Use of Remote Sensing Techniques to Estimate Plant Diversity within Ecological Networks: A Worked Example
Abstract
1. Introduction
2. Materials and Methods
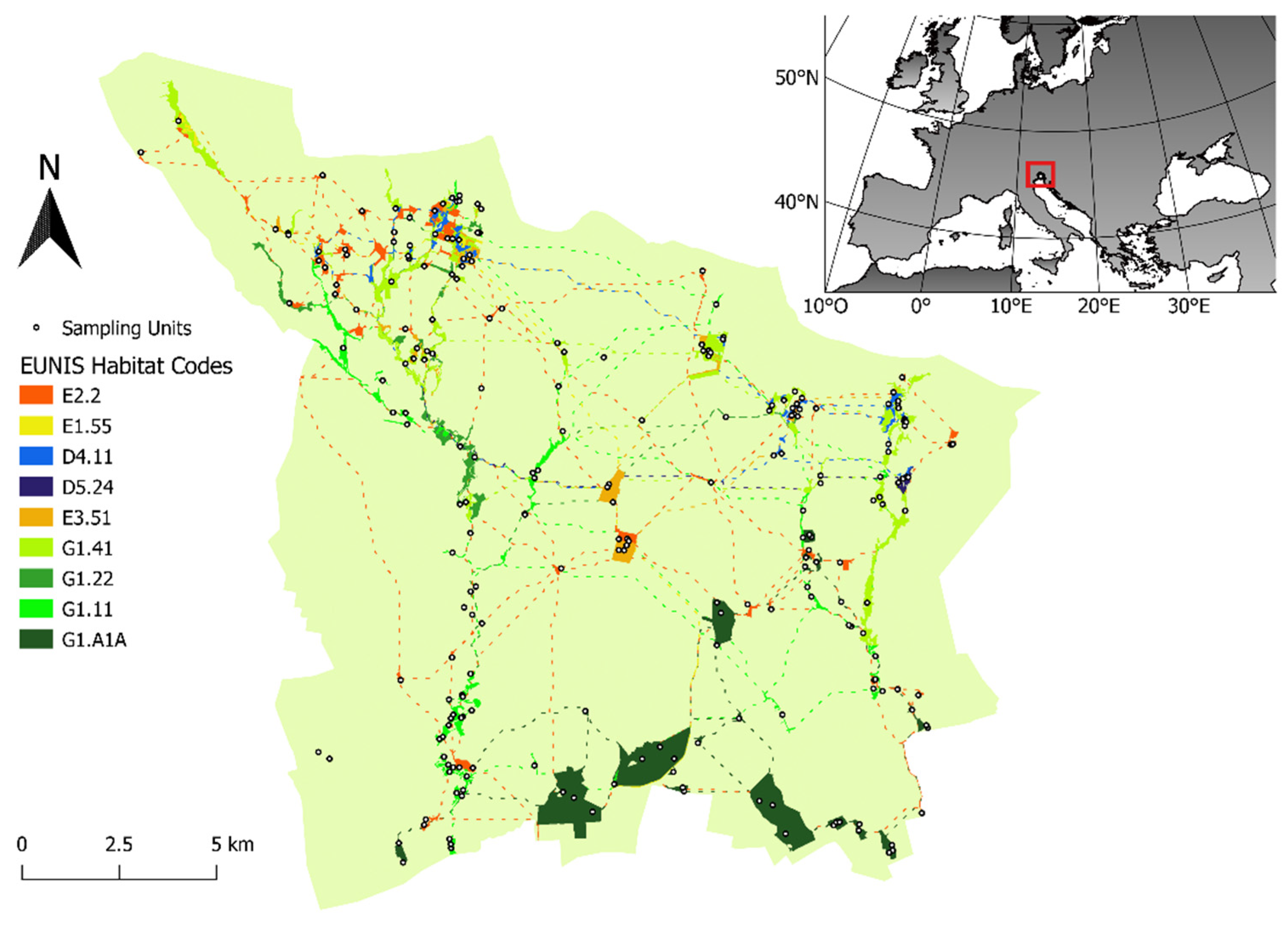
2.1. Study Site
2.2. Data Collection and Analysis
2.2.1. Sampling Units
2.2.2. Satellite Data
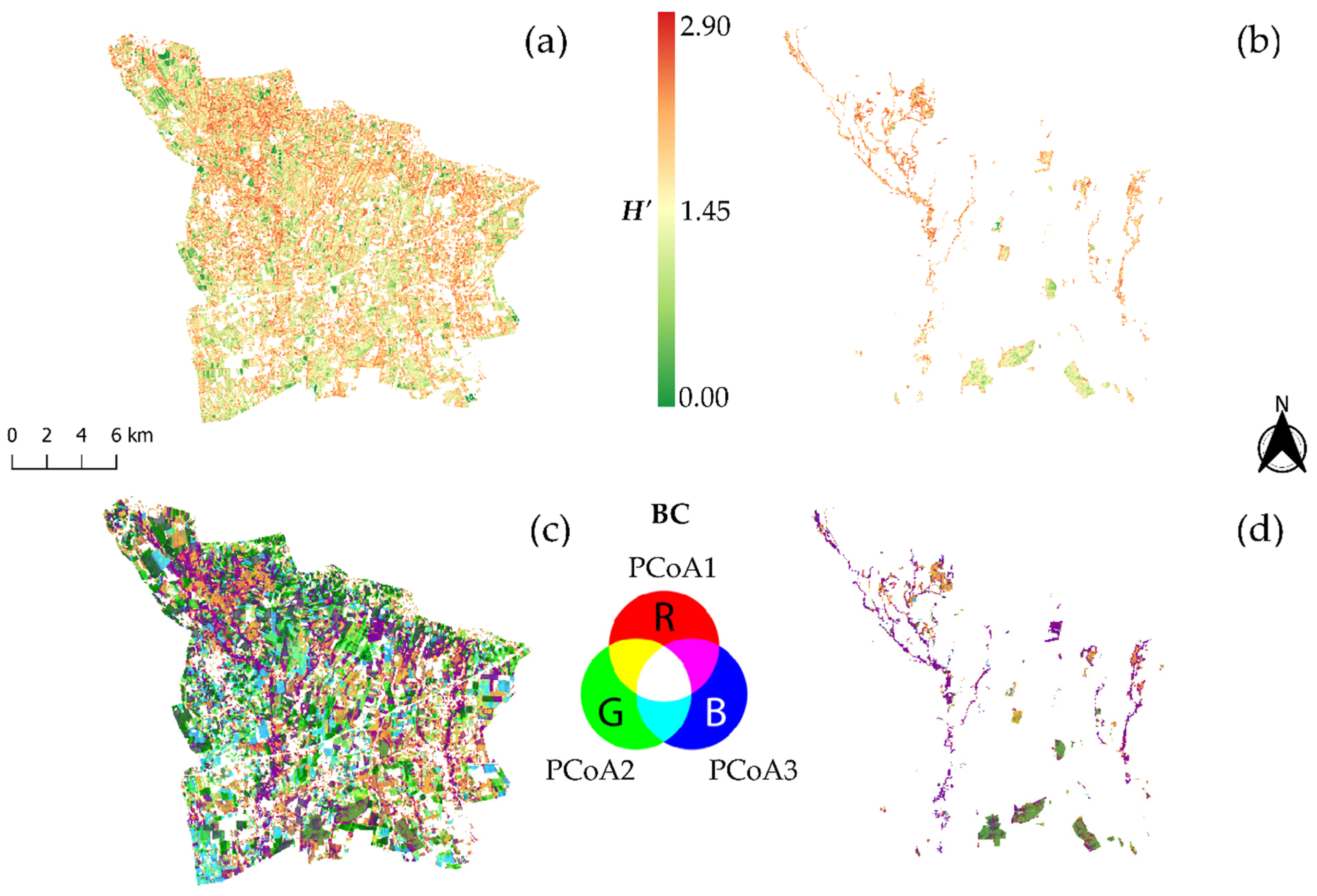
2.2.3. Spectral Diversity Estimation
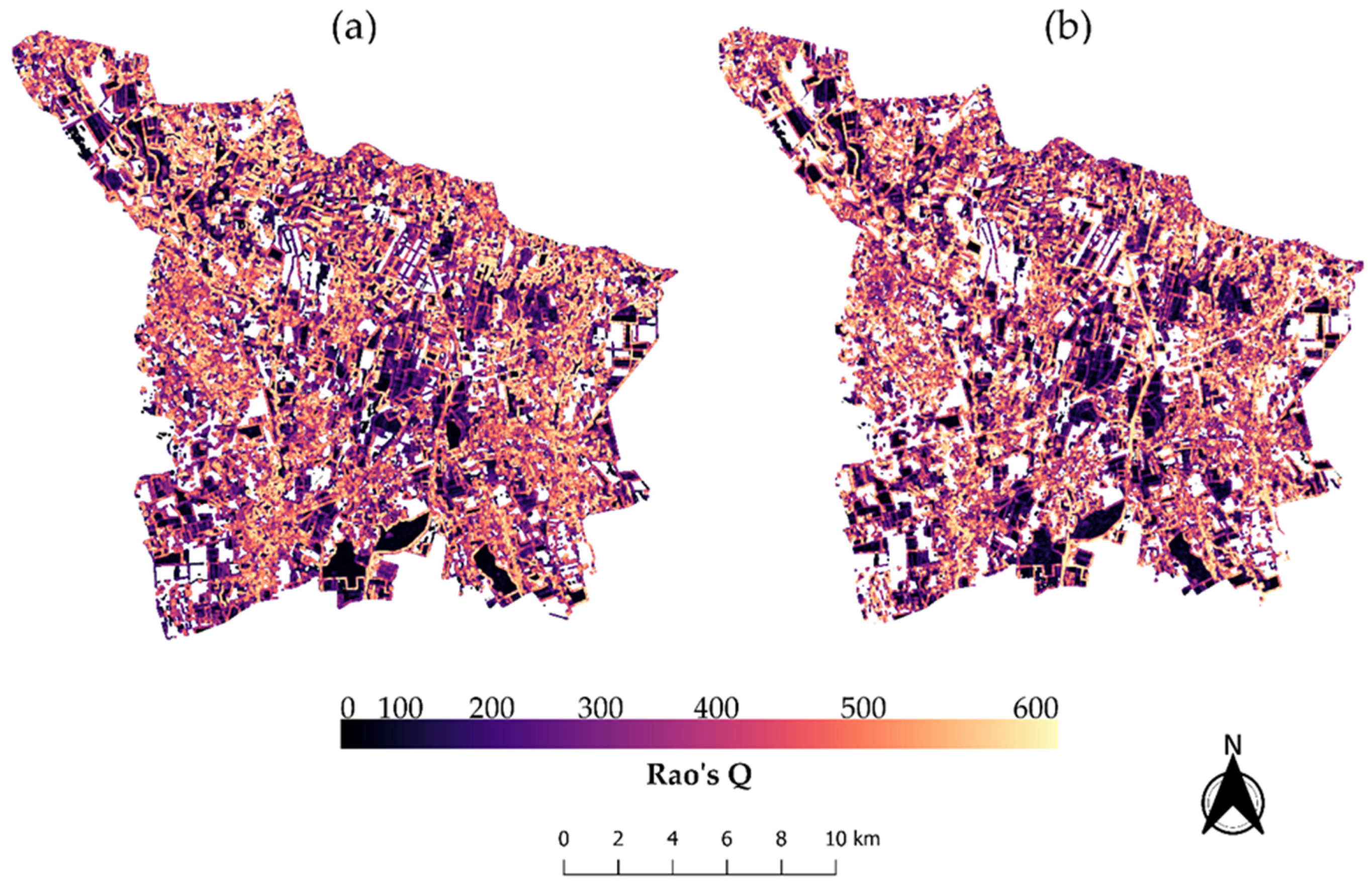
2.2.4. Spectral Ecosystem Heterogeneity Estimation
3. Results
3.1. Comparison of α and β Spectral Diversity vs. Measured Taxonomic Diversity
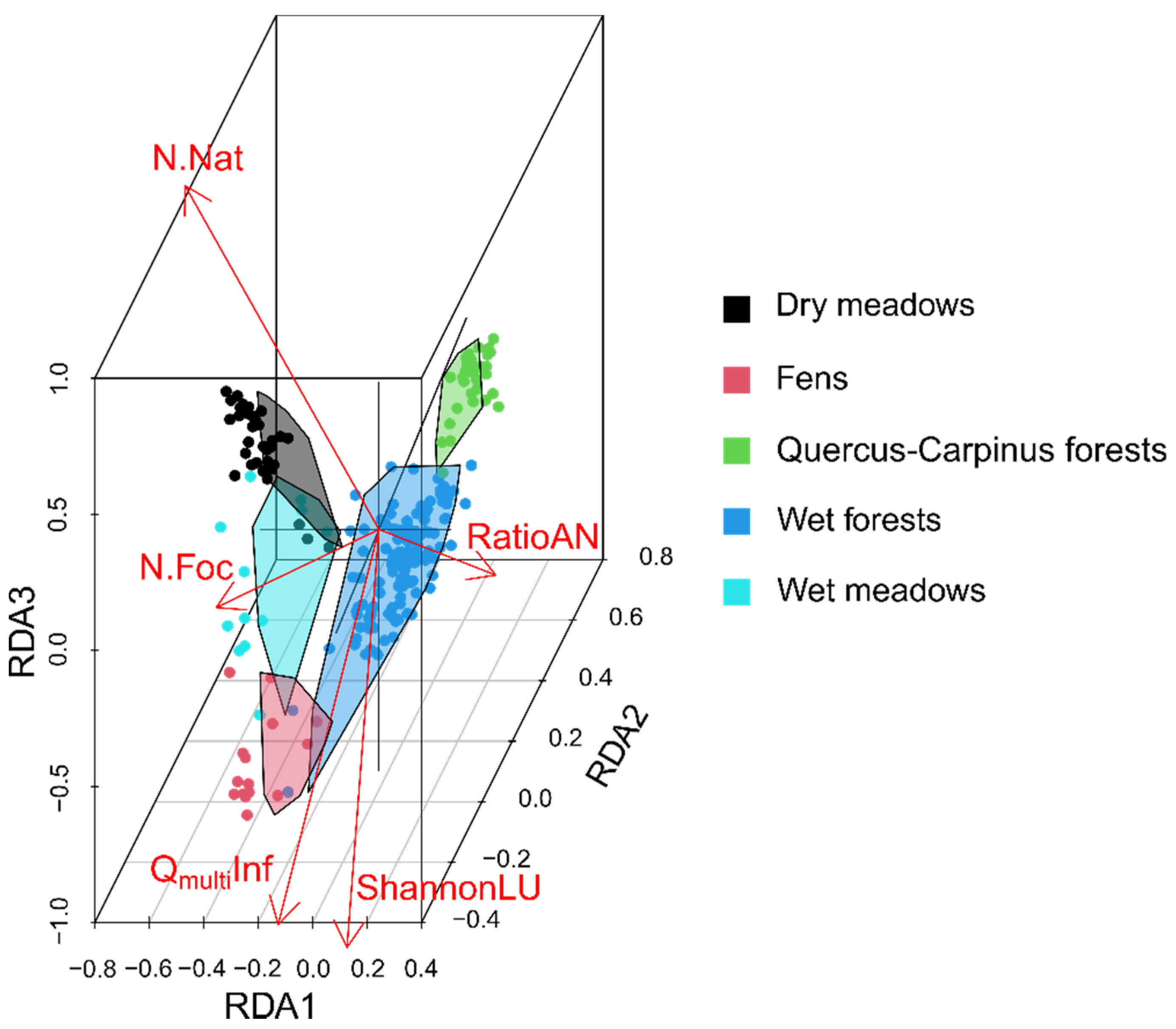
3.2. Spectral Heterogeneity vs. Landscape Heterogeneity and Taxonomic Plant Diversity
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- IPBES. Summary for Policymakers of the Global Assessment Report on Biodiversity and Ecosystem Services of the Intergovernmental Science-Policy Platform on Biodiversity and Ecosystem Services; Díaz, S., Settele, J., Brondízio, E.S., Ngo, H.T., Guèze, M., Agard, J., Arneth, A., Balvanera, P., Brauman, K.A., Butchart, S.H.M., et al., Eds.; IPBES Secretariat: Bonn, Germany, 2019; 56p. [Google Scholar] [CrossRef]
- Vihervaara, P.; Auvinen, A.P.; Mononen, L.; Törmä, M.; Ahlroth, P.; Anttila, S.; Böttcher, K.; Forsius, M.; Heino, J.; Heliola, J.; et al. How essential biodiversity variables and remote sensing can help national biodiversity monitoring. Glob. Ecol. Conserv. 2017, 10, 43–59. [Google Scholar] [CrossRef]
- Yoccoz, N.G.; Nichols, J.D.; Boulinier, T. Monitoring of biological diversity in space and time. Trends Ecol. Evol. 2001, 16, 446–453. [Google Scholar] [CrossRef]
