Estimating Community-Level Plant Functional Traits in a Species-Rich Alpine Meadow Using UAV Image Spectroscopy
Abstract
:1. Introduction
2. Materials and Methods
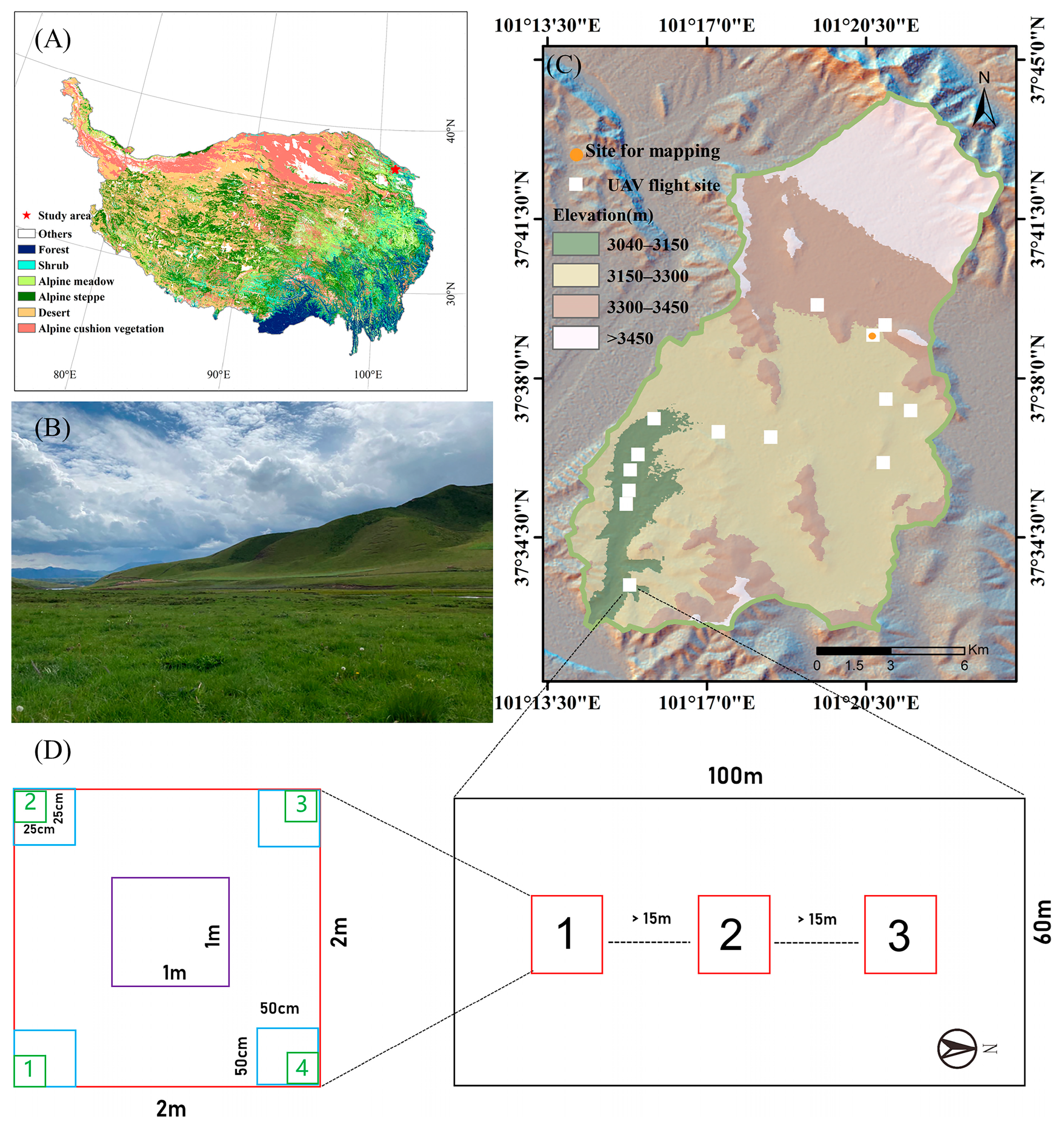
2.1. Study Area
2.2. Hyperspectral Data Collection and Pre-Processing
2.3. Field Data Collection
2.4. Foliar Trait Measurements and Plant Community Trait Calculation
2.5. Mapping Plant Community Traits
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Violle, C.; Navas, M.-L.; Vile, D.; Kazakou, E.; Fortunel, C.; Hummel, I.; Garnier, E. Let the Concept of Trait Be Functional! Oikos 2007, 116, 882–892. [Google Scholar] [CrossRef]
- Laughlin, D.C.; Messier, J. Fitness of Multidimensional Phenotypes in Dynamic Adaptive Landscapes. Trends Ecol. Evol. 2015, 30, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The Worldwide Leaf Economics Spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Díaz, S.; Lavorel, S.; de Bello, F.; Quétier, F.; Grigulis, K.; Robson, T.M. Incorporating Plant Functional Diversity Effects in Ecosystem Service Assessments. Proc. Natl. Acad. Sci. USA 2007, 104, 20684–20689. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bruelheide, H.; Dengler, J.; Purschke, O.; Lenoir, J.; Jiménez-Alfaro, B.; Hennekens, S.M.; Botta-Dukát, Z.; Chytrý, M.; Field, R.; Jansen, F.; et al. Global Trait–Environment Relationships of Plant Communities. Nat. Ecol. Evol. 2018, 2, 1906–1917. [Google Scholar] [CrossRef]
- Gillison, A.N. Plant Functional Types and Traits at the Community, Ecosystem and World Level. In Vegetation Ecology; van der Maarel, E., Franklin, J., Eds.; John Wiley & Sons, Ltd.: Oxford, UK, 2013; pp. 347–386. ISBN 978-1-118-45259-2. [Google Scholar]
- Muscarella, R.; Uriarte, M. Do Community-Weighted Mean Functional Traits Reflect Optimal Strategies? Proc. R. Soc. B Biol. Sci. 2016, 283, 20152434. [Google Scholar] [CrossRef]
- Bjorkman, A.D.; Myers-Smith, I.H.; Elmendorf, S.C.; Normand, S.; Rüger, N.; Beck, P.S.A.; Blach-Overgaard, A.; Blok, D.; Cornelissen, J.H.C.; Forbes, B.C.; et al. Plant Functional Trait Change across a Warming Tundra Biome. Nature 2018, 562, 57–62. [Google Scholar] [CrossRef]
- Tang, Z.; Xu, W.; Zhou, G.; Bai, Y.; Li, J.; Tang, X.; Chen, D.; Liu, Q.; Ma, W.; Xiong, G.; et al. Patterns of Plant Carbon, Nitrogen, and Phosphorus Concentration in Relation to Productivity in China’s Terrestrial Ecosystems. Proc. Natl. Acad. Sci. USA 2018, 115, 4033–4038. [Google Scholar] [CrossRef] [Green Version]
