Retrieving and Validating Leaf and Canopy Chlorophyll Content at Moderate Resolution: A Multiscale Analysis with the Sentinel-3 OLCI Sensor
Abstract
:1. Introduction
2. Methodology
2.1. Remote Sensing Data
2.2. In Situ Measurements
2.3. Retrieval Models
2.4. Scaling Analysis
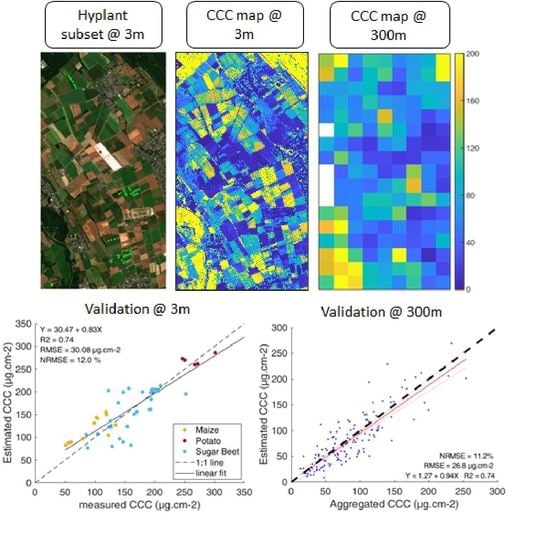
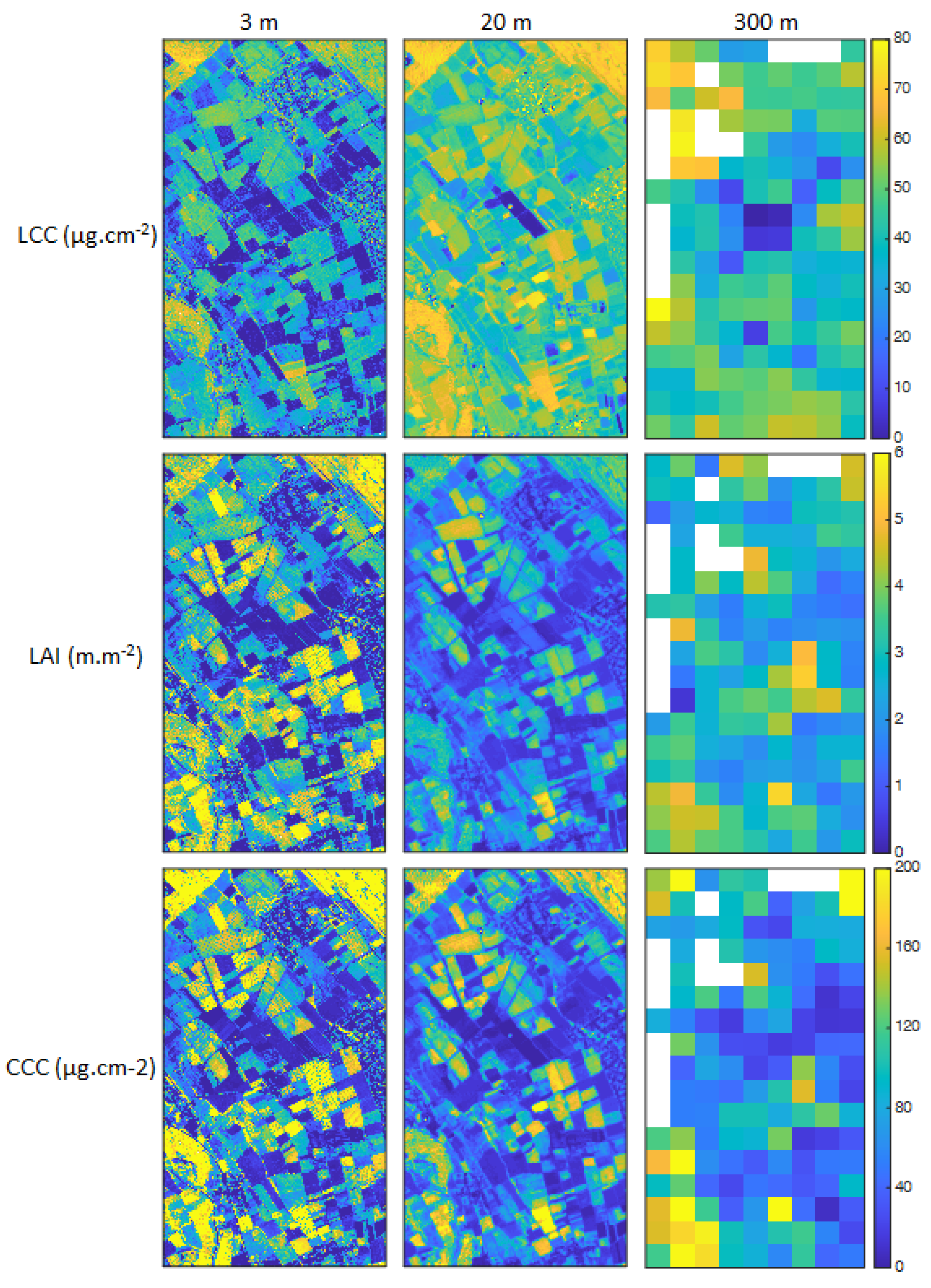
3. Results
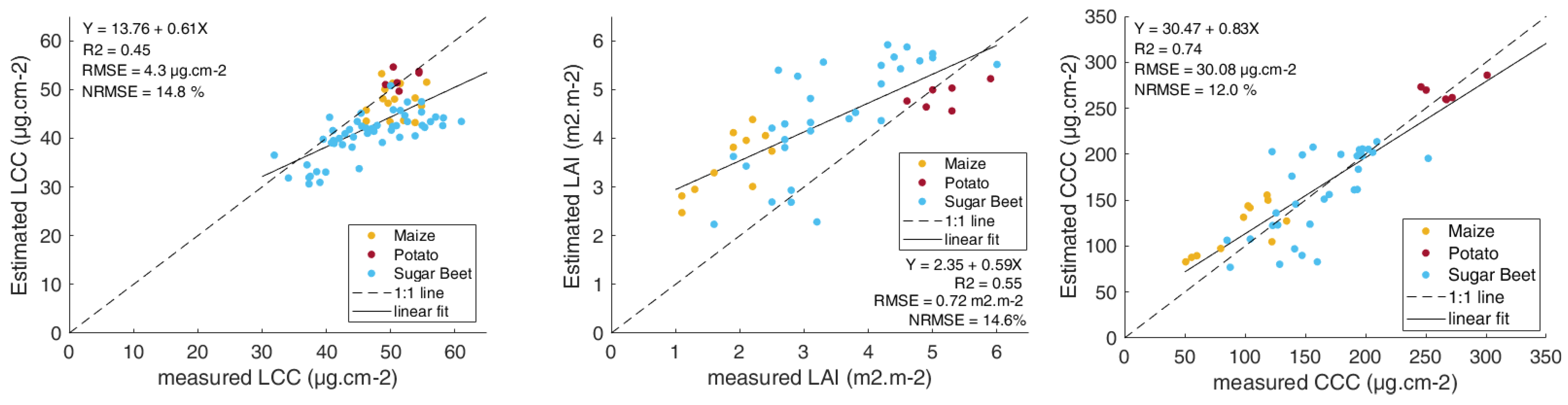
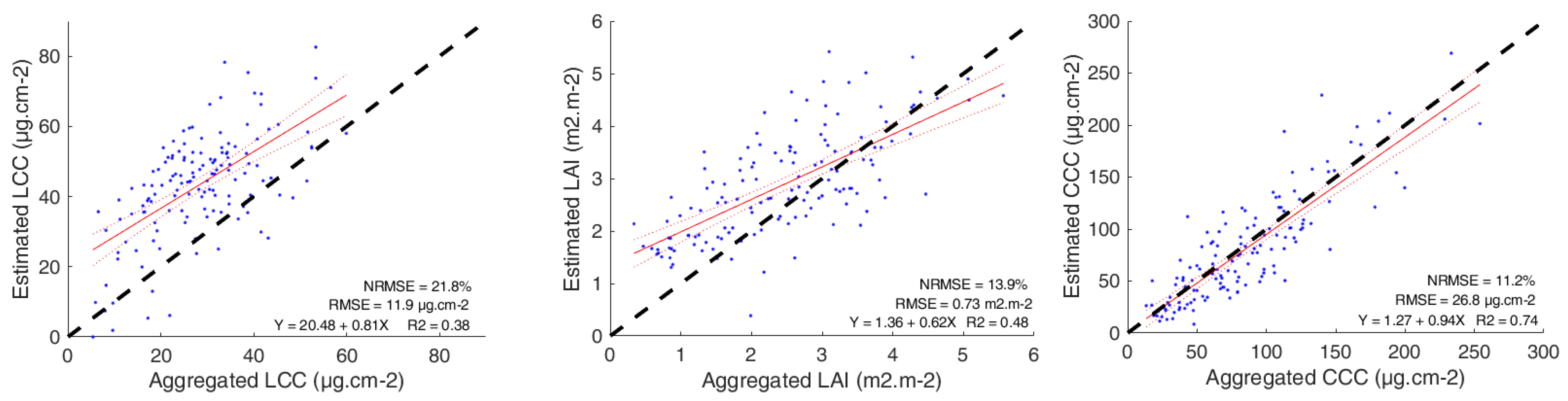
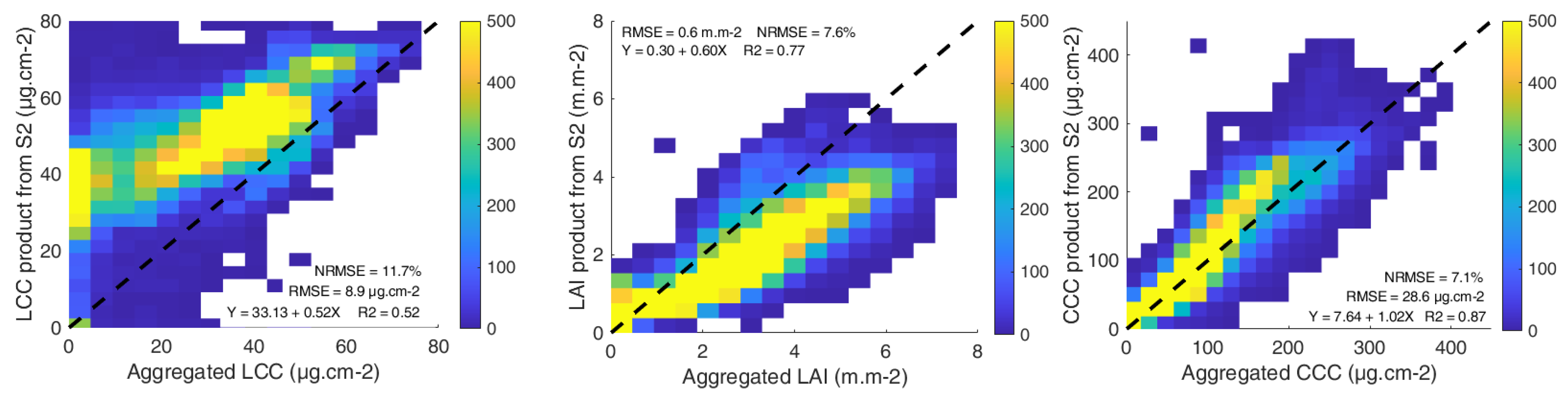
3.1. Model Validation
3.2. LCC Model Comparison
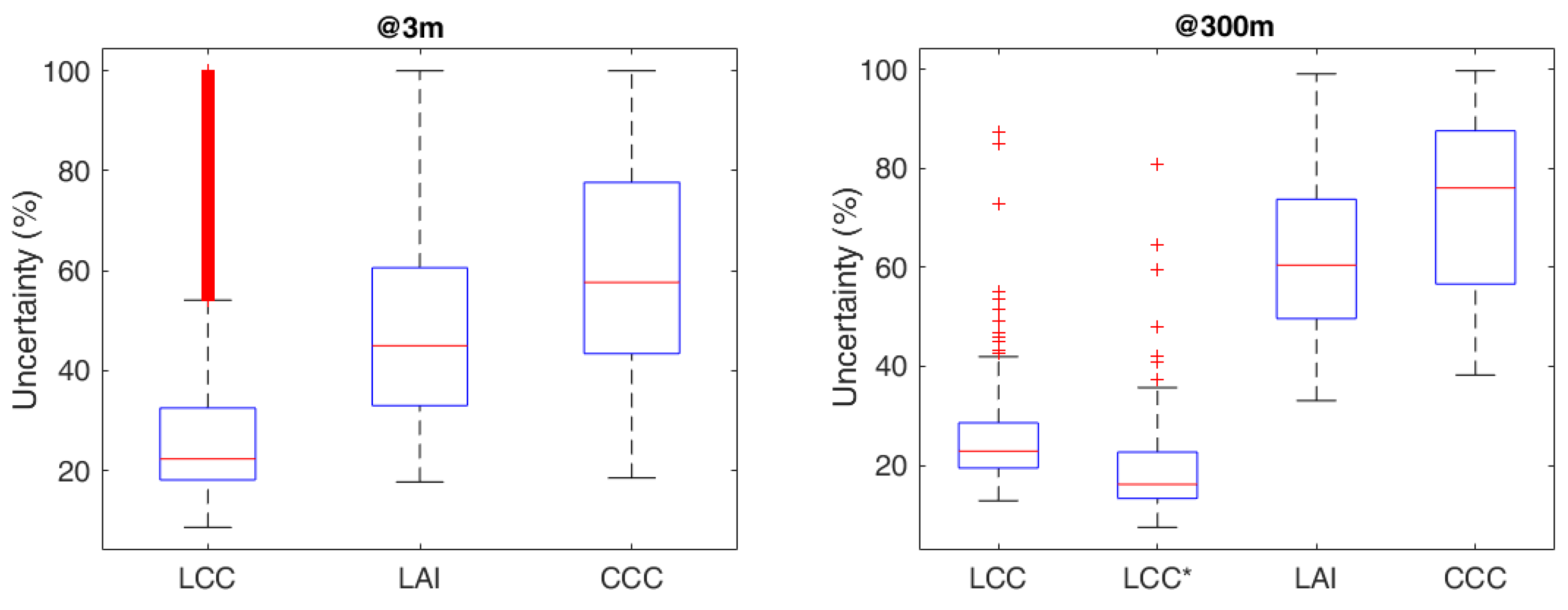
3.3. Analysis of Prediction Uncertainties
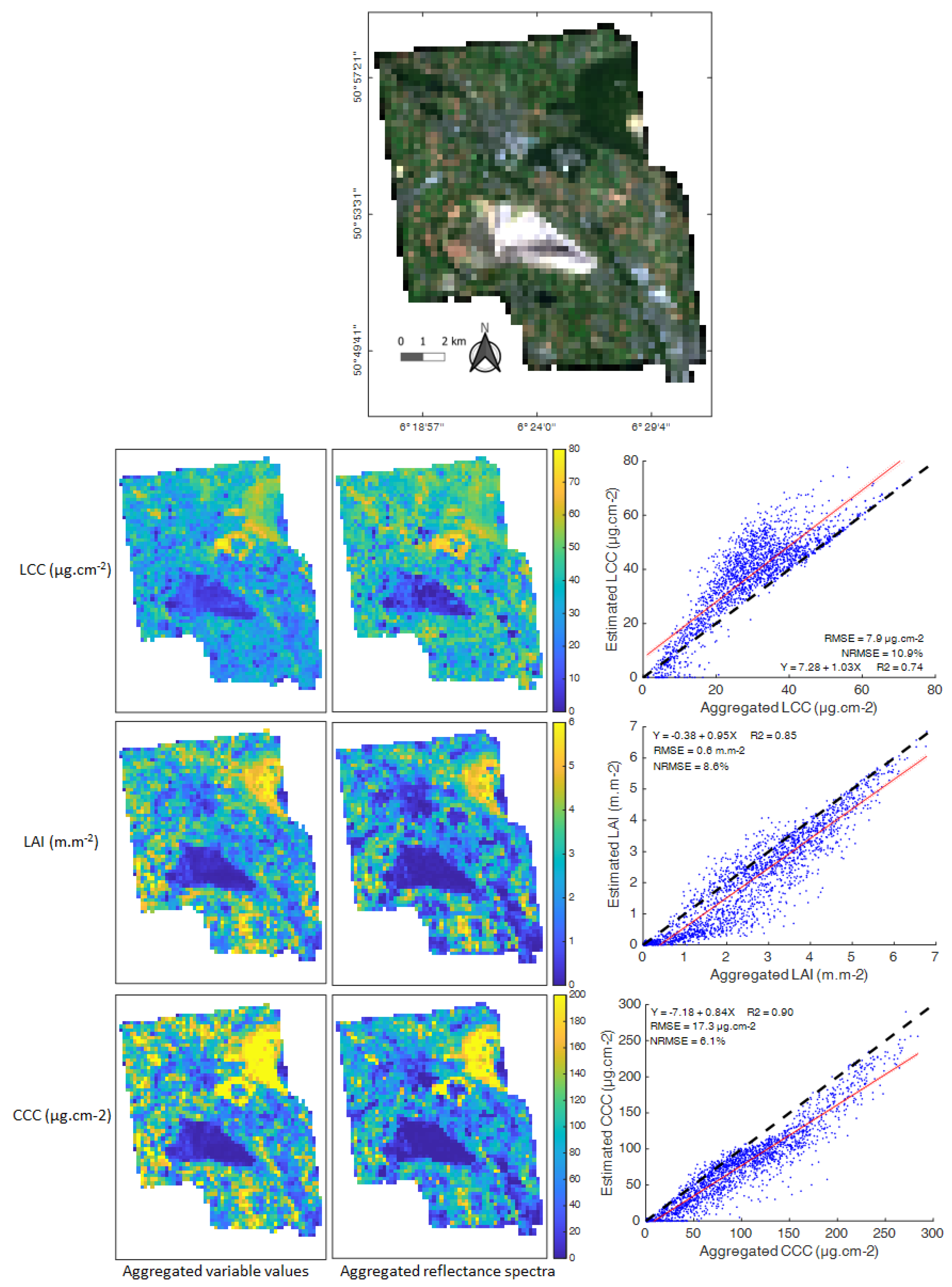
3.4. Scaling Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Variable Type | Variable | Distribution | Min | Max | Mean | SD |
|---|---|---|---|---|---|---|
| Leaf structure | N | Gaussian * | 1 | 2.7 | 1.5 | 0.5 |
| Cab (g·cm) | Uniform | 1 | 100 | |||
| Cca (g·cm) | Gaussian * | 0 | 30 | 10 | 5 | |
| Cdm (g·cm) ** | Gaussian * | 0.002 | 0.02 | 0.005 | 0.003 | |
| Cw (g·cm) ** | Gaussian * | 0.005 | 0.035 | 0.012 | 0.006 | |
| Canopy structure | LAI (m m) | Uniform | 0.1 | 10 | ||
| LIDFa *** | Uniform | −1 | 1 | |||
| LIDFb *** | Uniform | −1 | 1 | |||
| Soil | SMC (%) | Gaussian * | 5 | 55 | 25 | 12.5 |
| BSM Brightness | Gaussian * | 0.01 | 0.9 | 0.5 | 0.25 | |
| BSM lat () | Gaussian * | 20 | 40 | 25 | 12.5 | |
| BSM long () | Gaussian * | 45 | 65 | 50 | 10 | |
| Geometry | SZA () | Uniform | 0 | 80 | ||
| OZA () | Uniform | 0 | 25 | |||
| RAA () | Uniform | 0 | 180 |
References
- Fróna, D.; Szenderák, J.; Harangi-Rákos, M. The challenge of feeding the world. Sustainability 2019, 11, 5816. [Google Scholar] [CrossRef] [Green Version]
- Parry, M.; Rosenzweig, C.; Livermore, M. Climate change, global food supply and risk of hunger. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 2125–2138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hui, D. Food web: Concept and applications. Nat. Educ. Knowl. 2012, 3, 6. [Google Scholar]
- Gitelson, A.; Gritz, Y.; Merzlyak, M. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef] [PubMed]
