Multi-Temporal LiDAR and Hyperspectral Data Fusion for Classification of Semi-Arid Woody Cover Species
Abstract
:1. Introduction
2. Materials and Methods
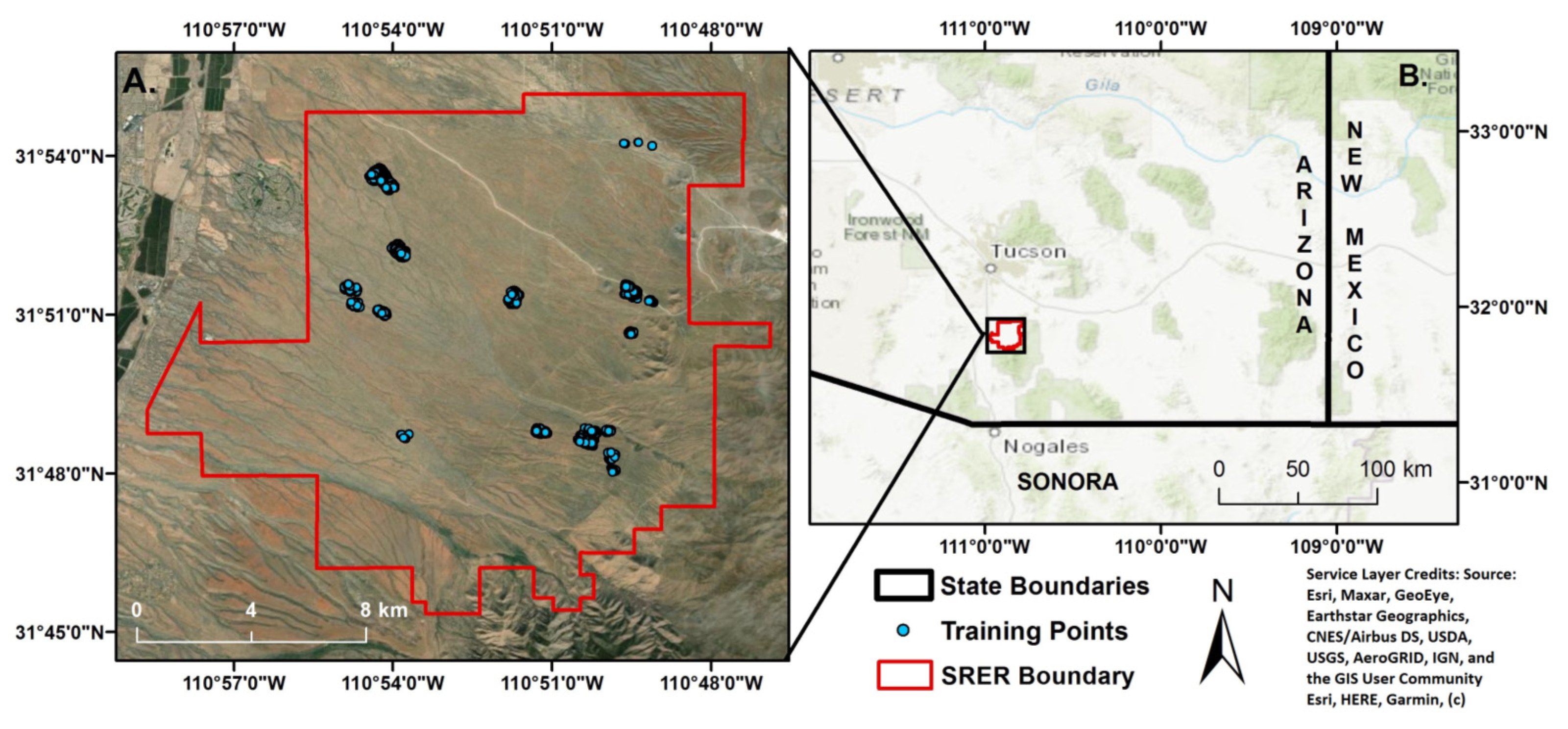
2.1. Study Area
2.2. Raw Data
2.2.1. Hyperspectral Data
2.2.2. Point Cloud Data
2.2.3. Drone Images
2.3. Data Processing
2.3.1. Indices
2.3.2. Canopy Height Model
2.3.3. Training Data
2.3.4. Classification
CART
SVM
RF
2.4. Accuracy Assessment
3. Results
3.1. Training Data Combinations
3.2. Classification Assessment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Briggs, J.M.; Schaafsma, H.; Trenkov, D. Woody vegetation expansion in a desert grassland: Prehistoric human impact? J. Arid Environ. 2007, 69, 458–472. [Google Scholar] [CrossRef]
- Barger, N.N.; Archer, S.R.; Campbell, J.L.; Huang, C.Y.; Morton, J.A.; Knapp, A.K. Woody plant proliferation in North American drylands: A synthesis of impacts on ecosystem carbon balance. J. Geophys. Res. Biogeosci. 2011, 116. [Google Scholar] [CrossRef]
- Huxman, T.E.; Wilcox, B.P.; Breshears, D.D.; Scott, R.L.; Snyder, K.A.; Small, E.E.; Hultine, K.; Pockman, W.T.; Jackson, R.B. Ecohydrological implications of woody plant encroachment. Ecology 2005, 86, 308–319. [Google Scholar] [CrossRef]
- Eldridge, D.J.; Bowker, M.A.; Maestre, F.T.; Roger, E.; Reynolds, J.F.; Whitford, W.G. Impacts of shrub encroachment on ecosystem structure and functioning: Towards a global synthesis. Ecol. Lett. 2011, 14, 709–722. [Google Scholar] [CrossRef]
- Grover, H.D.; Musick, H.B. Shrubland encroachment in southern New Mexico, USA: An analysis of desertification processes in the American Southwest. Clim. Chang. 1990, 17, 305–330. [Google Scholar] [CrossRef]
- Goudie, A.S. Human Impact on the Natural Environment; John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Pacala, S.W.; Hurtt, G.C.; Baker, D.; Peylin, P.; Houghton, R.A.; Birdsey, R.A.; Heath, L.; Sundquist, E.T.; Stallard, R.F.; Ciais, P.; et al. Consistent land-and atmosphere-based US carbon sink estimates. Science 2001, 292, 2316–2320. [Google Scholar] [CrossRef] [Green Version]
- Pan, Y.; Birdsey, R.A.; Fang, J.; Houghton, R.; Kauppi, P.E.; Kurz, W.A.; Phillips, O.L.; Shvidenko, A.; Lewis, S.L.; Canadell, J.G.; et al. A large and persistent carbon sink in the world’s forests. Science 2011, 333, 988–993. [Google Scholar] [CrossRef] [Green Version]
- Jackson, R.B.; Banner, J.L.; Jobbágy, E.G.; Pockman, W.T.; Wall, D.H. Ecosystem carbon loss with woody plant invasion of grasslands. Nature 2002, 418, 623–626. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setala, H.; Van Der Putten, W.H.; Wall, D.H. Ecological linkages between aboveground and belowground biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef]
- David, J.A.; Kari, E.V.; Jacob, R.G.; Corinna, R.; Truman, P.Y. Pathways for Positive Cattle–Wildlife Interactions in Semiarid Rangelands. In Conserving Wildlife in African Landscapes: Kenya’s Ewaso Ecosystem; Smithsonian Contributions to Zoology: Washington, DC, USA, 2011; pp. 55–71. [Google Scholar] [CrossRef]
- Mairs, J.W. The use of remote sensing techniques to identify potential natural areas in oregon. Biol. Conserv. 1976, 9, 259–266. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Franklin, S.E. Remote Sensing for Sustainable Forest Management; CRC Press: Boca Raton, FL, USA, 2001. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Fusion of hyperspectral and LIDAR remote sensing data for classification of complex forest areas. IEEE Trans. Geosci. Remote Sens. 2008, 46, 1416–1427. [Google Scholar] [CrossRef] [Green Version]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Chopping, M.; Su, L.; Rango, A.; Martonchik, J.V.; Peters, D.P.; Laliberte, A. Remote sensing of woody shrub cover in desert grasslands using MISR with a geometric-optical canopy reflectance model. Remote Sens. Environ. 2008, 112, 19–34. [Google Scholar] [CrossRef]
- Sankey, T.; Donager, J.; McVay, J.; Sankey, J.B. UAV lidar and hyperspectral fusion for forest monitoring in the southwestern USA. Remote Sens. Environ. 2017, 195, 30–43. [Google Scholar] [CrossRef]