- Maccherini, S.; Bacaro, G.; Tordoni, E.; Bertacchi, A.; Castagnini, P.; Foggi, B.; Gennai, M.; Mugnai, M.; Sarmati, S.; Angiolini, C. Enough Is Enough? Searching for the Optimal Sample Size to Monitor European Habitats: A Case Study from Coastal Sand Dunes. Diversity 2020, 12, 138. [Google Scholar] [CrossRef]
- Rocchini, D.; Salvatori, N.; Beierkuhnlein, C.; Chiarucci, A.; de Boissieu, F.; Förster, M.; Garzon-Lopez, C.X.; Gillespie, T.W.; Hauffe, H.C.; He, K.S.; et al. From local spectral species to global spectral communities: A benchmark for ecosystem diversity estimate by remote sensing. Ecol. Inform. 2021, 61, 101195. [Google Scholar] [CrossRef]
- Féret, J.B.; Rocchini, D.; He, K.S.; Nagendra, H.; Luque, S. Forest species mapping. In A Sourcebook of Methods and Procedures for Monitoring Essential Biodiversity Variables in Tropical Forests with Remote Sensing; GOFC‐GOLD, GEO BON, Ed.; Report Version UNCBD COP-13 GOFC-GOLD Land Cover Project Office; Wageningen University: Wageningen, The Netherlands, 2017. [Google Scholar]
- Rocchini, D.; Boyd, D.S.; Féret, J.B.; Foody, G.M.; He, K.S.; Lausch, A.; Nagendra, H.; Wegmann, M.; Pettorelli, N. Satellite remote sensing to monitor species diversity: Potential and pitfalls. Remote Sens. Ecol. Conserv. 2016, 2, 25–36. [Google Scholar] [CrossRef]
- Rocchini, D.; Luque, S.; Pettorelli, N.; Bastin, L.; Doktor, D.; Faedi, N.; Feilhauer, H.; Féret, J.B.; Foody, G.M.; Gavish, Y.; et al. Measuring β-diversity by remote sensing: A challenge for biodiversity monitoring. Methods Ecol. Evol. 2018, 9, 1787–1798. [Google Scholar] [CrossRef]