- Croft, H.; Chen, J.; Wang, R.; Mo, G.; Luo, S.; Luo, X.; He, L.; Gonsamo, A.; Arabian, J.; Zhang, Y.; et al. The global distribution of leaf chlorophyll content. Remote Sens. Environ. 2020, 236. [Google Scholar] [CrossRef]
- Luo, X.; Croft, H.; Chen, J.; He, L.; Keenan, T. Improved estimates of global terrestrial photosynthesis using information on leaf chlorophyll content. Glob. Chang. Biol. 2019, 25, 2499–2514. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.; Viña, A.; Ciganda, V.; Rundquist, D.; Arkebauer, T. Remote estimation of canopy chlorophyll in crops. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef] [Green Version]
- Jacquemoud, S.; Baret, F.; Andrieu, B.; Danson, F.; Jaggard, K. Extraction of vegetation biophysical parameters by inversion of the PROSPECT + SAIL models on sugar beet canopy reflectance data. Application to TM and AVIRIS sensors. Remote Sens. Environ. 1995, 52, 163–172. [Google Scholar] [CrossRef]
- Bacour, C.; Baret, F.; Béal, D.; Weiss, M.; Pavageau, K. Neural network estimation of LAI, fAPAR, fCover and LAIxCab, from top of canopy MERIS reflectance data: Principles and validation. Remote Sens. Environ. 2006, 105, 313–325. [Google Scholar] [CrossRef]
- Chen, J.; Black, T. Defining leaf area index for non-flat leaves. Plant Cell Environ. 1992, 15, 421–429. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C. Inversion of a radiative transfer model for estimating vegetation LAI and chlorophyll in a heterogeneous grassland. Remote Sens. Environ. 2008, 112, 2592–2604. [Google Scholar] [CrossRef]
- Clevers, J.; Kooistra, L.; van den Brande, M. Using Sentinel-2 data for retrieving LAI and leaf and canopy chlorophyll content of a potato crop. Remote Sens. 2017, 9, 405. [Google Scholar] [CrossRef] [Green Version]
- Jay, S.; Maupas, F.; Bendoula, R.; Gorretta, N. Retrieving LAI, chlorophyll and nitrogen contents in sugar beet crops from multi-angular optical remote sensing: Comparison of vegetation indices and PROSAIL inversion for field phenotyping. Field Crop. Res. 2017, 210, 33–46. [Google Scholar] [CrossRef] [Green Version]
- Bourg, L.; Blanot, L.; Lamquin, N.; Bruniquel, V.; Meskini, N.; Nieke, J.; Bouvet, M.; Fougnie, B. Sentinel-3 OLCI Radiometric and Spectral Performance Activities. In Sentinel-3 for Science Workshop; Ouwehand, L., Ed.; ESA Special Publication: Paris, France, 2015; Volume 734, p. 5. [Google Scholar]