- Manfreda, S.; McCabe, M.F.; Miller, P.E.; Lucas, R.; Pajuelo Madrigal, V.; Mallinis, G.; Ben Dor, E.; Helman, D.; Estes, L.; Ciraolo, G.; et al. On the use of unmanned aerial systems for environmental monitoring. Remote Sens. 2018, 10, 641. [Google Scholar] [CrossRef] [Green Version]
- Anderson, J.E.; Plourde, L.C.; Martin, M.E.; Braswell, B.H.; Smith, M.L.; Dubayah, R.O.; Hofton, M.A.; Blair, J.B. Integrating waveform lidar with hyperspectral imagery for inventory of a northern temperate forest. Remote Sens. Environ. 2008, 112, 1856–1870. [Google Scholar] [CrossRef]
- Dandois, J.P.; Ellis, E.C. High spatial resolution three-dimensional mapping of vegetation spectral dynamics using computer vision. Remote Sens. Environ. 2013, 136, 259–276. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of wetland vegetation: A review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Franklin, J. Mapping Species Distributions: Spatial Inference and Prediction; Cambridge University Press: Cambridge, UK, 2010. [Google Scholar] [CrossRef]
- Strecha, C.; Fletcher, A.; Lechner, A.; Erskine, P.; Fua, P. Developing species specific vegetation maps using multi-spectral hyperspatial imagery from unmanned aerial vehicles. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2012, 3, 311–316. [Google Scholar] [CrossRef] [Green Version]
- Gong, P.; Pu, R.; Yu, B. Conifer species recognition: An exploratory analysis of in situ hyperspectral data. Remote Sens. Environ. 1997, 62, 189–200. [Google Scholar] [CrossRef]
- Mather, P.M.; Koch, M. Computer Processing of Remotely-Sensed Images: An Introduction; John Wiley & Sons: Hoboken, NJ, USA, 2011. [Google Scholar] [CrossRef]
- Plourde, L.C.; Ollinger, S.V.; Smith, M.L.; Martin, M.E. Estimating species abundance in a northern temperate forest using spectral mixture analysis. Photogramm. Eng. Remote Sens. 2007, 73, 829–840. [Google Scholar] [CrossRef] [Green Version]
- Fassnacht, F.E.; Latifi, H.; Stereńczak, K.; Modzelewska, A.; Lefsky, M.; Waser, L.T.; Straub, C.; Ghosh, A. Review of studies on tree species classification from remotely sensed data. Remote Sens. Environ. 2016, 186, 64–87. [Google Scholar] [CrossRef]
- Jones, T.G.; Coops, N.C.; Sharma, T. Assessing the utility of airborne hyperspectral and LiDAR data for species distribution mapping in the coastal Pacific Northwest, Canada. Remote Sens. Environ. 2010, 114, 2841–2852. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Holmgren, P.; Thuresson, T. Satellite remote sensing for forestry planning—A review. Scand. J. For. Res. 1998, 13, 90–110. [Google Scholar] [CrossRef]
- Chlingaryan, A.; Sukkarieh, S.; Whelan, B. Machine learning approaches for crop yield prediction and nitrogen status estimation in precision agriculture: A review. Comput. Electron. Agric. 2018, 151, 61–69. [Google Scholar] [CrossRef]
- Liu, L.; Coops, N.C.; Aven, N.W.; Pang, Y. Mapping urban tree species using integrated airborne hyperspectral and LiDAR remote sensing data. Remote Sens. Environ. 2017, 200, 170–182. [Google Scholar] [CrossRef]
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps based on the fusion of very high geometrical resolution multispectral/hyperspectral images and LiDAR data. Remote Sens. Environ. 2012, 123, 258–270. [Google Scholar] [CrossRef]
- Kim, M.; Madden, M.; Warner, T.A. Forest type mapping using object-specific texture measures from multi-spectral Ikonos imagery. Photogramm. Eng. Remote Sens. 2009, 75, 819–829. [Google Scholar] [CrossRef] [Green Version]
- Cheng, G.; Han, J.; Lu, X. Remote sensing image scene classification: Benchmark and state of the art. Proc. IEEE 2017, 105, 1865–1883. [Google Scholar] [CrossRef] [Green Version]