- Féret, J.B.; de Boissieu, F. biodivMapR: An r package for α- and β-diversity mapping using remotely sensed images. Methods Ecol. Evol. 2020, 11, 64–70. [Google Scholar] [CrossRef]
- Rocchini, D.; Neteler, M. Let the four freedoms paradigm apply to ecology. Trends Ecol. Evol. 2012, 27, 310–311. [Google Scholar] [CrossRef]
- Palmer, M.W.; Earls, P.G.; Hoagland, B.W.; White, P.S.; Wohlgemuth, T. Quantitative tools for perfecting species lists. Environmetrics 2002, 13, 121–137. [Google Scholar] [CrossRef]
- Rocchini, D.; Hernández-Stefanoni, J.L.; He, K.S. Advancing species diversity estimate by remotely sensed proxies: A conceptual review. Ecol. Inform. 2015, 25, 22–28. [Google Scholar] [CrossRef]
- Lausch, A.; Heurich, M.; Magdon, P.; Rocchini, D.; Schulz, K.; Bumberger, J.; King, D.J. A Range of Earth Observation Techniques for Assessing Plant Diversity. In Remote Sensing of Plant Biodiversity, 1st ed.; Cavender-Bares, J., Gamon, J., Townsend, P., Eds.; Springer Nature: Cham, Switzerland, 2020; pp. 309–348. [Google Scholar]
- Rocchini, D.; Chiarucci, A.; Loiselle, S.A. Testing the spectral variation hypothesis by using satellite multispectral images. Acta Oecol. 2004, 26, 117–120. [Google Scholar] [CrossRef]
- Rocchini, D.; Balkenhol, N.; Carter, G.A.; Foody, G.M.; Gillespie, T.W.; He, K.S.; Kark, S.; Levin, N.; Lucas, K.; Luoto, M.; et al. Remotely sensed spectral heterogeneity as a proxy of species diversity: Recent advances and open challenges. Ecol. Inform. 2010, 5, 318–329. [Google Scholar] [CrossRef]
- Féret, J.B.; Asner, G.P. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecol. Appl. 2014, 24, 1289–1296. [Google Scholar] [CrossRef] [PubMed]
