A Comparison of Analytical Approaches for the Spectral Discrimination and Characterisation of Mite Infestations on Banana Plants
Abstract
:1. Introduction
2. Materials and Methods
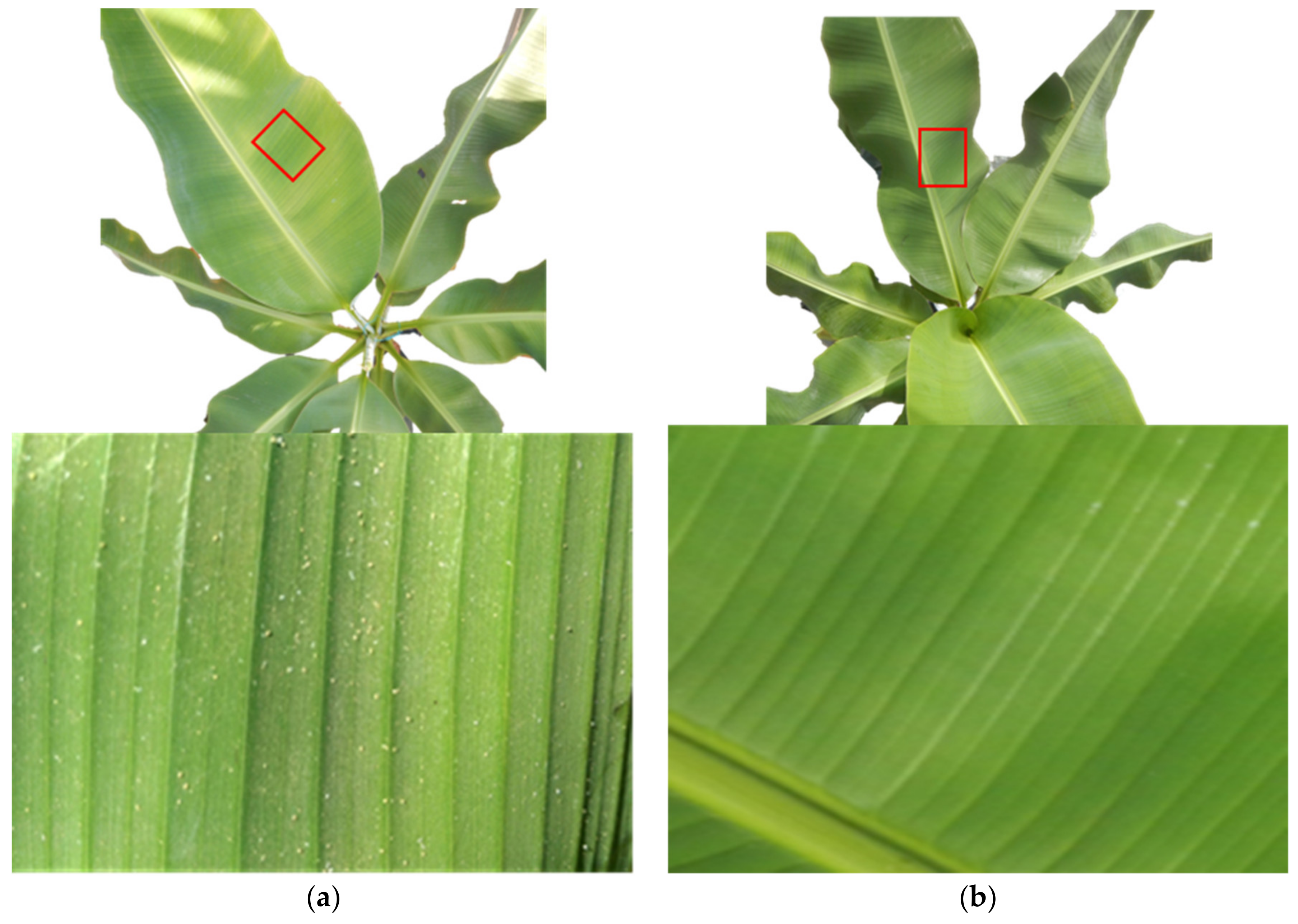
2.1. Experimental Design
2.2. Spectral Data Collection and Pre-Processing
2.3. Classification Models
2.4. Variable Selection of Key Spectral Wavelengths
2.5. UAV Multispectral Discrimination
3. Results and Discussion
3.1. Classification Model Results for Discriminating between Infested and Healthy Plants
3.2. Key Wavelength Selection
3.3. Key Wavelength Classification Model Accuracy
3.4. UAV Multispectral Discrimination of Healthy and Mite-Infested Banana Plants
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Scott, G.J. A review of root, tuber and banana crops in developing countries: Past, present and future. Int. J. Food Sci. Technol. 2021, 56, 1093–1114. [Google Scholar] [CrossRef] [PubMed]
- FAO. Banana Facts and Figures. Available online: https://www.fao.org/economic/est/est-commodities/oilcrops/bananas/bananafacts/en/ (accessed on 10 October 2022).