- Franklin, S.E.; Ahmed, O.S. Deciduous tree species classification using object-based analysis and machine learning with unmanned aerial vehicle multi-spectral data. Int. J. Remote Sens. 2018, 39, 5236–5245. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Caughlin, T.T.; Graves, S.J.; Asner, G.P.; Van Breugel, M.; Hall, J.S.; Martin, R.E.; Ashton, M.S.; Bohlman, S.A. A hyperspectral image can predict tropical tree growth rates in single-species stands. Ecol. Appl. 2016, 26, 2369–2375. [Google Scholar] [CrossRef]
- Dian, Y.; Li, Z.; Pang, Y. Spectral and texture features combined for forest tree species classification with airborne hyperspectral imagery. J. Indian Soc. Remote Sens. 2015, 43, 101–107. [Google Scholar] [CrossRef]
- Mitchell, J.J.; Shrestha, R.; Spaete, L.P.; Glenn, N.F. Combining airborne hyperspectral and LiDAR data across local sites for upscaling shrubland structural information: Lessons for HyspIRI. Remote Sens. Environ. 2015, 167, 98–110. [Google Scholar] [CrossRef]
- Shi, Y.; Skidmore, A.K.; Wang, T.; Holzwarth, S.; Heiden, U.; Pinnel, N.; Zhu, X.; Heurich, M. Tree species classification using plant functional traits from LiDAR and hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 207–219. [Google Scholar] [CrossRef]
- Mundt, J.T.; Streutker, D.R.; Glenn, N.F. Mapping sagebrush distribution using fusion of hyperspectral and lidar classifications. Photogramm. Eng. Remote Sens. 2006, 72, 47–54. [Google Scholar] [CrossRef] [Green Version]
- Debes, C.; Merentitis, A.; Heremans, R.; Hahn, J.; Frangiadakis, N.; van Kasteren, T.; Pacifici, F. Hyperspectral and LiDAR data fusion: Outcome of the 2013 GRSS data fusion contest. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 2405–2418. [Google Scholar] [CrossRef]
- Zhang, Z.; Kazakova, A.; Moskal, L.M.; Styers, D.M. Object-based tree species classification in urban ecosystems using LiDAR and hyperspectral data. Forests 2016, 7, 122. [Google Scholar] [CrossRef] [Green Version]
- Dashti, H.; Poley, A.; FGlenn, N.; Ilangakoon, N.; Spaete, L.; Roberts, D.; Enterkine, J.; Flores, A.N.; Ustin, S.L.; Mitchell, J.J. Regional scale dryland vegetation classification with an integrated lidar-hyperspectral approach. Remote Sens. 2019, 11, 2141. [Google Scholar] [CrossRef] [Green Version]
- Okin, G.S.; Roberts, D.A.; Murray, B.; Okin, W.J. Practical limits on hyperspectral vegetation discrimination in arid and semiarid environments. Remote Sens. Environ. 2001, 77, 212–225. [Google Scholar] [CrossRef]
- Myint, S.W.; Gober, P.; Brazel, A.; Grossman-Clarke, S.; Weng, Q. Per-pixel vs. object-based classification of urban land cover extraction using high spatial resolution imagery. Remote Sens. Environ. 2011, 115, 1145–1161. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Kennedy-Bowdoin, T.; Jones, M.O.; Martin, R.E.; Boardman, J.; Hughes, R.F. Invasive species detection in Hawaiian rainforests using airborne imaging spectroscopy and LiDAR. Remote Sens. Environ. 2008, 112, 1942–1955. [Google Scholar] [CrossRef]
- Stanturf, J.A.; Palik, B.J.; Dumroese, R.K. Contemporary forest restoration: A review emphasizing function. For. Ecol. Manag. 2014, 331, 292–323. [Google Scholar] [CrossRef]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree species classification using hyperspectral imagery: A comparison of two classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef] [Green Version]
- Modzelewska, A.; Kamińska, A.; Fassnacht, F.E.; Stereńczak, K. Multitemporal hyperspectral tree species classification in the Białowieża Forest World Heritage site. For. Int. J. For. Res. 2021, 94, 464–476. [Google Scholar] [CrossRef]
- Medina, A.L. The Santa Rita Experimental Range: History and Annotated Bibliography (1903–1988); DIANE Publishing: Collingdale, PA, USA, 1996. [Google Scholar]
- McClaran, M.P. Santa Rita Experimental Range: 100 Years (1903 to 2003) of Accomplishments and Contributions, Tucson, AZ, 30 October 2003–1 November 2003. In A Century of Vegetation Change on the Santa RITA Experimental Range; McClaran, M.P., Ffolliott, P.F., Edminster, C.B., Eds.; U.S. Department of Agriculture, Forest Service: Tucson, AZ, USA, 2003; pp. 16–33. Available online: https://www.fs.fed.us/rm/pubs/rmrs_p030/rmrs_p030_016_033.pdf (accessed on 1 June 2022).