- Ackerly, D.D.; Cornwell, W.K. A Trait-Based Approach to Community Assembly: Partitioning of Species Trait Values into within- and among-Community Components. Ecol. Lett. 2007, 10, 135–145. [Google Scholar] [CrossRef]
- Khalil, M.I.; Gibson, D.J.; Baer, S.G. Functional Response of Subordinate Species to Intraspecific Trait Variability within Dominant Species. J. Ecol. 2019, 107, 2040–2053. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Lyon, J.G.; Huete, A. (Eds.) Fundamentals, Sensor Systems, Spectral Libraries, and Data Mining for Vegetation: Hyperspectral Remote Sensing of Vegetation, 2nd ed.; CRC Press: Boca Raton, FL, USA, 2018; ISBN 978-1-315-16415-1. [Google Scholar]
- Pettorelli, N.; Wegmann, M.; Skidmore, A.; Mücher, S.; Dawson, T.P.; Fernandez, M.; Lucas, R.; Schaepman, M.E.; Wang, T.; O’Connor, B.; et al. Framing the Concept of Satellite Remote Sensing Essential Biodiversity Variables: Challenges and Future Directions. Remote Sens. Ecol. Conserv. 2016, 2, 122–131. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of Optical-Based Remote Sensing for Plant Trait Mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Singh, A.; Serbin, S.P.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Imaging Spectroscopy Algorithms for Mapping Canopy Foliar Chemical and Morphological Traits and Their Uncertainties. Ecol. Appl. 2015, 25, 2180–2197. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Gamon, J.A.; Emmerton, C.A.; Li, H.; Nestola, E.; Pastorello, G.Z.; Menzer, O. Integrated Analysis of Productivity and Biodiversity in a Southern Alberta Prairie. Remote Sens. 2016, 8, 214. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying Forest Canopy Traits: Imaging Spectroscopy versus Field Survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Zhao, Y.; Sun, Y.; Lu, X.; Zhao, X.; Yang, L.; Sun, Z.; Bai, Y. Hyperspectral Retrieval of Leaf Physiological Traits and Their Links to Ecosystem Productivity in Grassland Monocultures. Ecol. Indic. 2021, 122, 107267. [Google Scholar] [CrossRef]
- Wang, Z.; Townsend, P.A.; Schweiger, A.K.; Couture, J.J.; Singh, A.; Hobbie, S.E.; Cavender-Bares, J. Mapping Foliar Functional Traits and Their Uncertainties across Three Years in a Grassland Experiment. Remote Sens. Environ. 2019, 221, 405–416. [Google Scholar] [CrossRef]
- Li, C.; Wulf, H.; Schmid, B.; He, J.-S.; Schaepman, M.E. Estimating Plant Traits of Alpine Grasslands on the Qinghai-Tibetan Plateau Using Remote Sensing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 2263–2275. [Google Scholar] [CrossRef]