- Xiong, X.; Che, N.; Barnes, W. Terra MODIS on-orbit spatial characterization and performance. IEEE Trans. Geosci. Remote Sens. 2005, 43, 355–365. [Google Scholar] [CrossRef]
- Dierckx, W.; Sterckx, S.; Benhadj, I.; Livens, S.; Duhoux, G.; Achteren, T.V.; Francois, M.; Mellab, K.; Saint, G. PROBA-V mission for global vegetation monitoring: Standard products and image quality. Int. J. Remote Sens. 2014, 35, 2589–2614. [Google Scholar] [CrossRef]
- Garrigues, S.; Allard, D.; Baret, F.; Weiss, M. Quantifying spatial heterogeneity at the landscape scale using variogram models. Remote Sens. Environ. 2006, 103, 81–96. [Google Scholar] [CrossRef]
- Drusch, M.; Moreno, J.; Del Bello, U.; Franco, R.; Goulas, Y.; Huth, A.; Kraft, S.; Middleton, E.; Miglietta, F.; Mohammed, G.; et al. The FLuorescence EXplorer Mission Concept-ESA’s Earth Explorer 8. IEEE Trans. Geosci. Remote Sens. 2017, 55, 1273–1284. [Google Scholar] [CrossRef]
- De Grave, C.; Verrelst, J.; Morcillo-Pallarés, P.; Pipia, L.; Rivera-Caicedo, J.; Amin, E.; Belda, S.; Moreno, J. Quantifying vegetation biophysical variables from the Sentinel-3/FLEX tandem mission: Evaluation of the synergy of OLCI and FLORIS data sources. Remote Sens. Environ. 2020, 251. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhao, L.; Lin, A. Evaluating the performance of Sentinel-3A OLCI land products for gross primary productivity estimation using AmeriFlux data. Remote Sens. 2020, 12, 1927. [Google Scholar] [CrossRef]
- Pastor-Guzman, J.; Brown, L.; Morris, H.; Bourg, L.; Goryl, P.; Dransfeld, S.; Dash, J. The sentinel-3 OLCI terrestrial chlorophyll index (OTCI): Algorithm improvements, spatiotemporal consistency and continuity with the MERIS archive. Remote Sens. 2020, 12, 2652. [Google Scholar] [CrossRef]
- Van Der Tol, C.; Verhoef, W.; Timmermans, J.; Verhoef, A.; Su, Z. An integrated model of soil-canopy spectral radiances, photosynthesis, fluorescence, temperature and energy balance. Biogeosciences 2009, 6, 3109–3129. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, C.E.; Williams, C.K.I. Gaussian Processes for Machine Learning; MIT Press: Cambridge, MA, USA, 2006. [Google Scholar]