- Heumann, B.W.; Hackett, R.A.; Monfils, A.K. Testing the spectral diversity hypothesis using spectroscopy data in a simulated wetland community. Ecol. Inform. 2015, 25, 29–34. [Google Scholar] [CrossRef]
- Torresani, M.; Rocchini, D.; Sonnenschein, R.; Zebisch, M.; Marcantonio, M.; Ricotta, C.; Tonon, G. Estimating tree species diversity from space in an alpine conifer forest: The Rao’s Q diversity index meets the spectral variation hypothesis. Ecol. Inform. 2019, 52, 26–34. [Google Scholar] [CrossRef]
- Marzialetti, F.; Cascone, S.; Frate, L.; Di Febbraro, M.; Acosta, A.T.R.; Carranza, M.L. Measuring Alpha and Beta Diversity by Field and Remote-Sensing Data: A Challenge for Coastal Dunes Biodiversity Monitoring. Remote Sens. 2021, 13, 1928. [Google Scholar] [CrossRef]
- Whittaker, R.H. Vegetation of the Siskiyou mountains, Oregon and California. Ecol. Monogr. 1960, 30, 279–338. [Google Scholar] [CrossRef]
- Whittaker, R.H. Evolution and measurement of species diversity. Taxon 1972, 21, 213–215. [Google Scholar] [CrossRef]
- Fahrig, L. Landscape heterogeneity and metapopulation dynamics. In Key Topics in Landscape Ecology; Wu, J., Hobbs, R., Eds.; Cambridge University Press: Cambridge, UK, 2007; pp. 78–91. [Google Scholar]
- Malanson, G.P.; Cramer, B.E. Landscape heterogeneity, connectivity, and critical landscapes for conservation. Divers. Distrib. 1999, 5, 27–39. [Google Scholar] [CrossRef]
- Lozier, J.D.; Strange, J.P.; Koch, J.B. Landscape heterogeneity predicts gene flow in a widespread polymorphic bumble bee, Bombus bifarius (Hymenoptera: Apidae). Conserv. Genet. 2013, 14, 1099–1110. [Google Scholar] [CrossRef]