- Pajares, G. Overview and Current Status of Remote Sensing Applications Based on Unmanned Aerial Vehicles (UAVs). Photogramm. Eng. Remote Sens. 2015, 81, 281–330. [Google Scholar] [CrossRef] [Green Version]
- Rossi, C.; Kneubuehler, M.; Schuetz, M.; Schaepman, M.; Haller, R.; Risch, A. Spatial Resolution, Spectral Metrics and Biomass Are Key Aspects in Estimating Plant Species Richness from Spectral Diversity in Species-Rich Grasslands. Remote Sens. Ecol. Conserv. 2021, 8, 297–314. [Google Scholar] [CrossRef]
- de Castro, A.I.; Shi, Y.; Maja, J.M.; Peña, J.M. UAVs for Vegetation Monitoring: Overview and Recent Scientific Contributions. Remote Sens. 2021, 13, 2139. [Google Scholar] [CrossRef]
- Aasen, H.; Honkavaara, E.; Lucieer, A.; Zarco-Tejada, P.J. Quantitative Remote Sensing at Ultra-High Resolution with UAV Spectroscopy: A Review of Sensor Technology, Measurement Procedures, and Data Correction Workflows. Remote Sens. 2018, 10, 1091. [Google Scholar] [CrossRef] [Green Version]
- Jetz, W.; Cavender-Bares, J.; Pavlick, R.; Schimel, D.; Davis, F.W.; Asner, G.P.; Guralnick, R.; Kattge, J.; Latimer, A.M.; Moorcroft, P.; et al. Monitoring Plant Functional Diversity from Space. Nat. Plants 2016, 2, 16024. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Willis, C.G.; Klein, J.A.; Ma, Z.; Li, J.; Zhou, H.; Zhao, X. Recovery of Plant Species Diversity during Long-Term Experimental Warming of a Species-Rich Alpine Meadow Community on the Qinghai-Tibet Plateau. Biol. Conserv. 2017, 213, 218–224. [Google Scholar] [CrossRef]
- Hadley, B.C.; Garcia-Quijano, M.; Jensen, J.R.; Tullis, J.A. Empirical versus Model-based Atmospheric Correction of Digital Airborne Imaging Spectrometer Hyperspectral Data. Geocarto Int. 2005, 20, 21–28. [Google Scholar] [CrossRef]
- Arroyo-Mora, J.P.; Kalacska, M.; Løke, T.; Schläpfer, D.; Coops, N.C.; Lucanus, O.; Leblanc, G. Assessing the Impact of Illumination on UAV Pushbroom Hyperspectral Imagery Collected under Various Cloud Cover Conditions. Remote Sens. Environ. 2021, 258, 112396. [Google Scholar] [CrossRef]
- Jones, J.B., Jr. Laboratory Guide for Conducting Soil Tests and Plant Analysis; CRC press: New York, NY, USA, 2001; ISBN 1-4200-2529-5. [Google Scholar]
- Roelofsen, H.D.; Van Bodegom, P.M.; Kooistra, L.; Witte, J.-P.M. Trait Estimation in Herbaceous Plant Assemblages from in Situ Canopy Spectra. Remote Sens. 2013, 5, 6323–6345. [Google Scholar] [CrossRef]
- Wang, Z.; Skidmore, A.K.; Darvishzadeh, R.; Heiden, U.; Heurich, M.; Wang, T. Leaf Nitrogen Content Indirectly Estimated by Leaf Traits Derived From the PROSPECT Model. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 3172–3182. [Google Scholar] [CrossRef]