- Camps-Valls, G.; Verrelst, J.; Muñoz, J.; Laparra, V.; Mateo, F.; Gomez-Dans, J. A Survey on Gaussian Processes for Earth-Observation Data Analysis: A Comprehensive Investigation. IEEE Geosci. Remote Sens. Mag. 2016, 4, 58–78. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Malenovský, Z.; Van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.P.; Lewis, P.; North, P.; Moreno, J. Quantifying Vegetation Biophysical Variables from Imaging Spectroscopy Data: A Review on Retrieval Methods. Surv. Geophys. 2019, 40, 589–629. [Google Scholar] [CrossRef] [Green Version]
- Vicent, J.; Sabater Medina, N.; Tenjo, C.; Ramon Acarreta, J.; Manzano, M.; Rivera Caicedo, J.; Jurado Lozano, P.; Franco, R.; Alonso, L.; Verrelst, J.; et al. FLEX End-to-End Mission Performance Simulator. IEEE Trans. Geosci. Remote Sens. 2016, 54, 1–9. [Google Scholar] [CrossRef]
- Zuhlke, M.; Fomferra, N.; Brockmann, C.; Peters, M.; Veci, L.; Malik, J.; Regner, P. SNAP (sentinel application platform) and the ESA Sentinel 3 toolbox. In Proceedings of the Sentinel-3 for Science Workshop, Lido Palazzo del Casinò, Italy, 2–5 June 2015; Volume 734, p. 21. [Google Scholar]
- Malenovský, Z.; Homolová, L.; Lukeš, P.; Buddenbaum, H.; Verrelst, J.; Alonso, L.; Schaepman, M.; Lauret, N.; Gastellu-Etchegorry, J.P. Variability and Uncertainty Challenges in Scaling Imaging Spectroscopy Retrievals and Validations from Leaves up to Vegetation Canopies. Surv. Geophys. 2019, 40, 631–656. [Google Scholar] [CrossRef]
- Justice, C.; Belward, A.; Morisette, J.; Lewis, P.; Privette, J.; Baret, F. Developments in the ’validation’ of satellite sensor products for the study of the land surface. Int. J. Remote Sens. 2000, 21, 3383–3390. [Google Scholar] [CrossRef]
- Fuster, B.; Sánchez-Zapero, J.; Camacho, F.; García-Santos, V.; Verger, A.; Lacaze, R.; Weiss, M.; Baret, F.; Smets, B. Quality assessment of PROBA-V LAI, fAPAR and fCOVER collection 300 m products of copernicus global land service. Remote Sens. 2020, 12, 1017. [Google Scholar] [CrossRef] [Green Version]
- Fernandes, R.; Plummer, S.; Nightingale, J.; Baret, F.; Camacho de Coca, F.; Fang, H.; Garrigues, S.; Gobron, N.; Lang, M.; Lacaze, R.; et al. CEOS Global LAI Product Validation Good Practices; CEOS: Washington, DC, USA, 2014. [Google Scholar] [CrossRef]
- Baret, F.; Weiss, M.; Allard, D.; Garrigue, S.; Leroy, M.; Jeanjean, H.; Fernandes, R.; Myneni, R.; Privette, J.; Bohbot, H.; et al. VALERI: A network of sites and methodology for the validation of medium spatial resolution land products. Remote. Sens. Environ. 2013, 76, 36–39. [Google Scholar]
- Tan, B.; Hu, J.; Zhang, P.; Huang, D.; Shabanov, N.; Weiss, M.; Knyazikhin, Y.; Myneni, R. Validation of Moderate Resolution Imaging Spectroradiometer leaf area index product in croplands of Alpilles, France. J. Geophys. Res. D Atmos. 2005, 110, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Brown, L.; Meier, C.; Morris, H.; Pastor-Guzman, J.; Bai, G.; Lerebourg, C.; Gobron, N.; Lanconelli, C.; Clerici, M.; Dash, J. Evaluation of global leaf area index and fraction of absorbed photosynthetically active radiation products over North America using Copernicus Ground Based Observations for Validation data. Remote Sens. Environ. 2020, 247. [Google Scholar] [CrossRef]
- Morisette, J.; Baret, F.; Privette, J.; Myneni, R.; Nickeson, J.; Garrigues, S.; Shabanov, N.; Weiss, M.; Fernandes, R.; Leblanc, S.; et al. Validation of global moderate-resolution LAI products: A framework proposed within the CEOS land product validation subgroup. IEEE Trans. Geosci. Remote Sens. 2006, 44, 1804–1814. [Google Scholar] [CrossRef] [Green Version]
- Tian, Y.; Woodcock, C.; Wang, Y.; Privette, J.; Shabanov, N.; Zhou, L.; Zhang, Y.; Buermann, W.; Dong, J.; Veikkanen, B.; et al. Multiscale analysis and validation of the MODIS LAI product I. Uncertainty assessment. Remote Sens. Environ. 2002, 83, 414–430. [Google Scholar] [CrossRef]
- Siegmann, B.; Alonso, L.; Celesti, M.; Cogliati, S.; Colombo, R.; Damm, A.; Douglas, S.; Guanter, L.; Hanuš, J.; Kataja, K.; et al. The High-Performance Airborne Imaging Spectrometer HyPlant—From Raw Images to Top-of-Canopy Reflectance and Fluorescence Products: Introduction of an Automatized Processing Chain. Remote Sens. 2019, 11, 2760. [Google Scholar] [CrossRef] [Green Version]
- Henocq, C.; North, P.; Heckel, A.; Ferron, S.; Lamquin, N.; Dransfeld, S.; Bourg, L.; Tote, C.; Ramon, D. OLCI/SLSTR SYN L2 algorithm and products overview. In Proceedings of the International Geoscience and Remote Sensing Symposium (IGARSS), Valencia, Spain, 22–27 July 2018; pp. 8723–8726. [Google Scholar] [CrossRef]
- ESA. Sentinel-3 User Handbook, Sentinel-3 Team, 1.0 ed. 2013. Available online: https://sentinel.esa.int (accessed on 10 May 2020).
- Weiss, M.; Baret, F. S2 ToolBox Level 2 Products: LAI, FAPAR, FCOVER Version 1.1. 2016. Available online: https://step.esa.int/docs/extra/ATBD_S2ToolBox_L2B_V1.1.pdf (accessed on 15 June 2020).
- Delloye, C.; Weiss, M.; Defourny, P. Retrieval of the canopy chlorophyll content from Sentinel-2 spectral bands to estimate nitrogen uptake in intensive winter wheat cropping systems. Remote Sens. Environ. 2018, 216, 245–261. [Google Scholar] [CrossRef]
- Verrelst, J.; Camps-Valls, G.; Muñoz Marí, J.; Rivera, J.; Veroustraete, F.; Clevers, J.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties—A review. ISPRS J. Photogramm. Remote. Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Verger, A.; Baret, F.; Camacho, F. Optimal modalities for radiative transfer-neural network estimation of canopy biophysical characteristics: Evaluation over an agricultural area with CHRIS/PROBA observations. Remote Sens. Environ. 2011, 115, 415–426. [Google Scholar] [CrossRef]
- Baret, F.; Hagolle, O.; Geiger, B.; Bicheron, P.; Miras, B.; Huc, M.; Berthelot, B.; Niño, F.; Weiss, M.; Samain, O.; et al. LAI, fAPAR and fCover CYCLOPES global products derived from VEGETATION. Part 1: Principles of the algorithm. Remote Sens. Environ. 2007, 110, 275–286. [Google Scholar] [CrossRef] [Green Version]
- García-Haro, F.; Campos-Taberner, M.; Muñoz Marí, J.; Laparra, V.; Camacho, F.; Sánchez-Zapero, J.; Camps-Valls, G. Derivation of global vegetation biophysical parameters from EUMETSAT Polar System. ISPRS J. Photogramm. Remote Sens. 2018, 139, 57–74. [Google Scholar] [CrossRef]
- QGIS Development Team. QGIS Geographic Information System. Open Source Geospatial Foundation. Available online: http://qgis.org (accessed on 25 July 2020).