- Marcantonio, M.; Iannacito, M.; Thouverai, E.; Da Re, D.; Tattoni, C.; Bacaro, G.; Vicario, S.; Ricotta, C.; Rocchini, D. rasterdiv: Diversity Indices for Numerical Matrices. 2021, R Package Version 0.2-3. Available online: https://CRAN.R-project.org/package=rasterdiv (accessed on 12 October 2021).
- Rocchini, D.; Thouverai, E.; Marcantonio, M.; Iannacito, M.; Da Re, D.; Torresani, M.; Bacaro, G.; Bazzichetto, M.; Bernardi, A.; Foody, G.M.; et al. rasterdiv—An Information Theory tailored R package for measuring ecosystem heterogeneity from space: To the origin and back. Methods Ecol. Evol. 2021, 12, 1093–1102. [Google Scholar] [CrossRef] [PubMed]
- Sigura, M.; Boscutti, F.; Buccheri, M.; Dorigo, L.; Glerean, P.; Lapini, L. La Rel dei Paesaggi di Pianura, di Area Montana e Urbanizzati. Piano Paesaggistico Regionale del Friuli-Venezia Giulia (Parte Strategica) E1 -Allegato alla Scheda di RER. Regione Friuli-Venezia Giulia. 2017. Available online: http://www.regione.fvg.it/rafvg/cms/RAFVG/ambiente-territorio/pianificazione-gestione-territorio/FOGLIA21/#id9 (accessed on 12 October 2021).
- Jones, T.A.; Hughes, J.M.R. Wetland inventories and wetland loss studies: A European perspective. In Waterfowl and Wetland Conservation in the 1990s: A Global Perspective, Proceedings of the IWRB Symposium, St Petersburg Beach, FL, USA, 12–19 November 1993; Moser, M., Prentice, R.C., van Vessem, J., Eds.; IWRB Spec. Publ. No. 26; IWRB: Slimbridge, UK, 1993; pp. 164–169. [Google Scholar]
- European Commission. Life and Europe’s Wetlands. Restoring a Vital Ecosystem; European Commission: Luxembourg, 2007.