- Schweiger, A.K.; Cavender-Bares, J.; Townsend, P.A.; Hobbie, S.E.; Madritch, M.D.; Wang, R.; Tilman, D.; Gamon, J.A. Plant Spectral Diversity Integrates Functional and Phylogenetic Components of Biodiversity and Predicts Ecosystem Function. Nat. Ecol. Evol. 2018, 2, 976–982. [Google Scholar] [CrossRef]
- Durán, S.M.; Martin, R.E.; Díaz, S.; Maitner, B.S.; Malhi, Y.; Salinas, N.; Shenkin, A.; Silman, M.R.; Wieczynski, D.J.; Asner, G.P.; et al. Informing Trait-Based Ecology by Assessing Remotely Sensed Functional Diversity across a Broad Tropical Temperature Gradient. Sci. Adv. 2019, 5, eaaw8114. [Google Scholar] [CrossRef] [Green Version]
- Rosipal, R.; Krämer, N. Overview and Recent Advances in Partial Least Squares. In Subspace, Latent Structure and Feature Selection; Saunders, C., Grobelnik, M., Gunn, S., Shawe-Taylor, J., Eds.; Lecture Notes in Computer Science; Springer: Berlin, Heidelberg, Germany, 2006; Volume 3940, pp. 34–51. ISBN 978-3-540-34137-6. [Google Scholar]
- Mehmood, T.; Liland, K.H.; Snipen, L.; Sæbø, S. A Review of Variable Selection Methods in Partial Least Squares Regression. Chemom. Intell. Lab. Syst. 2012, 118, 62–69. [Google Scholar] [CrossRef]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A Review of Methods to Deal with It and a Simulation Study Evaluating Their Performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Hasegawa, K.; Miyashita, Y.; Funatsu, K. GA Strategy for Variable Selection in QSAR Studies: GA-Based PLS Analysis of Calcium Channel Antagonists. J. Chem. Inf. Comput. Sci. 1997, 37, 306–310. [Google Scholar] [CrossRef]
- Santos-Rufo, A.; Mesas-Carrascosa, F.-J.; García-Ferrer, A.; Meroño-Larriva, J.E. Wavelength Selection Method Based on Partial Least Square from Hyperspectral Unmanned Aerial Vehicle Orthomosaic of Irrigated Olive Orchards. Remote Sens. 2020, 12, 3426. [Google Scholar] [CrossRef]
- Mehmood, T.; Sæbø, S.; Liland, K.H. Comparison of Variable Selection Methods in Partial Least Squares Regression. J. Chemom. 2020, 34, e3226. [Google Scholar] [CrossRef] [Green Version]
- Moreno-Martínez, Á.; Camps-Valls, G.; Kattge, J.; Robinson, N.; Reichstein, M.; van Bodegom, P.; Kramer, K.; Cornelissen, J.H.C.; Reich, P.; Bahn, M.; et al. A Methodology to Derive Global Maps of Leaf Traits Using Remote Sensing and Climate Data. Remote Sens. Environ. 2018, 218, 69–88. [Google Scholar] [CrossRef] [Green Version]