- Schreiner-McGraw, A.P.; Vivoni, E.R.; Ajami, H.; Sala, O.E.; Throop, H.L.; Peters, D.P. Woody Plant encroachment has a larger impact than climate change on Dryland water budgets. Sci. Rep. 2020, 10, 8112. [Google Scholar] [CrossRef]
- NEON (National Ecological Observatory Network). Spectrometer Orthorectified Surface Directional Reflectance-Mosaic (DP3.30006.001), RELEASE-2022. Available online: https://data.neonscience.org/data-products/DP3.30006.001 (accessed on 3 December 2021).
- NEON (National Ecological Observatory Network). Discrete Return LiDAR Point Cloud (DP1.30003.001), RELEASE-2022. Available online: https://data.neonscience.org/data-products/DP1.30003.001 (accessed on 3 December 2021).
- Gillan, J.; Ponce-Campos, G.E.; Swetnam, T.L.; Gorlier, A.; Heilman, P.; McClaran, M.P. Innovations to expand drone data collection and analysis for rangeland monitoring. Ecosphere 2021, 12, e03649. [Google Scholar] [CrossRef]
- RStudio Team. RStudio: Integrated Development Environment for R; RStudio Team: Boston, MA, USA, 2015; Available online: http://www.rstudio.com/ (accessed on 1 January 2021).
- van Leeuwen, W.J. Visible, Near-IR, and Shortwave IR Spectral Characteristics of Terrestrial Surfaces; SAGE Publications Ltd.: London, UK, 2009; pp. 33–50. [Google Scholar] [CrossRef]
- Farella, M.M.; Barnes, M.L.; Breshears, D.D.; Mitchell, J.; van Leeuwen, W.J.; Gallery, R.E. Evaluation of vegetation indices and imaging spectroscopy to estimate foliar nitrogen across disparate biomes. Ecosphere 2022, 13, e3992. [Google Scholar] [CrossRef]
- Morsy, S.; Shaker, A.; El-Rabbany, A.; LaRocque, P.E. Airborne Multi-Spectral Lidar Data for Land-Cover Classification and Land/Water Mapping Using Different Spectral Indexes. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 3, 217–224. [Google Scholar] [CrossRef] [Green Version]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Bannari, A.; Morin, D.; Bonn, F.; Huete, A. A review of vegetation indices. Remote Sens. Rev. 1995, 13, 95–120. [Google Scholar] [CrossRef]
- Shakhatreh, H.; Sawalmeh, A.H.; Al-Fuqaha, A.; Dou, Z.; Almaita, E.; Khalil, I.; Othman, N.S.; Khreishah, A.; Guizani, M. Unmanned aerial vehicles (UAVs): A survey on civil applications and key research challenges. IEEE Access 2019, 7, 48572–48634. [Google Scholar] [CrossRef]
- Serrano, L.; Penuelas, J.; Ustin, S.L. Remote sensing of nitrogen and lignin in Mediterranean vegetation from AVIRIS data: Decomposing biochemical from structural signals. Remote Sens. Environ. 2002, 81, 355–364. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef] [Green Version]
- Gamon, J.; Serrano, L.; Surfus, J. The photochemical reflectance index: An optical indicator of photosynthetic radiation use efficiency across species, functional types, and nutrient levels. Oecologia 1997, 112, 492–501. [Google Scholar] [CrossRef]
- Sims, D.A.; Gamon, J.A. Relationships between leaf pigment content and spectral reflectance across a wide range of species, leaf structures and developmental stages. Remote Sens. Environ. 2002, 81, 337–354. [Google Scholar] [CrossRef]
- Suárez, L.; Zarco-Tejada, P.J.; Sepulcre-Cantó, G.; Pérez-Priego, O.; Miller, J.R.; Jiménez-Muñoz, J.C.; Sobrino, J. Assessing canopy PRI for water stress detection with diurnal airborne imagery. Remote Sens. Environ. 2008, 112, 560–575. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; González-Dugo, V.; Berni, J.A. Fluorescence, temperature and narrow-band indices acquired from a UAV platform for water stress detection using a micro-hyperspectral imager and a thermal camera. Remote Sens. Environ. 2012, 117, 322–337. [Google Scholar] [CrossRef]
- Hartfield, K.; Gillan, J.K.; Norton, C.L.; Conley, C.; van Leeuwen, W.J.D. A Novel Spectral Index to Identify Cacti in the Sonoran Desert at Multiple Scales Using Multi-Sensor Hyperspectral Data Acquisitions. Land 2022, 11, 786. [Google Scholar] [CrossRef]
- Dash, J.; Curran, P.J. MTCI: The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Viña, A.; Gitelson, A.A.; Nguy-Robertson, A.L.; Peng, Y. Comparison of different vegetation indices for the remote assessment of green leaf area index of crops. Remote Sens. Environ. 2011, 115, 3468–3478. [Google Scholar] [CrossRef]
- McMorrow, J.M.; Cutler, M.E.J.; Evans, M.G.; Al-Roichdi, A. Hyperspectral indices for characterizing upland peat composition. Int. J. Remote Sens. 2004, 25, 313–325. [Google Scholar] [CrossRef]