- FAO. Banana Market Review February 2020 Snapshot. Available online: http://www.fao.org/3/ca9212en/ca9212en.pdf (accessed on 30 November 2020).
- Picq, C.; Fouré, E.; Frison, E.A. Bananas and Food Security; Bioversity International: Rome, Italy, 1999. [Google Scholar]
- FAO. Banana Market Review: Preliminary Results 2021. Available online: https://www.fao.org/3/cb9411en/cb9411en.pdf (accessed on 1 September 2022).
- Horticulture Innovation. Australian Horticulture Statistics Handbook; Hort Innovation: Sydney, Australia, 2020. [Google Scholar]
- Pinese, B.; Piper, R. Bananas: Insect & Mite Management; Queensland, Department of Primary Industries: Brisbane, Australia, 1994.
- Campbell, R.; Randolph, G.; Marini, R. Surface and Ultrastructural Feeding Injury to Strawberry Leaves by the Twospotted Spider Mite. HortScience 1990, 25, 948–951. [Google Scholar] [CrossRef] [Green Version]
- Kielkiewicz, M. Ultrastructural cell modification in tomato (Lycopersicon esculentum) leaf tissue in response to the carmine spider mite (Tetranychus cinnabarinus) feeding. In Ecology and Evolution of the Acari, Proceedings of the 3rd Symposium of the European Association of Acarologists, Amsterdam, The Netherlands, 1–5 July 1996; Bruin, J., van der Geest, L.P.S., Sabelis, M.W., Eds.; Springer: Dordrecht, The Netherlands, 1999; pp. 603–615. [Google Scholar]
- Agut, B.; Pastor, V.; Jaques, J.A.; Flors, V. Can Plant Defence Mechanisms Provide New Approaches for the Sustainable Control of the Two-Spotted Spider Mite Tetranychus urticae? Int. J. Mol. Sci. 2018, 19, 614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Robinson, J.C.; Saúco, V.G. Bananas and Plantains; Cabi: Wallingford, UK, 2010; Volume 19.
- Lindsay, S.; Campagnolo, D.; Daniells, J.; Lemin, C.; Goebel, R.; Pinese, B.; Peterson, R.; Evanas, D.; Pattison, T. Tropical Banana Information Kit; Department of Primary Industries, Queensland Horticulture Institute: Brisbane, Australia, 1998.
- Queensland Department of Agriculture Fisheries and Forestry [QDAFF]. A-Z List of Horticultural Insect Pests. Available online: https://www.daf.qld.gov.au/business-priorities/agriculture/plants/fruit-vegetable/insect-pests (accessed on 10 May 2022).
- Alatawi, F.; Margolies, D.; Nechols, J. Aesthetic damage thresholds for twospotted spider mites (Acari: Tetranychidae) on impatiens: Effect of plant age and level of infestation. J. Econ. Entomol. 2007, 100, 1904–1909. [Google Scholar] [CrossRef] [PubMed]
- Lan, Y.; Zhang, H.; Hoffmann, W.; Lopez, J.J.D. Spectral response of spider mite infested cotton: Mite density and miticide rate study. Int. J. Agric. Biol. Eng. 2013, 6, 48–52. [Google Scholar]
- Hort Innovation. Australian Banana Best Practice. Available online: https://www.horticulture.com.au/globalassets/hort-innovation/resource-assets/ba13004-banana-spider-mites-pdf.pdf (accessed on 10 May 2022).
- Migeon, A.; Nouguier, E.; Dorkeld, F. Spider Mites Web: A comprehensive database for the Tetranychidae. In Trends in Acarology; Springer: Berlin, Germany, 2010; pp. 557–560. [Google Scholar]
- Martin, D.; Latheef, M. Remote Sensing Evaluation of Two-spotted Spider Mite Damage on Greenhouse Cotton. J. Vis. Exp. 2017, 122, e54314. [Google Scholar] [CrossRef] [Green Version]
- Fraulo, A.B.; Cohen, M.; Liburd, O.E. Visible/Near Infrared Reflectance (Vnir) Spectroscopy for Detecting Twospotted Spider Mite (Acari: Tetranychidae) Damage in Strawberries. Environ. Entomol. 2009, 38, 137–142. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, I.; Berenstein, M.; Paz-Kagan, T.; Sade, A.; Karnieli, A. Spectral assessment of two-spotted spider mite damage levels in the leaves of greenhouse-grown pepper and bean. Biosyst. Eng. 2017, 157, 72–85. [Google Scholar] [CrossRef]
- Luedeling, E.; Hale, A.; Zhang, M.; Bentley, W.J.; Dharmasri, L.C. Remote sensing of spider mite damage in California peach orchards. Int. J. Appl. Earth Obs. Geoinf. 2009, 11, 244–255. [Google Scholar] [CrossRef]
- Uygun, T.; Ozguven, M.M.; Yanar, D. A new approach to monitor and assess the damage caused by two-spotted spider mite. Exp. Appl. Acarol. 2020, 82, 335–346. [Google Scholar] [CrossRef]
- Wang, J.; Shen, C.; Liu, N.; Jin, X.; Fan, X.; Dong, C.; Xu, Y. Non-Destructive Evaluation of the Leaf Nitrogen Concentration by In-Field Visible/Near-Infrared Spectroscopy in Pear Orchards. Sensors 2017, 17, 538. [Google Scholar] [CrossRef] [PubMed]
- Sinha, P.; Robson, A.; Schneider, D.; Kilic, T.; Mugera, H.K.; Ilukor, J.; Tindamanyire, J.M. The potential of in-situ hyperspectral remote sensing for differentiating 12 banana genotypes grown in Uganda. ISPRS J. Photogramm. Remote Sens. 2020, 167, 85–103. [Google Scholar] [CrossRef]
- Usha, K.; Singh, B. Potential applications of remote sensing in horticulture—A review. Sci. Hortic. 2013, 153, 71–83. [Google Scholar] [CrossRef]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Zhao, J.; Fang, Y.; Chu, G.; Yan, H.; Hu, L.; Huang, L. Identification of Leaf-Scale Wheat Powdery Mildew (Blumeria graminis f. sp. Tritici) Combining Hyperspectral Imaging and an SVM Classifier. Plants 2020, 9, 936. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Hu, T.; Luo, L.; He, Q.; Zhang, S.; Li, M.; Cui, X.; Li, H. Rapid estimation of leaf nitrogen content in apple-trees based on canopy hyperspectral reflectance using multivariate methods. Infrared Phys. Technol. 2020, 111, 103542. [Google Scholar] [CrossRef]
- Jiang, H.; Ye, L.; Li, X.; Shi, M. Variety Identification of Chinese Walnuts Using Hyperspectral Imaging Combined with Chemometrics. Appl. Sci. 2021, 11, 9124. [Google Scholar] [CrossRef]
- Khan, I.H.; Liu, H.; Cheng, T.; Tian, Y.; Cao, Q.; Zhu, Y.; Cao, W.; Yao, X. Detection of wheat powdery mildew based on hyperspectral reflectance through SPA and PLS-LDA. Int. J. Precis. Agric. Aviat. 2020, 3, 13–22. [Google Scholar]
- Wei, L.; Guozhu, F.; Wentao, D.; Hailiang, Z.; Baishao, Z.; Liu, X. Non-destructive determination of four tea polyphenols in fresh tea using visible and near-infrared spectroscopy. Infrared Phys. Technol. 2022, 123, 104037. [Google Scholar]
- Bao, Y.; Mi, C.; Wu, N.; Liu, F.; He, Y. Rapid Classification of Wheat Grain Varieties Using Hyperspectral Imaging and Chemometrics. Appl. Sci. 2019, 9, 4119. [Google Scholar] [CrossRef] [Green Version]
- Torres-Sánchez, J.; López-Granados, F.; Serrano, N.; Arquero, O.; Peña, J.M. High-throughput 3-D monitoring of agricultural-tree plantations with unmanned aerial vehicle (UAV) technology. PLoS ONE 2015, 10, e0130479. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, W.H.; Steppe, K. Perspectives for remote sensing with unmanned aerial vehicles in precision agriculture. Trends Plant Sci. 2019, 24, 152–164. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.; Johansen, K.; Phinn, S.; Robson, A.; Tu, Y.-H. Inter-comparison of remote sensing platforms for height estimation of mango and avocado tree crowns. Int. J. Appl. Earth Obs. Geoinf. 2020, 89, 102091. [Google Scholar] [CrossRef]
- Dlamini, S.N.; Beloconi, A.; Mabaso, S.; Vounatsou, P.; Impouma, B.; Fall, I.S. Review of remotely sensed data products for disease mapping and epidemiology. Remote Sens. Appl. Soc. Environ. 2019, 14, 108–118. [Google Scholar] [CrossRef]
- Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Deng, X.; Zhang, L.; Wen, S.; Jiang, Y.; Suo, G.; Chen, P. A two-stage classification approach for the detection of spider mite-infested cotton using UAV multispectral imagery. Remote Sens. Lett. 2018, 9, 933–941. [Google Scholar] [CrossRef]
- Roderick, H.; Mbiru, E.; Coyne, D.; Tripathi, L.; Atkinson, H. Quantitative Digital Imaging of Banana Growth Suppression by Plant Parasitic Nematodes. PLoS ONE 2012, 7, e53355. [Google Scholar] [CrossRef]
- Selvaraj, M.G.; Vergara, A.; Ruiz, H.; Safari, N.; Elayabalan, S.; Ocimati, W.; Blomme, G. AI-powered banana diseases and pest detection. Plant Methods 2019, 15, 92. [Google Scholar] [CrossRef] [Green Version]
- Robinson, J.C.; Sáuco, V.G. Nursery hardening of in vitro-produced banana plants. Fruits 2009, 64, 383–392. [Google Scholar] [CrossRef]
- Hunter, M.; Scattini, W. The ANOVApot® and Twinpot reduce root escape and save water. In Proceedings of the XXIX International Horticultural Congress on Horticulture: Sustaining Lives, Livelihoods and Landscapes (IHC2014), Brisbane, Australia, 17–22 August 2014; Volume 1112, pp. 23–30. [Google Scholar]
- Poorter, H.; Fiorani, F.; Stitt, M.; Schurr, U.; Finck, A.; Gibon, Y.; Usadel, B.; Munns, R.; Atkin, O.K.; Tardieu, F.; et al. The art of growing plants for experimental purposes: A practical guide for the plant biologist. Funct. Plant Biol. 2012, 39, 821–838. [Google Scholar] [CrossRef] [Green Version]
- Hatchell, D.C. ASD Technical Guide; Analytical Spectral Devices, Inc.: Boulder, CO, USA, 1999; Volume 5335. [Google Scholar]
- ASD Inc. FieldSpec 4™ User Manual; ASD Inc.: Boulder, CO, USA, 2008. [Google Scholar]
- Hennessy, A.; Clarke, K.; Lewis, M. Hyperspectral Classification of Plants: A Review of Waveband Selection Generalisability. Remote Sens. 2020, 12, 113. [Google Scholar] [CrossRef] [Green Version]
- Rinnan, Å; Berg, F.V.D.; Engelsen, S.B. Review of the most common pre-processing techniques for near-infrared spectra. TrAC Trends Anal. Chem. 2009, 28, 1201–1222. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods 2013, 5, 3790–3798. [Google Scholar] [CrossRef]
- Li, H.-D.; Xu, Q.-S.; Liang, Y.-Z. libPLS: An integrated library for partial least squares regression and linear discriminant analysis. Chemom. Intell. Lab. Syst. 2018, 176, 34–43. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Pérez-Roncal, C.; López-Maestresalas, A.; Lopez-Molina, C.; Jarén, C.; Urrestarazu, J.; Santesteban, L.G.; Arazuri, S. Hyperspectral imaging to assess the presence of powdery mildew (Erysiphe necator) in cv. Carignan Noir grapevine bunches. Agronomy 2020, 10, 88. [Google Scholar] [CrossRef] [Green Version]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. A J. Chemom. Soc. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Lee, L.C.; Liong, C.-Y.; Jemain, A.A. Partial least squares-discriminant analysis (PLS-DA) for classification of high-dimensional (HD) data: A review of contemporary practice strategies and knowledge gaps. Analyst 2018, 143, 3526–3539. [Google Scholar] [CrossRef]
- Taunk, K.; De, S.; Verma, S.; Swetapadma, A. A brief review of nearest neighbor algorithm for learning and classification. In Proceedings of the 2019 International Conference on Intelligent Computing and Control Systems (ICCS), Madurai, India, 15–17 May 2019; pp. 1255–1260. [Google Scholar]
- Cortes, C.; Vapnik, V. Support-vector networks. Mach. Learn. 1995, 20, 273–297. [Google Scholar] [CrossRef]
- Rumelhart, D.E.; Hinton, G.E.; Williams, R.J. Learning representations by back-propagating errors. Nature 1986, 323, 533–536. [Google Scholar] [CrossRef]
- Wythoff, B.J. Backpropagation neural networks: A tutorial. Chemom. Intell. Lab. Syst. 1993, 18, 115–155. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef] [PubMed]
- Li, H.-D.; Xu, Q.-S.; Liang, Y.-Z. Random frog: An efficient reversible jump Markov Chain Monte Carlo-like approach for variable selection with applications to gene selection and disease classification. Anal. Chim. Acta 2012, 740, 20–26. [Google Scholar] [CrossRef]
- Mehmood, T.; Sæbø, S.; Liland, K.H. Comparison of variable selection methods in partial least squares regression. J. Chemom. 2020, 34, e3226. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and photographic infrared linear combinations for monitoring vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Deng, J.; Lan, Y.; Yang, A.; Jiang, Y.; Suo, G.; Chen, P. Automatic difference vegetation index generator for spider mite-infested cotton detection using hyperspectral reflectance. Int. J. Precis. Agric. Aviat. 2020, 3, 83–88. [Google Scholar] [CrossRef]
- Knipling, E.B. Physical and physiological basis for the reflectance of visible and near-infrared radiation from vegetation. Remote Sens. Environ. 1970, 1, 155–159. [Google Scholar] [CrossRef]
- Karabourniotis, G.; Liakopoulos, G.; Bresta, P.; Nikolopoulos, D. The Optical Properties of Leaf Structural Elements and Their Contribution to Photosynthetic Performance and Photoprotection. Plants 2021, 10, 1455. [Google Scholar] [CrossRef]
- Adar, E.; Inbar, M.; Gal, S.; Issman, L.; Palevsky, E. Plant cell piercing by a predatory mite: Evidence and implications. Exp. Appl. Acarol. 2015, 65, 181–193. [Google Scholar] [CrossRef]
- Fürstenberg-Hägg, J.; Zagrobelny, M.; Bak, S. Plant Defense against Insect Herbivores. Int. J. Mol. Sci. 2013, 14, 10242–10297. [Google Scholar] [CrossRef] [Green Version]
- Huang, J.; Wei, C.; Zhang, Y.; Blackburn, G.A.; Wang, X.; Wei, C.; Wang, J. Meta-analysis of the detection of plant pigment concentrations using hyperspectral remotely sensed data. PLoS ONE 2015, 10, e0137029. [Google Scholar] [CrossRef] [Green Version]
- Gausman, H.; Allen, W. Optical parameters of leaves of 30 plant species. Plant Physiol. 1973, 52, 57–62. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Ma, J.; Zhong, N. Fourier transform near-infrared spectroscopy coupled with variable selection methods for fast determination of salmon fillets storage time. J. Mol. Struct. 2022, 1264, 133223. [Google Scholar] [CrossRef]
- Reisig, D.; Godfrey, L. Spectral response of cotton aphid–(Homoptera: Aphididae) and spider mite–(Acari: Tetranychidae) infested cotton: Controlled studies. Environ. Entomol. 2014, 36, 1466–1474. [Google Scholar] [CrossRef]
- Tu, Y.-H.; Phinn, S.; Johansen, K.; Robson, A. Assessing Radiometric Correction Approaches for Multi-Spectral UAS Imagery for Horticultural Applications. Remote Sens. 2018, 10, 1684. [Google Scholar] [CrossRef]




| Band | Centre (nm) | FWHM (nm) |
|---|---|---|
| Green | 550 | 40 |
| Red | 660 | 40 |
| Red edge | 735 | 10 |
| Near-infrared | 790 | 40 |
| Modelling Method | Accuracy | |||
|---|---|---|---|---|
| Calibration Set | Validation Set | