Potential of Hybrid CNN-RF Model for Early Crop Mapping with Limited Input Data
Abstract
:1. Introduction
2. Study Areas and Datasets
2.1. Study Areas
2.2. Datasets
3. Methodology
3.1. Classification Model
3.1.1. RF
3.1.2. CNN
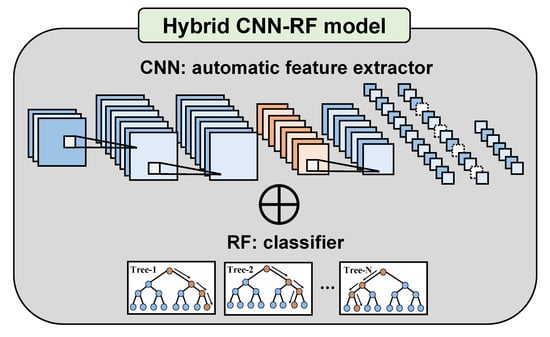
3.1.3. Hybrid CNN-RF Model
3.2. Training and Reference Data Sampling
3.3. Optimization of Model Parameters
3.4. Incremental Classification
3.5. Analysis Procedures and Implementation
4. Results
4.1. Time-Series Analysis of Vegetation Index
4.2. Comparison of Class Separability in the Feature Space
4.3. Incremental Classification Results
4.3.1. Results in Anbandegi
4.3.2. Results in Hapcheon
5. Discussion
6. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Kim, N.; Ha, K.J.; Park, N.W.; Cho, J.; Hong, S.; Lee, Y.W. A comparison between major artificial intelligence models for crop yield prediction: Case study of the Midwestern United States, 2006–2105. ISPRS Int. J. Geo-Inf. 2019, 8, 240. [Google Scholar] [CrossRef] [Green Version]
- Na, S.I.; Park, C.W.; So, K.H.; Ahn, H.Y.; Lee, K.D. Application method of unmanned aerial vehicle for crop monitoring in Korea. Korean J. Remote Sens. 2018, 34, 829–846, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Kwak, G.H.; Park, N.W. Impact of texture information on crop classification with machine learning and UAV images. Appl. Sci. 2019, 9, 643. [Google Scholar] [CrossRef] [Green Version]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First experience with Sentinel-2 data for crop and tree species classifications in central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Böhler, J.E.; Schaepman, M.E.; Kneubühler, M. Crop classification in a heterogeneous arable landscape using uncalibrated UAV data. Remote Sens. 2018, 10, 1282. [Google Scholar] [CrossRef] [Green Version]
- Villa, P.; Stroppiana, D.; Fontanelli, G.; Azar, R.; Brivio, P.A. In-season mapping of crop type with optical and X-band SAR data: A classification tree approach using synoptic seasonal features. Remote Sens. 2015, 7, 12859–12886. [Google Scholar] [CrossRef] [Green Version]
- Hao, P.; Zhan, Y.; Wang, L.; Niu, Z.; Shakir, M. Feature selection of time series MODIS data for early crop classification using random forest: A case study in Kansas, USA. Remote Sens. 2015, 7, 5347–5369. [Google Scholar] [CrossRef] [Green Version]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D convolutional neural networks for crop classification with multi-temporal remote sensing images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Wei, S.; Zhang, H.; Wang, C.; Wang, Y.; Xu, L. Multi-temporal SAR data large-scale crop mapping based on U-Net model. Remote Sens. 2019, 11, 68. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Chen, Z.; Jiang, H.; Jing, W.; Sun, L.; Feng, M. Evaluation of three deep learning models for early crop classification using Sentinel-1A imagery time series—A case study in Zhanjiang, China. Remote Sens. 2019, 11, 2673. [Google Scholar] [CrossRef] [Green Version]
- Skakun, S.; Franch, B.; Vermote, E.; Roger, J.C.; Becker-Reshef, I.; Justice, C.; Kussul, N. Early season large-area winter crop mapping using MODIS NDVI data, growing degree days information and a Gaussian mixture model. Remote Sens. Environ. 2017, 195, 244–258. [Google Scholar] [CrossRef]
- Foody, G.M.; Mathur, A. The use of small training sets containing mixed pixels for accurate hard image classification: Training on mixed spectral responses for classification by a SVM. Remote Sens. Environ. 2006, 103, 179–189. [Google Scholar] [CrossRef]
- Deng, F.; Pu, S.; Chen, X.; Shi, Y.; Yuan, T.; Pu, S. Hyperspectral image classification with capsule network using limited training samples. Sensors 2018, 18, 3153. [Google Scholar] [CrossRef] [Green Version]
- Mas, J.F.; Flores, J.J. The application of artificial neural networks to the analysis of remotely sensed data. Int. J. Remote Sens. 2008, 29, 617–663. [Google Scholar] [CrossRef]