- Lichtenthaler, H.; Gitelson, A.; Lang, M. Non-destructive determination of chlorophyll content of leaves of a green and an aurea mutant of tobacco by reflectance measurements. J. Plant Physiol. 1996, 148, 483–493. [Google Scholar] [CrossRef]
- Sims, D.; Gamon, J. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Widlowski, J.L.; Pinty, B.; Lavergne, T.; Verstraete, M.; Gobron, N. Horizontal radiation transport in 3-D forest canopies at multiple spatial resolutions: Simulated impact on canopy absorption. Remote Sens. Environ. 2006, 103, 379–397. [Google Scholar] [CrossRef]
- Wu, H.; Li, Z.L. Scale issues in remote sensing: A review on analysis, processing and modeling. Sensors 2009, 9, 1768–1793. [Google Scholar] [CrossRef] [PubMed]
- Hardiman, B.; LaRue, E.; Atkins, J.; Fahey, R.; Wagner, F.; Gough, C. Spatial variation in canopy structure across forest landscapes. Forests 2018, 9, 474. [Google Scholar] [CrossRef] [Green Version]
- Dufrêne, E.; Bréda, N. Estimation of deciduous forest leaf area index using direct and indirect methods. Oecologia 1995, 104, 156–162. [Google Scholar] [CrossRef] [PubMed]
- Yan, G.; Hu, R.; Luo, J.; Weiss, M.; Jiang, H.; Mu, X.; Xie, D.; Zhang, W. Review of indirect optical measurements of leaf area index: Recent advances, challenges, and perspectives. Agric. For. Meteorol. 2019, 265, 390–411. [Google Scholar] [CrossRef]
- Chen, J. Spatial scaling of a remotely sensed surface parameter by contexture. Remote Sens. Environ. 1999, 69, 30–42. [Google Scholar] [CrossRef]
- Jacob, F.; Weiss, M. Mapping biophysical variables from solar and thermal infrared remote sensing: Focus on agricultural landscapes with spatial heterogeneity. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1844–1848. [Google Scholar] [CrossRef]
- Garrigues, S.; Allard, D.; Baret, F.; Weiss, M. Influence of landscape spatial heterogeneity on the non-linear estimation of leaf area index from moderate spatial resolution remote sensing data. Remote Sens. Environ. 2006, 105, 286–298. [Google Scholar] [CrossRef]
- Sentinel-3 MPC. Sentinel-3 Mission Performance Center Optical Annual Performance Report—Year 2018. Technical Report, ACRI-ST. 2019. Available online: https://sentinel.esa.int/web/sentinel/user-guides/sentinel-3-olci/document-library/-/asset_publisher/hkf7sg9Ny1d5/content/sentinel-3-optical-annual-performance-report-year-2 (accessed on 20 April 2020).
- Bell, S.A. A Beginner’s Guide to Uncertainty of Measurement; National Physical Laboratory: Teddington, UK, 2001. [Google Scholar]
- Verrelst, J.; Rivera, J.P.; Moreno, J.; Camps-Valls, G. Gaussian processes uncertainty estimates in experimental Sentinel-2 LAI and leaf chlorophyll content retrieval. ISPRS J. Photogramm. Remote Sens. 2013, 86, 157–167. [Google Scholar] [CrossRef]
- Lázaro-Gredilla, M.; Titsias, M.; Verrelst, J.; Camps-Valls, G. Retrieval of biophysical parameters with heteroscedastic Gaussian processes. IEEE Geosci. Remote Sens. Lett. 2014, 11, 838–842. [Google Scholar] [CrossRef]
- Estévez, J.; Vicent, J.; Rivera-Caicedo, J.; Morcillo-Pallarés, P.; Vuolo, F.; Sabater, N.; Camps-Valls, G.; Moreno, J.; Verrelst, J. Gaussian processes retrieval of LAI from Sentinel-2 top-of-atmosphere radiance data. ISPRS J. Photogramm. Remote Sens. 2020, 167, 289–304. [Google Scholar] [CrossRef]
- Berger, K.; Verrelst, J.; Féret, J.B.; Hank, T.; Wocher, M.; Mauser, W.; Camps-Valls, G. Retrieval of aboveground crop nitrogen content with a hybrid machine learning method. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102174. [Google Scholar] [CrossRef]
- Verhoef, W.; van der Tol, C.; Middleton, E. Hyperspectral radiative transfer modeling to explore the combined retrieval of biophysical parameters and canopy fluorescence from FLEX–Sentinel-3 tandem mission multi-sensor data. Remote Sens. Environ. 2018, 204, 942–963. [Google Scholar] [CrossRef]

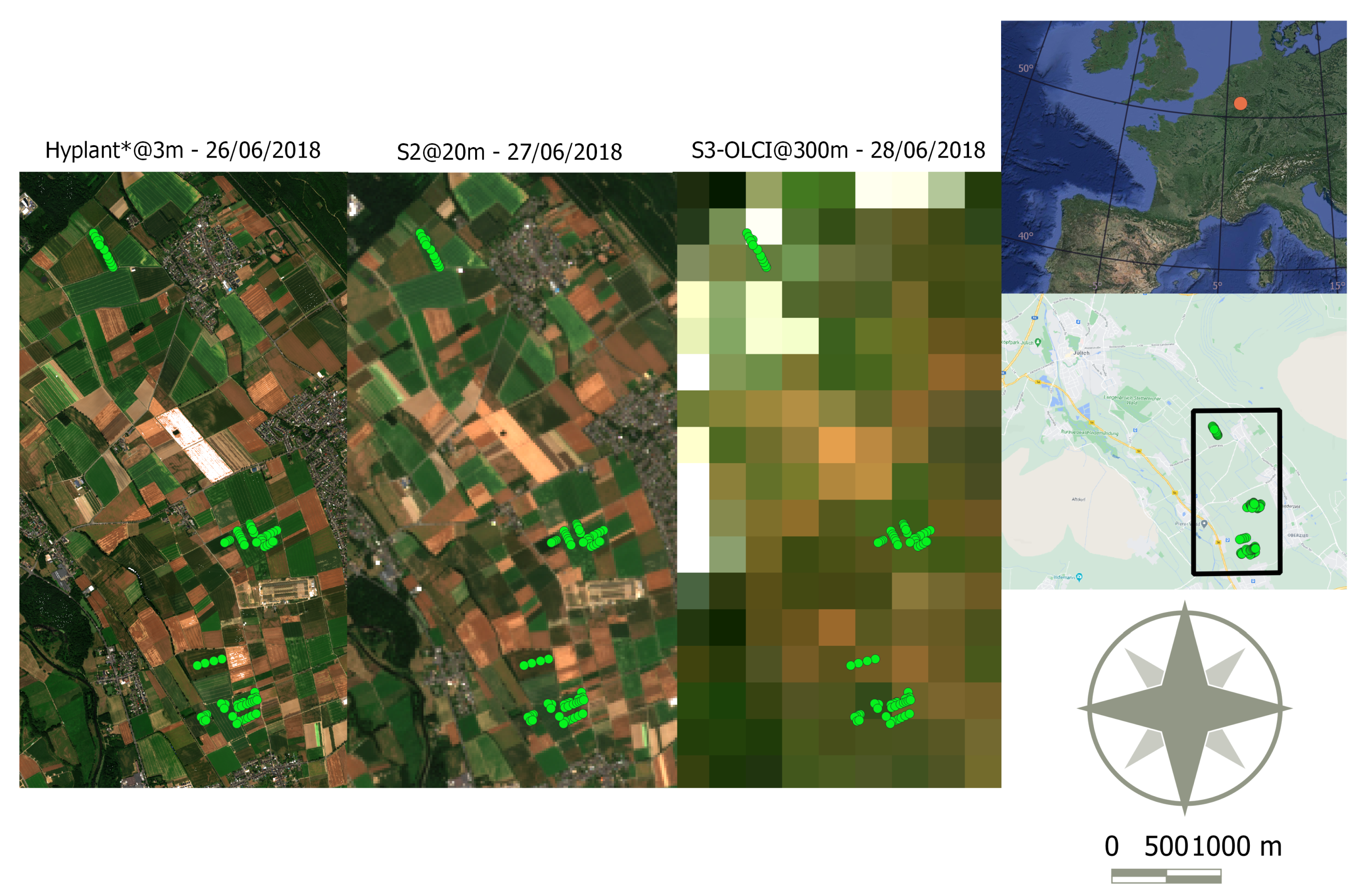

| Sensor | Acquisition
Date | Spatial Resolution [m] | Bands | Spectral Range | Continuous Sampling |
|---|---|---|---|---|---|
| HyPlant* DUAL | 26 June 2018 | 3 | 626 | 380–2500 nm | True |
| S2-MSI | 27 June 2018 | 20 | 10 | 400–2200 nm | False |
| S3-OLCI | 28 June 2018 | 300 | 16 | 400–1020 nm | False |
| LCC (g cm) | LAI (m m) | CCC (g cm) | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| # Samples | Mean | Min | Max | # Samples | Mean | Min | Max | # Samples | Mean | Min | Max | |
| Maize | 15 | 50.7 | 46.2 | 55.6 | 11 | 1.8 | 1.1 | 2.5 | 11 | 95.1 | 50.8 | 134.5 |
| Potato | 6 | 51.8 | 49.2 | 54.4 | 6 | 5.2 | 4.6 | 5.9 | 6 | 267.1 | 246.0 | 300.9 |
| Sugar Beet | 51 | 46.6 | 31.9 | 61.0 | 30 | 3.5 | 1.6 | 6.0 | 30 | 161.2 | 85.1 | 252.0 |
| Total | 72 | 47.9 | 31.9 | 61.0 | 47 | 3.3 | 1.1 | 6.0 | 47 | 159.3 | 50.8 | 300.9 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
De Grave, C.; Pipia, L.; Siegmann, B.; Morcillo-Pallarés, P.; Rivera-Caicedo, J.P.; Moreno, J.; Verrelst, J. Retrieving and Validating Leaf and Canopy Chlorophyll Content at Moderate Resolution: A Multiscale Analysis with the Sentinel-3 OLCI Sensor. Remote Sens. 2021, 13, 1419. https://doi.org/10.3390/rs13081419
De Grave C, Pipia L, Siegmann B, Morcillo-Pallarés P, Rivera-Caicedo JP, Moreno J, Verrelst J. Retrieving and Validating Leaf and Canopy Chlorophyll Content at Moderate Resolution: A Multiscale Analysis with the Sentinel-3 OLCI Sensor. Remote Sensing. 2021; 13(8):1419. https://doi.org/10.3390/rs13081419
Chicago/Turabian StyleDe Grave, Charlotte, Luca Pipia, Bastian Siegmann, Pablo Morcillo-Pallarés, Juan Pablo Rivera-Caicedo, José Moreno, and Jochem Verrelst. 2021. "Retrieving and Validating Leaf and Canopy Chlorophyll Content at Moderate Resolution: A Multiscale Analysis with the Sentinel-3 OLCI Sensor" Remote Sensing 13, no. 8: 1419. https://doi.org/10.3390/rs13081419
APA StyleDe Grave, C., Pipia, L., Siegmann, B., Morcillo-Pallarés, P., Rivera-Caicedo, J. P., Moreno, J., & Verrelst, J. (2021). Retrieving and Validating Leaf and Canopy Chlorophyll Content at Moderate Resolution: A Multiscale Analysis with the Sentinel-3 OLCI Sensor. Remote Sensing, 13(8), 1419. https://doi.org/10.3390/rs13081419