- Loozen, Y.; Rebel, K.T.; de Jong, S.M.; Lu, M.; Ollinger, S.V.; Wassen, M.J.; Karssenberg, D. Mapping Canopy Nitrogen in European Forests Using Remote Sensing and Environmental Variables with the Random Forests Method. Remote Sens. Environ. 2020, 247, 111933. [Google Scholar] [CrossRef]
- Chemura, A.; Mutanga, O.; Odindi, J.; Kutywayo, D. Mapping Spatial Variability of Foliar Nitrogen in Coffee (Coffea arabica L.) Plantations with Multispectral Sentinel-2 MSI Data. ISPRS J. Photogramm. Remote Sens. 2018, 138, 1–11. [Google Scholar] [CrossRef]
- Yang, X.; Yang, R.; Ye, Y.; Yuan, Z.; Wang, D.; Hua, K. Winter Wheat SPAD Estimation from UAV Hyperspectral Data Using Cluster-Regression Methods. Int. J. Appl. Earth Obs. Geoinf. 2021, 105, 102618. [Google Scholar] [CrossRef]
- Mevik, B.-H.; Wehrens, R. The Pls Package: Principal Component and Partial Least Squares Regression in R. J. Stat. Softw. 2007, 18, 1–23. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; Hong, X.; Harris, C.J.; Sharkey, P.M. Sparse Modeling Using Orthogonal Forward Regression with PRESS Statistic and Regularization. IEEE Trans. Syst. Man Cybern. Part B 2004, 34, 898–911. [Google Scholar] [CrossRef] [PubMed]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; He, T.; Benesty, M.; Khotilovich, V.; Tang, Y.; Cho, H.; Chen, K.; Mitchell, R.; Cano, I.; Zhou, T.; et al. xgboost: Extreme Gradient Boosting; R Package Version 1.6.0.1; 2022; Available online: https://CRAN.R-project.org/package=xgboost (accessed on 24 May 2022).
- R Core Team. R: A Language and Environment for Statistical Computing; R Core Team: Vienna, Austria, 2019. [Google Scholar]
- Ferwerda, J.G.; Skidmore, A.K.; Mutanga, O. Nitrogen Detection with Hyperspectral Normalized Ratio Indices across Multiple Plant Species. Int. J. Remote Sens. 2005, 26, 4083–4095. [Google Scholar] [CrossRef]
- Ollinger, S.V. Sources of Variability in Canopy Reflectance and the Convergent Properties of Plants. New Phytol. 2011, 189, 375–394. [Google Scholar] [CrossRef] [PubMed]
- Ramoelo, A.; Skidmore, A.K.; Cho, M.A.; Schlerf, M.; Mathieu, R.; Heitkönig, I.M.A. Regional Estimation of Savanna Grass Nitrogen Using the Red-Edge Band of the Spaceborne RapidEye Sensor. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 151–162. [Google Scholar] [CrossRef]
- Zhang, J.; Cheng, T.; Guo, W.; Xu, X.; Qiao, H.; Xie, Y.; Ma, X. Leaf Area Index Estimation Model for UAV Image Hyperspectral Data Based on Wavelength Variable Selection and Machine Learning Methods. Plant Methods 2021, 17, 49. [Google Scholar] [CrossRef]