- Jantke, K.; Schleupner, C.; Schneider, U.A. Gap analysis of European wetland species: Priority regions for expanding the Natura 2000 network. Biodivers. Conserv. 2011, 20, 581–605. [Google Scholar] [CrossRef]
- Liccari, F.; Castello, M.; Poldini, L.; Altobelli, A.; Tordoni, E.; Sigura, M.; Bacaro, G. Do Habitats Show a Different Invasibility Pattern by Alien Plant Species? A Test on a Wetland Protected Area. Diversity 2020, 12, 267. [Google Scholar] [CrossRef]
- Davies, C.E.; Moss, D.; Hill, M.O. EUNIS Habitat Classification Revised 2004; Report to the European Topic Centre on Nature Protection and Biodiversity; European Environment Agency: Copenhagen, Denmark, 2004.
- Chytrý, M.; Tichý, L.; Hennekens, S.M.; Knollová, I.; Janssen, J.A.M.; Rodwell, J.S.; Peterka, T.; Marcenò, C.; Landucci, F.; Danihelka, J.; et al. EUNIS habitat classification: Expert system, characteristic species combinations and distribution maps of European habitats. Appl. Veg. Sci. 2020, 23, 648–675. [Google Scholar] [CrossRef]
- Liccari, F.; Sigura, M.; Tordoni, E.; Boscutti, F.; Bacaro, G. Determining plant diversity within interconnected natural habitat remnants (ecological network) in an agricultural landscape: A matter of sampling design? Diversity 2022, 14, 12. [Google Scholar] [CrossRef]
- Liccari, F.; Boscutti, F.; Bacaro, G.; Sigura, M. Connectivity, landscape structure, and plant diversity across agricultural landscapes: Novel insight into effective ecological network planning. J. Environ. Manag. 2022, 317, 115358. [Google Scholar] [CrossRef]
- Bartolucci, F.; Peruzzi, L.; Galasso, G.; Albano, A.; Alessandrini, A.; Ardenghi, N.M.G.; Astuti, G.; Bacchetta, G.; Ballelli, S.; Banfi, E.; et al. An updated checklist of the vascular flora native to Italy. Plant. Biosyst. 2018, 152, 179–303. [Google Scholar] [CrossRef]
- Galasso, G.; Conti, F.; Peruzzi, L.; Ardenghi, N.M.G.; Banfi, E.; Celesti-Grapow, L.; Albano, A.; Alessandrini, A.; Bacchetta, G.; Ballelli, S.; et al. An updated checklist of the vascular flora alien to Italy. Plant. Biosyst. 2018, 152, 556–592. [Google Scholar] [CrossRef]
- Copernicus Open Access Hub. Sentinel-2 Data. Available online: https://scihub.copernicus.eu/ (accessed on 15 September 2021).
- SNAP-ESA Sentinel Application Platform v7.0. Available online: http://step.esa.int (accessed on 15 September 2021).
- Ustin, S.L.; Gamon, J.A. Remote sensing of plant functional types. New Phytol. 2010, 186, 795–816. [Google Scholar] [CrossRef]
- Homolová, L.; Malenovský, Z.; Clevers, J.G.P.W.; García-Santos, G.; Schaepman, M.E. Review of optical-based remote sensing for plant trait mapping. Ecol. Complex. 2013, 15, 1–16. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell. Syst. Tech. J. 1948, 23, 379–423. [Google Scholar] [CrossRef]
- Bray, J.R.; Curtis, J.T. An ordination of the upland forest communities of Southern Wisconsin. Ecol. Monogr. 1957, 27, 325–349. [Google Scholar] [CrossRef]
- Hamner, B.; Frasco, M. Metrics: Evaluation Metrics for Machine Learning. R package version 0.1.4. 2018. Available online: https://CRAN.R-project.org/package=Metrics (accessed on 12 October 2021).
- Rao, C.R. Diversity and dissimilarity coefficients: A unified approach. Theor. Popul. Biol. 1982, 21, 24–43. [Google Scholar] [CrossRef]
- Wood, S.N. Fast stable restricted maximum likelihood and marginal likelihood estimation of semiparametric generalized linear models. J. R. Stat. Soc. 2011, 73, 3–36. [Google Scholar] [CrossRef]
- Legendre, P.; Gallagher, E. Ecologically meaningful transformations for ordination of species data. Oecologia 2001, 129, 271–280. [Google Scholar] [CrossRef] [PubMed]
- Legendre, P.; Legendre, L. Numerical Ecology, 3rd ed.; Elsevier: Amsterdam, The Netherlands, 2012. [Google Scholar]
- Blanchet, F.G.; Legendre, P.; Bergeron, J.A.C.; He, F. Consensus RDA across dissimilarity coefficients for canonical ordination of community composition data. Ecol. Monogr. 2014, 84, 491–511. [Google Scholar] [CrossRef]
- Nagendra, H.; Rocchini, D.; Ghate, R.; Sharma, B.; Pareeth, S. Assessing plant diversity in a dry tropical forest: Comparing the utility of Landsat and Ikonos satellite images. Remote Sens. 2010, 2, 478–496. [Google Scholar] [CrossRef]
- Hall, K.; Reitalu, T.; Sykes, M.T.; Prentice, H.C. Spectral heterogeneity of QuickBird satellite data is related to fine-scale plant species spatial turnover in semi-natural grasslands. Appl. Veg. Sci. 2012, 15, 145–157. [Google Scholar] [CrossRef]
- Warren, S.D.; Alt, M.; Olson, K.D.; Irl, S.D.H.; Steinbauer, M.J.; Jentsch, A. The relationship between the spectral diversity of satellite imagery habitat heterogeneity, and plant species richness. Ecol. Inform. 2014, 24, 160–168. [Google Scholar] [CrossRef]
- Arekhi, M.; Yilmaz, O.Y.; Yilmaz, H.; Akyüz, Y.F. Can tree species diversity be assessed with Landsat data in a temperate forest? Environ. Monit. Assess. 2017, 189, 586. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Ramoelo, A.; Mutanga, O. Remote sensing of species diversity using Landsat 8 spectral variables. ISPRS J. Photogramm. 2017, 133, 116–127. [Google Scholar] [CrossRef]
- Rossi, C.; Kneubühler, M.; Schütz, M.; Schaepman, M.E.; Haller, R.M.; Risch, A.C. Spatial resolution, spectral metrics and biomass are key aspects in estimating plant species richness from spectral diversity in species-rich grasslands. Remote Sens. Ecol. Conserv. 2021, 8, 297–314. [Google Scholar] [CrossRef]
- Porensky, L.M.; Young, T.P. Edge-Effect Interactions in Fragmented and Patchy Landscapes. Conserv. Biol. 2013, 27, 509–519. [Google Scholar] [CrossRef] [PubMed]
- Amici, V.; Rocchini, D.; Filibeck, G.; Bacaro, G.; Santi, E.; Geri, F.; Landi, S.; Scoppola, A.; Chiarucci, A. Landscape structure effects on forest plant diversity at local scale: Exploring the role of spatial extent. Ecol. Complex. 2015, 21, 44–52. [Google Scholar] [CrossRef]
- Oldeland, J.; Wesuls, D.; Rocchini, D.; Schmidt, M.; Jurgens, N. Does using species abundance data improve estimates of species diversity from remotely sensed spectral heterogeneity? Ecol. Indic. 2010, 10, 390–396. [Google Scholar] [CrossRef]
- Jelinski, D.E.; Wu, J. The modifiable areal unit problem and implications for landscape ecology. Landsc. Ecol. 1996, 11, 129–140. [Google Scholar] [CrossRef]
- Schmidtlein, S.; Fassnacht, F.E. The spectral variability hypothesis does not hold across landscapes. Remote Sens. Environ. 2017, 192, 114–125. [Google Scholar] [CrossRef]
- Hutchinson, G. Homage to Santa Rosalia or why are there so many kinds of animals? Am. Nat. 1959, 93, 145–159. [Google Scholar] [CrossRef]
- Blonder, B. Hypervolume concepts in niche and trait based ecology. Ecography 2018, 41, 1441–1455. [Google Scholar] [CrossRef]
- Thouverai, E.; Marcantonio, M.; Bacaro, G.; Da Re, D.; Iannacito, M.; Marchetto, E.; Ricotta, C.; Tattoni, C.; Vicario, S.; Rocchini, D. Measuring diversity from space: A global view of the free and open source rasterdiv R package under a coding perspective. Community Ecol. 2021, 22, 1–11. [Google Scholar] [CrossRef]
- Rocchini, D.; Andreo, V.; Förster, M.; Garzon-Lopez, C.X.; Gutierrez, A.P.; Gillespie, T.W.; Hauffe, H.C.; He, K.S.; Kleinschmit, B.; Mairota, P.; et al. Potential of remote sensing to predict species invasions: A modelling perspective. Prog. Phys. Geogr. Earth Environ. 2015, 39, 283–309. [Google Scholar] [CrossRef]
- Müllerová, J.; Brůna, J.; Dvořák, P.; Bartaloš, T.; Vítková, M. Does the data resolution/origin matter? satellite, airborne and Uav imagery to tackle plant invasions. ISPRS—Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2016, 41B7, 903–908. [Google Scholar] [CrossRef]
- Skowronek, S.; Asner, G.P.; Feilhauer, H. Performance of one-class classifiers for invasive species mapping using airborne imaging spectroscopy. Ecol. Inform. 2017, 37, 66–76. [Google Scholar] [CrossRef]
- Skowronek, S.; Ewald, M.; Isermann, M.; Kerchove, R.V.D.; Lenoir, J.; Aerts, R.; Warrie, J.; Hattab, T.; Honnay, O.; Schmidtlein, S.; et al. Mapping an invasive bryophyte species using hyperspectral remote sensing data. Biol. Invasions 2017, 19, 239–254. [Google Scholar] [CrossRef]
- Vaz, A.S.; Alcaraz-Segura, D.; Campos, J.C.; Vicente, J.R.; Honrado, J.P. Managing plant invasions through the lens of remote sensing: A review of progress and the way forward. Sci. Total Environ. 2018, 642, 1328–1339. [Google Scholar] [CrossRef]
- Lopatin, J.; Dolos, K.; Kattenborn, T.; Fassnacht, F.E. How canopy shadow affects invasive plant species classification in high spatial resolution remote sensing. Remote Sens. Ecol. Conserv. 2019, 5, 302–317. [Google Scholar] [CrossRef]
- Ewald, M.; Skowronek, S.; Aerts, R.; Lenoir, J.; Feilhauer, H.; Van De Kerchove, R.; Honnay, O.; Somers, B.; Garzón-López, C.X.; Rocchini, D.; et al. Assessing the impact of an invasive bryophyte on plant species richness using high resolution imaging spectroscopy. Ecol. Indic. 2020, 110, 105882. [Google Scholar] [CrossRef]
- Paton, P. The effect of edge on avian nest success: How strong is the evidence? Conserv. Biol. 1994, 8, 17–26. [Google Scholar] [CrossRef]
- Fagan, W.; Cantrell, R.; Cosner, C. How habitat edges change species interaction. Am. Nat. 1999, 153, 165–182. [Google Scholar] [CrossRef]
- Loveridge, A.; Hemson, G.; Davidson, Z.; Macdonald, D. African lions on the edge: Reserve boundaries as “attractive sinks”. Biol. Conserv. Wild Felids 2010, 283, 283–304. [Google Scholar]
- An, Y.; Liu, S.; Sun, Y.; Beazley, R. Construction and optimization of an ecological network based on morphological spatial pattern analysis and circuit theory. Landsc. Ecol. 2021, 36, 2059–2076. [Google Scholar] [CrossRef]
| QNDVI1 ~ RatioAN + s(ShannonLU) | Est. ± SE | p-Value | Edf | R2 = 0.21 |
|---|---|---|---|---|
| Terms | p-Value | |||
| Intercept | 45.43 ± 1.95 | <0.001 | - | - |
| RatioAN | 51.82 ± 19.83 | 0.010 | - | - |
| Smooth (ShannonLU) | - | - | 2.25 | <0.001 |
| QNDVI5 ~ RatioAN + s(ShannonLU) | R2 = 0.25 | |||
| Intercept | 76.36 ± 3.49 | <0.001 | - | - |
| RatioAN | 99.34 ± 35.60 | 0.006 | - | - |
| Smooth (ShannonLU) | - | - | 3.37 | <0.001 |
| QNDVIInf ~ RatioAN + s(ShannonLU) | R2 = 0.27 | |||
| Intercept | 282.50 ± 12.34 | <0.001 | - | - |
| RatioAN | 347.65 ± 125.88 | 0.006 | - | - |
| Smooth (ShannonLU) | - | - | 3.67 | <0.001 |
| Qmulti1 ~ RatioAN + s(ShannonLU) | R2 = 0.25 | |||
| Intercept | 51.92 ± 1.59 | <0.001 | - | - |
| RatioAN | 18.02 ± 16.20 | NS | - | - |
| Smooth (ShannonLU) | - | - | 3.34 | < 0.001 |
| Qmulti5 ~ RatioAN + s(ShannonLU) | R2 = 0.41 | |||
| Intercept | 92.52 ± 2.40 | <0.001 | - | - |
| RatioAN | 72.55 ± 24.55 | 0.003 | - | - |
| Smooth (ShannonLU) | - | - | 4.64 | <0.001 |
| QmultiInf ~ RatioAN + s(ShannonLU) | R2 = 0.43 | |||
| Intercept | 368.62 ± 9.64 | <0.001 | - | - |
| RatioAN | 401.20 ± 98.43 | <0.001 | - | - |
| Smooth (ShannonLU) | - | - | 4.51 | <0.001 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liccari, F.; Sigura, M.; Bacaro, G. Use of Remote Sensing Techniques to Estimate Plant Diversity within Ecological Networks: A Worked Example. Remote Sens. 2022, 14, 4933. https://doi.org/10.3390/rs14194933
Liccari F, Sigura M, Bacaro G. Use of Remote Sensing Techniques to Estimate Plant Diversity within Ecological Networks: A Worked Example. Remote Sensing. 2022; 14(19):4933. https://doi.org/10.3390/rs14194933
Chicago/Turabian StyleLiccari, Francesco, Maurizia Sigura, and Giovanni Bacaro. 2022. "Use of Remote Sensing Techniques to Estimate Plant Diversity within Ecological Networks: A Worked Example" Remote Sensing 14, no. 19: 4933. https://doi.org/10.3390/rs14194933
APA StyleLiccari, F., Sigura, M., & Bacaro, G. (2022). Use of Remote Sensing Techniques to Estimate Plant Diversity within Ecological Networks: A Worked Example. Remote Sensing, 14(19), 4933. https://doi.org/10.3390/rs14194933