- Tatsumi, K.; Yamashiki, Y.; Morante, A.K.; Fernández, L.R.; Nalvarte, R.A. Pixel-based crop classification in Peru from Landsat 7 ETM+ images using a random forest model. J. Agric. Meteorol. 2016, 72, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Fu, T.; Blaschke, T.; Li, M.; Tiede, D.; Zhou, Z.; Ma, X.; Chen, D. Evaluation of feature selection methods for object-based land cover mapping of unmanned aerial vehicle imagery using random forest and support vector machine classifiers. ISPRS Int. J. Geo-Inf. 2017, 6, 51. [Google Scholar] [CrossRef]
- Kim, Y.; Park, N.W.; Lee, K.D. Self-learning based land-cover classification using sequential class patterns from past land-cover maps. Remote Sens. 2017, 9, 921. [Google Scholar] [CrossRef] [Green Version]
- Löw, F.; Michel, U.; Dech, S.; Conrad, C. Impact of feature selection on the accuracy and spatial uncertainty of per-field crop classification using support vector machines. ISPRS J. Photogramm. Remote Sens. 2013, 85, 102–119. [Google Scholar] [CrossRef]
- Sidike, P.; Sagan, V.; Maimaitijiang, M.; Maimaitiyiming, M.; Shakoor, N.; Burken, J.; Mockler, T.; Fritschi, F.B. dPEN: Deep Progressively Expanded Network for mapping heterogeneous agricultural landscape using WorldView-3 satellite imagery. Remote Sens. Environ. 2019, 221, 756–772. [Google Scholar] [CrossRef]
- Zhao, W.; Du, S. Spectral–spatial feature extraction for hyperspectral image classification: A dimension reduction and deep learning approach. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4544–4554. [Google Scholar] [CrossRef]
- Yang, X.; Ye, Y.; Li, X.; Lau, R.Y.; Zhang, X.; Huang, X. Hyperspectral image classification with deep learning models. IEEE Trans. Geosci. Remote Sens. 2018, 56, 5408–5423. [Google Scholar] [CrossRef]
- Song, H.; Kim, Y.; Kim, Y. A patch-based light convolutional neural network for land-cover mapping using Landsat-8 images. Remote Sens. 2019, 11, 114. [Google Scholar] [CrossRef] [Green Version]
- Kwak, G.H.; Park, C.W.; Lee, K.D.; Na, S.I.; Ahn, H.Y.; Park, N.W. Combining 2D CNN and bidirectional LSTM to consider spatio-temporal features in crop classification. Korean J. Remote Sens. 2019, 35, 681–692, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Park, M.G.; Kwak, G.H.; Park, N.W. A convolutional neural network model with weighted combination of multi-scale spatial features for crop classification. Korean J. Remote Sens. 2019, 35, 1273–1283, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Chen, X.; Xiang, S.; Liu, C.L.; Pan, C.H. Vehicle detection in satellite images by hybrid deep convolutional neural networks. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1797–1801. [Google Scholar] [CrossRef]
- Li, K.; Cheng, G.; Bu, S.; You, X. Rotation-insensitive and context-augmented object detection in remote sensing images. IEEE Trans. Geosci. Remote Sens. 2018, 56, 2337–2348. [Google Scholar] [CrossRef]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef] [Green Version]
- Zhong, L.; Hu, L.; Zhou, H. Deep learning based multi-temporal crop classification. Remote Sens. Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Crisóstomo de Castro Filho, H.; Abílio de Carvalho Júnior, O.; Ferreira de Carvalho, O.L.; Pozzobon de Bem, P.; dos Santos de Moura, R.; Olino de Albuquerque, A.; Rosa Silva, C.; Guimaraes Ferreira, P.H.; Fontes Guimaraes, R.; Trancoso Gomes, R.A. Rice crop detection using LSTM, Bi-LSTM, and machine learning models from sentinel-1 time series. Remote Sens. 2020, 12, 2655. [Google Scholar] [CrossRef]
- Kussul, N.; Lavreniuk, M.; Skakun, S.; Shelestov, A. Deep learning classification of land cover and crop types using remote sensing data. IEEE Geosci. Remote Sens. Lett. 2017, 14, 778–782. [Google Scholar] [CrossRef]