- Lowe, J.J.; Walker, M. Reconstructing Quaternary Environments; Routledge: London, UK, 2014. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Lyon, J.G. (Eds.) Hyperspectral Remote Sensing of Vegetation; CRC Press: Boca Raton, FL, USA, 2016. [Google Scholar] [CrossRef]
- Hively, W.D.; Lamb, B.T.; Daughtry, C.S.; Serbin, G.; Dennison, P.; Kokaly, R.F.; Wu, Z.; Masek, J.G. Evaluation of SWIR Crop Residue Bands for the Landsat Next Mission. Remote Sens. 2021, 13, 3718. [Google Scholar] [CrossRef]
- Jensen, J.R. Introductory Digital Image Processing: A Remote Sensing Perspective; Prentice Hall Press: Hoboken, NJ, USA, 2015. [Google Scholar]
- Congalton, R.G.; Green, K. Assessing the Accuracy of Remotely Sensed Data: Principles and Practices; CRC Press: Boca Raton, FL, USA, 2019. [Google Scholar] [CrossRef]
- Brantley, S.T.; Zinnert, J.C.; Young, D.R. Application of hyperspectral vegetation indices to detect variations in high leaf area index temperate shrub thicket canopies. Remote Sens. Environ. 2011, 115, 514–523. [Google Scholar] [CrossRef] [Green Version]
- Galvão, L.S.; Roberts, D.A.; Formaggio, A.R.; Numata, I.; Breunig, F.M. View angle effects on the discrimination of soybean varieties and on the relationships between vegetation indices and yield using off-nadir Hyperion data. Remote Sens. Environ. 2009, 113, 846–856. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plümer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Teluguntla, P.; Gumma, M.K.; Dheeravath, V. Hyperspectral Remote Sensing for Terrestrial Applications. In Land Resources Monitoring, Modeling, and Mapping with Remote Sensing; CRC Press: Boca Raton, FL, USA, 2015; pp. 201–233. ISBN 9781482217957. Available online: http://oar.icrisat.org/id/eprint/8611 (accessed on 1 June 2022).
- Adão, T.; Hruška, J.; Pádua, L.; Bessa, J.; Peres, E.; Morais, R.; Sousa, J.J. Hyperspectral imaging: A review on UAV-based sensors, data processing and applications for agriculture and forestry. Remote Sens. 2017, 9, 1110. [Google Scholar] [CrossRef] [Green Version]
- Roussel, J.R.; Auty, D.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.; Meador, A.S.; Bourdon, J.-F.; de Boissieu, F.; Achim, A. lidR: An R package for analysis of Airborne Laser Scanning (ALS) data. Remote Sens. Environ. 2020, 251, 112061. [Google Scholar] [CrossRef]
- Breiman, L.; Friedman, J.H.; Olshen, R.A.; Stone, C.J. Classification and Regression Trees; Routledge: London, UK, 2017. [Google Scholar] [CrossRef]
- Morgan, J.N.; Sonquist, J.A. Problems in the analysis of survey data, and a proposal. J. Am. Stat. Assoc. 1963, 58, 415–434. [Google Scholar] [CrossRef]
- Hastie, T.J.; Tibshirani, R.J. Generalized Additive Models; Routledge: London, UK, 2017. [Google Scholar] [CrossRef] [Green Version]
- Therneau, T.M.; Atkinson, E.J. An Introduction to Recursive Partitioning Using the RPART Routines; Technical Report; Mayo Foundation: Rochester, UK, 1997; Volume 61, p. 452. Available online: https://stat.ethz.ch/R-manual/R-patched/library/rpart/doc/longintro.pdf (accessed on 1 August 2021).
- Therneau, T.M.; Atkinson, E.J. An Introduction to Recursive Partitioning Using the RPART Routines; Mayo Foundation: Rochester, UK, 2015; Available online: https://cran.r-project.org/web/packages/rpart/vignettes/longintro.pdf (accessed on 1 August 2021).
- Ngai, E.W.; Xiu, L.; Chau, D.C. Application of data mining techniques in customer relationship management: A literature review and classification. Expert Syst. Appl. 2009, 36, 2592–2602. [Google Scholar] [CrossRef]
- Wamba, S.F.; Akter, S.; Edwards, A.; Chopin, G.; Gnanzou, D. How ‘big data’can make big impact: Findings from a systematic review and a longitudinal case study. Int. J. Prod. Econ. 2015, 165, 234–246. [Google Scholar] [CrossRef]
- Therneau, T.M.; Atkinson, B.; Ripley, M.B. The Rpart Package; R Foundation for Statistical Computing: Oxford, UK, 2010; Available online: https://cran.r-project.org/web/packages/rpart/index.html (accessed on 1 August 2021).
- Vapnik, V.N. The Nature of Statistical Learning Theory; Springer: New York, NY, USA, 1995; 188p. [Google Scholar] [CrossRef]
- Goodfellow, I.; Bengio, Y.; Courville, A. Deep Learning; MIT Press: Cambridge, MA, USA, 2016; Available online: http://www.deeplearningbook.org (accessed on 1 June 2022).
