.67 | 67.64 | 63.88 | 64.38 | 84.02 | 81.69 | 83.33 | 81.10 | 86.30 | |
| IP | Class | Method | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| No. | k-means | FCM | GSC | LSC | SSC | LRSC | GLRSC | HGLRSC | DHLR | UDHLR | |
| User’s accuracy (%) | C1 | 97.39 | 97.13 | 56.68 | 72.17 | 63.15 | 95.50 | 78.07 | 78.44 | 95.13 | 92.38 |
| C2 | 59.14 | 56.22 | 99.87 | 99.21 | 97.92 | 67.55 | 90.10 | 90.01 | 91.91 | 100 | |
| C3 | 90.07 | 93.28 | 42.17 | 79.65 | 71.49 | 100 | 97.40 | 97.32 | 97.65 | 84.22 | |
| C4 | 0.00 | 0.00 | 99.46 | 0.00 | 100 | 0.13 | 2.52 | 2.52 | 1.19 | 87.38 | |
| Producer’s accuracy (%) | C1 | 93.35 | 95.38 | 100 | 100 | 99.63 | 99.88 | 99.92 | 100 | 99.81 | 92.91 |
| C2 | 100 | 100 | 98.83 | 99.90 | 99.54 | 100 | 99.96 | 99.89 | 100 | 95.08 | |
| C3 | 71.19 | 71.49 | 40.94 | 69.76 | 58.29 | 71.94 | 60.23 | 60.43 | 72.13 | 88.60 | |
| C4 | 0.00 | 0.00 | 34.70 | 0.00 | 49.02 | 0.01 | 5.47 | 5.38 | 2.75 | 91.26 | |
| OA (%) | 75.56 | 75.28 | 70.75 | 77.55 | 79.52 | 80.12 | 81.08 | 81.16 | 82.89 | 92.56 | |
| AA (%) | 61.65 | 61.66 | 74.55 | 62.76 | 83.14 | 65.79 | 67.02 | 67.07 | 68.33 | 90.99 | |
| 0.662 | 0.659 | 0.606 | 0.690 | 0.719 | 0.723 | 0.732 | 0.733 | 0.757 | 0.894 | ||
| NMI (%) | 73.56 | 74.45 | 70.24 | 74.48 | 69.25 | 78.43 | 68.72 | 68.82 | 71.46 | 77.18 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, J.; Xiao, L.; Yang, J. Unified Low-Rank Subspace Clustering with Dynamic Hypergraph for Hyperspectral Image. Remote Sens. 2021, 13, 1372. https://doi.org/10.3390/rs13071372
Xu J, Xiao L, Yang J. Unified Low-Rank Subspace Clustering with Dynamic Hypergraph for Hyperspectral Image. Remote Sensing. 2021; 13(7):1372. https://doi.org/10.3390/rs13071372
Chicago/Turabian StyleXu, Jinhuan, Liang Xiao, and Jingxiang Yang. 2021. "Unified Low-Rank Subspace Clustering with Dynamic Hypergraph for Hyperspectral Image" Remote Sensing 13, no. 7: 1372. https://doi.org/10.3390/rs13071372