- Xie, B.; Zhang, H.K.; Xue, J. Deep convolutional neural network for mapping smallholder agriculture using high spatial resolution satellite image. Sensors 2019, 19, 2398. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.; Kwak, G.H.; Lee, K.D.; Na, S.I.; Park, C.W.; Park, N.W. Performance evaluation of machine learning and deep learning algorithms in crop classification: Impact of hyper-parameters and training sample size. Korean J. Remote Sens. 2018, 34, 811–827, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Du, P.; Xia, J.; Zhang, W.; Tan, K.; Liu, Y.; Liu, S. Multiple classifier system for remote sensing image classification: A review. Sensors 2012, 12, 4764–4792. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Gu, L.; Li, X.; Jiang, T.; Ren, R. Crop classification method based on optimal feature selection and hybrid CNN-RF networks for multi-temporal remote sensing imagery. Remote Sens. 2020, 12, 3119. [Google Scholar] [CrossRef]
- Wang, A.; Wang, Y.; Chen, Y. Hyperspectral image classification based on convolutional neural network and random forest. Remote Sens. Lett. 2019, 10, 1086–1094. [Google Scholar] [CrossRef]
- Dong, L.; Du, H.; Mao, F.; Han, N.; Li, X.; Zhou, G.; Zhu, D.; Zheng, J.; Zhang, M.; Xing, L.; et al. Very high resolution remote sensing imagery classification using a fusion of random forest and deep learning technique—Subtropical area for example. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 113–128. [Google Scholar] [CrossRef]
- Li, T.; Leng, J.; Kong, L.; Guo, S.; Bai, G.; Wang, K. DCNR: Deep cube CNN with random forest for hyperspectral image classification. Multimed. Tools Appl. 2019, 78, 3411–3433. [Google Scholar] [CrossRef]
- Inglada, J.; Vincent, A.; Arias, M.; Marais-Sicre, C. Improved early crop type identification by joint use of high temporal resolution SAR and optical image time series. Remote Sens. 2016, 8, 362. [Google Scholar] [CrossRef] [Green Version]
- Yoo, H.Y.; Lee, K.D.; Na, S.I.; Park, C.W.; Park, N.W. Field crop classification using multi-temporal high-resolution satellite imagery: A case study on garlic/onion field. Korean J. Remote Sens. 2017, 33, 621–630, (In Korean with English Abstract). [Google Scholar] [CrossRef]
- Environmental Geographic Information Service (EGIS). Available online: http://egis.me.go.kr (accessed on 9 January 2021).
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Guidici, D.; Clark, M.L. One-Dimensional convolutional neural network land-cover classification of multi-seasonal hyperspectral imagery in the San Francisco Bay Area, California. Remote Sens. 2017, 9, 629. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Yoo, C.; Lee, Y.; Cho, D.; Im, J.; Han, D. Improving local climate zone classification using incomplete building data and Sentinel 2 images based on convolutional neural networks. Remote Sens. 2020, 12, 3552. [Google Scholar] [CrossRef]
- Feng, Q.; Zhu, D.; Yang, J.; Li, B. Multisource hyperspectral and LiDAR data fusion for urban land-use mapping based on a modified two-branch convolutional neural network. ISPRS Int. J. Geo-Inf. 2019, 8, 28. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Wang, H.; Xu, F.; Jin, Y.Q. Target classification using the deep convolutional networks for SAR images. IEEE Trans. Geosci. Remote Sens. 2016, 54, 4806–4817. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Van der Maaten, L.; Hinton, G. Visualizing data using t-SNE. J. Mach. Learn. Res. 2008, 9, 2579–2605. [Google Scholar]
- Jiao, L.; Liang, M.; Chen, H.; Yang, S.; Liu, H.; Cao, X. Deep fully convolutional network-based spatial distribution prediction for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 5585–5599. [Google Scholar] [CrossRef]
- Scikit-Learn: Machine Learning in Python. Available online: https://scikit-learn.org (accessed on 9 January 2021).
- TensorFlow. Available online: https://tensorflow.org (accessed on 9 January 2021).
- Keras Documentation. Available online: https://keras.io (accessed on 9 January 2021).