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Kuncheva, L.I. Combining Pattern Classifiers: Methods and Algorithms; John Wiley & Sons: Hoboken, NJ, USA, 2014. [Google Scholar] [CrossRef]
- Ge, G.; Shi, Z.; Zhu, Y.; Yang, X.; Hao, Y. Land use/cover classification in an arid desert-oasis mosaic landscape of China using remote sensed imagery: Performance assessment of four machine learning algorithms. Glob. Ecol. Conserv. 2020, 22, e00971. [Google Scholar] [CrossRef]
- Colgan, M.S.; Baldeck, C.A.; Féret, J.B.; Asner, G.P. Mapping savanna tree species at ecosystem scales using support vector machine classification and BRDF correction on airborne hyperspectral and LiDAR data. Remote Sens. 2012, 4, 3462–3480. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.B.; Sinca, F.; Vaughn, N.R.; Llactayo, W. Airborne laser-guided imaging spectroscopy to map forest trait diversity and guide conservation. Science 2017, 355, 385–389. [Google Scholar] [CrossRef]
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F.; Chang, C.-C.; Lin, C.-C.; Meyer, M.D. Package ‘e1071’. R J. 2019. Available online: https://cran.r-project.org/web/packages/e1071/e1071.pdf (accessed on 1 August 2021).
- Dimitriadou, E.; Hornik, K.; Leisch, F.; Meyer, D.; Weingessel, A.; Leisch, M.F. The e1071 Package. Misc Functions of Department of Statistics (e1071), TU Wien. 2006, pp. 297–304. Available online: https://www.researchgate.net/profile/Friedrich-Leisch-2/publication/221678005_E1071_Misc_Functions_of_the_Department_of_Statistics_E1071_TU_Wien/links/547305880cf24bc8ea19ad1d/E1071-Misc-Functions-of-the-Department-of-Statistics-E1071-TU-Wien.pdf (accessed on 1 June 2022).
- Torgo, L. Data Mining with R: Learning with CASE Studies; Chapman and Hall/CRC: Boca Raton, FL, USA, 2011. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Hsiao, C. Analysis of Panel Data; Cambridge University Press: Cambridge, UK, 2022. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Guan, H.; Li, J.; Chapman, M.; Deng, F.; Ji, Z.; Yang, X. Integration of orthoimagery and lidar data for object-based urban thematic mapping using random forests. Int. J. Remote Sens. 2013, 34, 5166–5186. [Google Scholar] [CrossRef]
- Ma, L.; Li, M.; Ma, X.; Cheng, L.; Du, P.; Liu, Y. A review of supervised object-based land-cover image classification. ISPRS J. Photogramm. Remote Sens. 2017, 130, 277–293. [Google Scholar] [CrossRef]
- Lawrence, R.L.; Wood, S.D.; Sheley, R.L. Mapping invasive plants using hyperspectral imagery and Breiman Cutler classifications (RandomForest). Remote Sens. Environ. 2006, 100, 356–362. [Google Scholar] [CrossRef]
- Rodriguez-Galiano, V.F.; Ghimire, B.; Rogan, J.; Chica-Olmo, M.; Rigol-Sanchez, J.P. An assessment of the effectiveness of a random forest classifier for land-cover classification. ISPRS J. Photogramm. Remote Sens. 2012, 67, 93–104. [Google Scholar] [CrossRef]
- James, G.; Witten, D.; Hastie, T.; Tibshirani, R. An Introduction to Statistical Learning; Springer: New York, NY, USA, 2013; Volume 112, p. 18. [Google Scholar]
- Lohr, S.L. Sampling: Design and Analysis; Chapman and Hall/CRC: Boca Raton, FL, USA, 2021. [Google Scholar] [CrossRef]
- Stone, M. Cross-validatory choice and assessment of statistical predictions. J. R. Stat. Soc. Ser. B (Methodol.) 1974, 36, 111–133. [Google Scholar] [CrossRef]
- Silverman, B.W. Density Estimation for Statistics and Data Analysis; Routledge: London, UK, 2018. [Google Scholar] [CrossRef]
- Ramezan, C.A.; Warner, T.A.; Maxwell, A.E. Evaluation of sampling and cross-validation tuning strategies for regional-scale machine learning classification. Remote Sens. 2019, 11, 185. [Google Scholar] [CrossRef] [Green Version]
- Ilia, I.; Loupasakis, C.; Tsangaratos, P. Land subsidence phenomena investigated by spatiotemporal analysis of groundwater resources, remote sensing techniques, and random forest method: The case of Western Thessaly, Greece. Environ. Monit. Assess. 2018, 190, 623. [Google Scholar] [CrossRef]
- Immitzer, M.; Atzberger, C.; Koukal, T. Tree species classification with random forest using very high spatial resolution 8-band WorldView-2 satellite data. Remote Sens. 2012, 4, 2661–2693. [Google Scholar] [CrossRef] [Green Version]
- Thanh Noi, P.; Kappas, M. Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using Sentinel-2 imagery. Sensors 2017, 18, 18. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Chehbouni, A.; Huete, A.R.; Kerr, Y.H.; Sorooshian, S. A modified soil adjusted vegetation index. Remote Sens. Environ. 1994, 48, 119–126. [Google Scholar] [CrossRef]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Shafri, H.Z.; Hamdan, N. Hyperspectral imagery for mapping disease infection in oil palm plantationusing vegetation indices and red edge techniques. Am. J. Appl. Sci. 2009, 6, 1031. [Google Scholar] [CrossRef] [Green Version]
- Sankaran, S.; Mishra, A.; Ehsani, R.; Davis, C. A review of advanced techniques for detecting plant diseases. Comput. Electron. Agric. 2010, 72, 1–13. [Google Scholar] [CrossRef]
- Alonzo, M.; Bookhagen, B.; Roberts, D.A. Urban tree species mapping using hyperspectral and lidar data fusion. Remote Sens. Environ. 2014, 148, 70–83. [Google Scholar] [CrossRef]
- Matsuki, T.; Yokoya, N.; Iwasaki, A. Hyperspectral tree species classification of Japanese complex mixed forest with the aid of LiDAR data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2177–2187. [Google Scholar] [CrossRef]
- Weil, G.; Lensky, I.M.; Resheff, Y.S.; Levin, N. Optimizing the timing of unmanned aerial vehicle image acquisition for applied mapping of woody vegetation species using feature selection. Remote Sens. 2017, 9, 1130. [Google Scholar] [CrossRef] [Green Version]
- Grybas, H.; Congalton, R.G. A Comparison of Multi-Temporal RGB and Multi-spectral UAS Imagery for Tree Species Classification in Heterogeneous New Hampshire Forests. Remote Sens. 2021, 13, 2631. [Google Scholar] [CrossRef]
- Wolter, P.T.; Mladenoff, D.J.; Host, G.E.; Crow, T.R. Using multi-temporal landsat imagery. Photogramm. Eng. Remote Sens. 1995, 61, 1129–1143. Available online: https://www.mnatlas.org/metadata/arrow95_14.pdf (accessed on 1 June 2022).