- Rivera-Caicedo, J.P.; Verrelst, J.; Muñoz-Marí, J.; Camps-Valls, G.; Moreno, J. Hyperspectral Dimensionality Reduction for Biophysical Variable Statistical Retrieval. ISPRS J. Photogramm. Remote Sens. 2017, 132, 88–101. [Google Scholar] [CrossRef]
- Zhao, X.; Pan, X.; Ma, Y.; Yan, W. Research on Forage Hyperspectral Image Recognition Based on F-SVD and XGBoost. MATEC Web Conf. 2021, 336, 06027. [Google Scholar] [CrossRef]





| Trait | PLSR | GA-PLSR | RF | XGBoost | ||||
|---|---|---|---|---|---|---|---|---|
| R2 | nRMSE | R2 | nRMSE | R2 | nRMSE | R2 | nRMSE | |
| chlorophyll a | 0.58 | 20.3% | 0.79 | 10.7% | 0.28 | 38.7% | 0.51 | 22.0% |
| chlorophyll b | 0.60 | 18.5% | 0.83 | 10.7% | 0.25 | 37.9% | 0.39 | 32.0% |
| carotenoid | 0.51 | 22.2% | 0.64 | 16.0% | 0.25 | 40.7% | 0.62 | 15.8% |
| specific leaf area | 0.52 | 15.1% | 0.70 | 12.8% | 0.26 | 41.3% | 0.34 | 25.6% |
| leaf thickness | 0.34 | 24.5% | 0.68 | 13.5% | 0.07 | 55.3% | 0.20 | 50.0% |
| plant height | 0.20 | 31.4% | 0.44 | 25.3% | 0.32 | 39.3% | 0.40 | 95.7% |
| phosphorus content | 0.31 | 20.7% | 0.54 | 19.5% | 0.20 | 40.3% | 0.29 | 28.7% |
| nitrogen content | 0.03 | 62.0% | 0.50 | 22.3% | 0.06 | 47.3% | 0.14 | 44.1% |
| starch content | 0.47 | 18.4% | 0.68 | 13.5% | 0.04 | 51.9% | 0.07 | 37.6% |
| leaf dry matter content | 0.05 | 59.8% | 0.30 | 24.2% | 0.07 | 74.4% | 0.09 | 44.9% |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Y.-W.; Wang, T.; Guo, Y.; Skidmore, A.; Zhang, Z.; Tang, R.; Song, S.; Tang, Z. Estimating Community-Level Plant Functional Traits in a Species-Rich Alpine Meadow Using UAV Image Spectroscopy. Remote Sens. 2022, 14, 3399. https://doi.org/10.3390/rs14143399
Zhang Y-W, Wang T, Guo Y, Skidmore A, Zhang Z, Tang R, Song S, Tang Z. Estimating Community-Level Plant Functional Traits in a Species-Rich Alpine Meadow Using UAV Image Spectroscopy. Remote Sensing. 2022; 14(14):3399. https://doi.org/10.3390/rs14143399
Chicago/Turabian StyleZhang, Yi-Wei, Tiejun Wang, Yanpei Guo, Andrew Skidmore, Zhenhua Zhang, Rong Tang, Shanshan Song, and Zhiyao Tang. 2022. "Estimating Community-Level Plant Functional Traits in a Species-Rich Alpine Meadow Using UAV Image Spectroscopy" Remote Sensing 14, no. 14: 3399. https://doi.org/10.3390/rs14143399
APA StyleZhang, Y.-W., Wang, T., Guo, Y., Skidmore, A., Zhang, Z., Tang, R., Song, S., & Tang, Z. (2022). Estimating Community-Level Plant Functional Traits in a Species-Rich Alpine Meadow Using UAV Image Spectroscopy. Remote Sensing, 14(14), 3399. https://doi.org/10.3390/rs14143399