- Foody, G.M. Thematic map comparison: Evaluating the statistical significance of differences in classification accuracy. Photogramm. Eng. Remote Sens. 2004, 70, 627–633. [Google Scholar] [CrossRef]
- Yi, Z.; Jia, L.; Chen, Q. Crop classification using multi-temporal Sentinel-2 data in the Shiyang River Basin of China. Remote Sens. 2020, 12, 4052. [Google Scholar] [CrossRef]
- Ren, T.; Liu, Z.; Zhang, L.; Liu, D.; Xi, X.; Kang, Y.; Zhao, Y.; Zhang, C.; Li, S.; Zhang, X. Early identification of seed maize and common maize production fields using Sentinel-2 images. Remote Sens. 2020, 12, 2140. [Google Scholar] [CrossRef]
- Li, W.; Chen, C.; Zhang, M.; Li, H.; Du, Q. Data augmentation for hyperspectral image classification with deep CNN. IEEE Geosci. Remote Sens. Lett. 2018, 16, 593–597. [Google Scholar] [CrossRef]
- Zhu, X. Semi-Supervised Learning Literature Survey; Technical Report 1530; Department of Computer Sciences, University of Wisconsin: Madison, WI, USA, 2005. [Google Scholar]
- Settles, B. Active Learning Literature Survey; Technical Report 1648; Department of Computer Sciences, University of Wisconsin: Madison, WI, USA, 2010. [Google Scholar]
- Hu, J.; Shen, L.; Sun, G. Squeeze-And-Excitation Networks. In Proceedings of the 2018 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Salt Lake City, UT, USA, 18–22 June 2018; pp. 7132–7141. [Google Scholar]
- Shi, C.; Lv, Z.; Shen, H.; Fang, L.; You, Z. Improved metric learning with the CNN for very-high-resolution remote sensing image classification. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2021, 14, 631–644. [Google Scholar] [CrossRef]
- Park, S.; Park, N.W. Effects of class purity of training patch on classification performance of crop classification with convolutional neural network. Appl. Sci. 2020, 10, 3773. [Google Scholar] [CrossRef]















| Area | Acquisition Date | Image Size | |
|---|---|---|---|
| Anbandegi | 7 June 2018 | (A1) | 1314 × 1638 |
| 16 June 2018 | (A2) | ||
| 28 June 2018 | (A3) | ||
| 16 July 2018 | (A4) | ||
| 1 August 2018 | (A5) | ||
| 15 August 2018 | (A6) | ||
| 4 September 2018 | (A7) | ||
| 19 September 2018 | (A8) | ||
| Hapcheon | 4 April 2019 | (H1) | 1866 × 1717 |
| 18 April 2019 | (H2) | ||
| 2 May 2019 | (H3) | ||
| Study Area | Class | Training Data Size | Reference Data | ||||
|---|---|---|---|---|---|---|---|
| T1 | T2 | T3 | T4 | T5 | |||
| Anbandegi | Highland Kimchi cabbage | 38 | 76 | 152 | 304 | 608 | 5000 |
| Cabbage | 19 | 38 | 76 | 152 | 304 | 5000 | |
| Potato | 10 | 20 | 40 | 80 | 160 | 5000 | |
| Fallow | 13 | 26 | 52 | 104 | 208 | 5000 | |
| Total | 80 | 160 | 320 | 640 | 1280 | 20,000 | |
| Hapcheon | Garlic | 33 | 66 | 132 | 264 | 528 | 5000 |
| Onion | 32 | 64 | 128 | 256 | 512 | 5000 | |
| Barley | 7 | 14 | 28 | 56 | 112 | 5000 | |
| Fallow | 8 | 16 | 32 | 64 | 128 | 5000 | |
| Total | 80 | 160 | 320 | 640 | 1280 | 20,000 | |
| Model | Hyper-Parameter (Layer Description) | Tested Hyper-Parameters |
|---|---|---|
| CNN and CNN-RF | Image patch size | 3 to 17 (interval of 2) |
| Convolution layer 1 | 32, 64, 128, 256 | |
| Convolution layer 2 | ||
| Convolution layer 3 | ||
| Dropout | 0.3, 0.5, 0.7 | |
| Model epochs | 1 to 500 (interval of 1) | |
| Learning rate | 0.0001, 0.0005, 0.001, 0.005 | |
| RF and CNN-RF | The number of trees to be grown in the forest (ntree) | 100, 500, 1000, 1500 |
| The number of variables for node partitioning (mtry) | , , |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kwak, G.-H.; Park, C.-w.; Lee, K.-d.; Na, S.-i.; Ahn, H.-y.; Park, N.-W. Potential of Hybrid CNN-RF Model for Early Crop Mapping with Limited Input Data. Remote Sens. 2021, 13, 1629. https://doi.org/10.3390/rs13091629
Kwak G-H, Park C-w, Lee K-d, Na S-i, Ahn H-y, Park N-W. Potential of Hybrid CNN-RF Model for Early Crop Mapping with Limited Input Data. Remote Sensing. 2021; 13(9):1629. https://doi.org/10.3390/rs13091629
Chicago/Turabian StyleKwak, Geun-Ho, Chan-won Park, Kyung-do Lee, Sang-il Na, Ho-yong Ahn, and No-Wook Park. 2021. "Potential of Hybrid CNN-RF Model for Early Crop Mapping with Limited Input Data" Remote Sensing 13, no. 9: 1629. https://doi.org/10.3390/rs13091629
APA StyleKwak, G.-H., Park, C.-w., Lee, K.-d., Na, S.-i., Ahn, H.-y., & Park, N.-W. (2021). Potential of Hybrid CNN-RF Model for Early Crop Mapping with Limited Input Data. Remote Sensing, 13(9), 1629. https://doi.org/10.3390/rs13091629