- Mickelson, J.G.; Civco, D.L.; Silander, J.A. Delineating forest canopy species in the northeastern United States using multi-temporal TM imagery. Photogramm. Eng. Remote Sens. 1998, 64, 891–904. Available online: https://www.asprs.org/wp-content/uploads/pers/1998journal/sep/1998_sep_891-904.pdf (accessed on 1 June 2022).
- Key, T.; Warner, T.A.; McGraw, J.B.; Fajvan, M.A. A comparison of multi-spectral and multitemporal information in high spatial resolution imagery for classification of individual tree species in a temperate hardwood forest. Remote Sens. Environ. 2001, 75, 100–112. Available online: http://www.as.wvu.edu/~jmcgraw/JBMPersonalSite/2001KeyEtAl.pdf (accessed on 1 June 2022). [CrossRef]
- Clark, M.L.; Roberts, D.A.; Clark, D.B. Hyperspectral discrimination of tropical rain forest tree species at leaf to crown scales. Remote Sens. Environ. 2005, 96, 375–398. [Google Scholar] [CrossRef]
- Yang, H.; Du, J. Classification of desert steppe species based on unmanned aerial vehicle hyperspectral remote sensing and continuum removal vegetation indices. Optik 2021, 247, 167877. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.E.; Shimabukuro, Y.E.; de Souza Filho, C.R. Tree species classification in tropical forests using visible to shortwave infrared WorldView-3 images and texture analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]
- Nezami, S.; Khoramshahi, E.; Nevalainen, O.; Pölönen, I.; Honkavaara, E. Tree species classification of drone hyperspectral and RGB imagery with deep learning convolutional neural networks. Remote Sens. 2020, 12, 1070. [Google Scholar] [CrossRef] [Green Version]
- Takahashi Miyoshi, G.; Imai, N.N.; Garcia Tommaselli, A.M.; Antunes de Moraes, M.V.; Honkavaara, E. Evaluation of hyperspectral multitemporal information to improve tree species identification in the highly diverse atlantic forest. Remote Sensing 2020, 12, 244. [Google Scholar] [CrossRef] [Green Version]
- Laliberte, A.S.; Rango, A.; Havstad, K.M.; Paris, J.F.; Beck, R.F.; McNeely, R.; Gonzalez, A.L. Object-oriented image analysis for mapping shrub encroachment from 1937 to 2003 in southern New Mexico. Remote Sens. Environ. 2004, 93, 198–210. [Google Scholar] [CrossRef]
- Blaschke, T. Object based image analysis for remote sensing. ISPRS J. Photogramm. Remote Sens. 2010, 65, 2–16. [Google Scholar] [CrossRef] [Green Version]
- Hantson, W.; Kooistra, L.; Slim, P.A. Mapping invasive woody species in coastal dunes in the N etherlands: A remote sensing approach using LIDAR and high-resolution aerial photographs. Appl. Veg. Sci. 2012, 15, 536–547. [Google Scholar] [CrossRef]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual tree crown segmentation and classification of 13 tree species using airborne hyperspectral data. Remote Sens. 2018, 10, 1218. [Google Scholar] [CrossRef] [Green Version]
- Oddi, L.; Cremonese, E.; Ascari, L.; Filippa, G.; Galvagno, M.; Serafino, D.; Cella, U.M.d. Using UAV Imagery to Detect and Map Woody Species Encroachment in a Subalpine Grassland: Advantages and Limits. Remote Sens. 2021, 13, 1239. [Google Scholar] [CrossRef]




| HDF5 Band | TIFF Band | Wavelength (nm) | Indices | Equation |
|---|---|---|---|---|
| 31 | 4 | 531 | NDVI | |
| 35 | 5 | 551 | NDWI | |
| 38 | 6 | 564 | PRI | |
| 57 | 10 | 661 | PRI2 | |
| 59 | 11 | 671 | SWIRI | |
| 67 | 13 | 711 | SAVI | |
| 75 | 14 | 751 | CACTI | |
| 97 | 16 | 862 | CACTI2 | |
| 119 | 17 | 970 | MTCI | |
| 139 | 18 | 1070 | CI | |
| 173 | 20 | 1242 | CAI | |
| 226 | 22 | 1508 | NDNI | |
| 234 | 23 | 1548 | ||
| 254 | 25 | 1648 | ||
| 260 | 27 | 1678 | ||
| 324 | 32 | 2000 | ||
| 351 | 34 | 2134 | ||
| 366 | 36 | 2210 |
| All Years Indices + Lidar | SVM | RF | CART |
|---|---|---|---|
| OA | 77.48 | 95.28 | 88.55 |
| Kappa | 72.14 | 94.17 | 85.88 |
| Processing Time (min) | 55.46 | 52.30 | 46.27 |
| Training Data Combinations | OA | Kappa |
|---|---|---|
| All Years Indices + Lidar | 95.28 | 94.17 |
| 2019 Indices + Lidar | 92.62 | 90.88 |
| 2018 Indices + Lidar | 92.62 | 90.88 |
| 2017 Indices + Lidar | 89.11 | 86.90 |
| All Years Indices | 93.19 | 91.57 |
| 2019 Indices | 90.61 | 88.41 |
| 2018 Indices | 88.76 | 86.99 |
| 2017 Indices | 87.78 | 84.87 |
| All Years Indices + LiDAR | Bare Ground | Grass | Mesquite | Cactus | Lotebush | Paloverde | Creosote | Sum | UA |
| Bare ground | 365 | 2 | 0 | 1 | 0 | 0 | 0 | 368 | 99.18 |
| Grass | 5 | 287 | 0 | 1 | 2 | 0 | 2 | 297 | 96.63 |
| Mesquite | 0 | 0 | 397 | 1 | 32 | 5 | 2 | 437 | 90.85 |
| Cactus | 1 | 2 | 2 | 1068 | 1 | 3 | 1 | 1078 | 99.07 |
| Lotebush | 0 | 2 | 43 | 1 | 242 | 2 | 7 | 297 | 81.48 |
| Paloverde | 0 | 0 | 8 | 10 | 2 | 285 | 1 | 306 | 93.14 |
| Creosote | 0 | 1 | 1 | 5 | 2 | 0 | 350 | 359 | 97.49 |
| Sum | 371 | 294 | 451 | 1087 | 281 | 295 | 363 | 3142 | |
| PA | 98.38 | 97.62 | 88.03 | 98.25 | 86.12 | 96.61 | 96.42 | ||
| 2017 Indices | Bare Ground | Grass | Mesquite | Cactus | Lotebush | Paloverde | Creosote | Sum | UA |
| Bare ground | 351 | 4 | 0 | 10 | 0 | 0 | 3 | 368 | 95.38 |
| Grass | 8 | 249 | 5 | 14 | 6 | 1 | 14 | 297 | 83.84 |
| Mesquite | 0 | 3 | 370 | 1 | 50 | 8 | 5 | 437 | 84.67 |
| Cactus | 12 | 21 | 10 | 1022 | 0 | 9 | 4 | 1078 | 94.81 |
| Lotebush | 0 | 5 | 105 | 2 | 175 | 1 | 9 | 297 | 58.92 |
| Paloverde | 0 | 4 | 13 | 16 | 4 | 266 | 3 | 306 | 86.93 |
| Creosote | 13 | 5 | 2 | 10 | 3 | 1 | 325 | 359 | 90.53 |
| Sum | 384 | 291 | 505 | 1075 | 238 | 286 | 363 | 3142 | |
| PA | 91.41 | 85.57 | 73.27 | 95.07 | 73.53 | 93.01 | 89.53 |
| All Years Indices + LiDAR | MDG | 2017 Indices | MDG |
|---|---|---|---|
| CACTI_2019 | 131.3818 | CACTI_2017 | 291.5153 |
| CACTI_2018 | 128.0493 | NDVI_2017 | 266.4296 |
| CHM_2018 | 118.9476 | CACTI2_2017 | 260.4622 |
| CACTI2_2019 | 105.366 | CI_2017 | 240.1875 |
| CACTI2_2018 | 97.37399 | NDWI_2017 | 168.1216 |
| NDVI_2018 | 83.49702 | SWIRI_2017 | 151.0334 |
| CI_2018 | 75.12076 | NDNI_2017 | 140.9909 |
| NDVI_2019 | 74.52218 | SAVI_2017 | 124.6892 |
| NDWI_2019 | 71.98403 | PRI2_2017 | 115.7335 |
| NDWI_2018 | 61.93623 | MTCI_2017 | 114.6043 |
| NDNI_2018 | 58.84034 | CAI_2017 | 86.10462 |
| SWIRI_2019 | 55.42875 | PRI_2017 | 72.31956 |
| CI_2019 | 52.13248 | CACTI_2017 | 291.5153 |
| CAI_2019 | 51.43631 | NDVI_2017 | 266.4296 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Norton, C.L.; Hartfield, K.; Collins, C.D.H.; van Leeuwen, W.J.D.; Metz, L.J. Multi-Temporal LiDAR and Hyperspectral Data Fusion for Classification of Semi-Arid Woody Cover Species. Remote Sens. 2022, 14, 2896. https://doi.org/10.3390/rs14122896
Norton CL, Hartfield K, Collins CDH, van Leeuwen WJD, Metz LJ. Multi-Temporal LiDAR and Hyperspectral Data Fusion for Classification of Semi-Arid Woody Cover Species. Remote Sensing. 2022; 14(12):2896. https://doi.org/10.3390/rs14122896
Chicago/Turabian StyleNorton, Cynthia L., Kyle Hartfield, Chandra D. Holifield Collins, Willem J. D. van Leeuwen, and Loretta J. Metz. 2022. "Multi-Temporal LiDAR and Hyperspectral Data Fusion for Classification of Semi-Arid Woody Cover Species" Remote Sensing 14, no. 12: 2896. https://doi.org/10.3390/rs14122896








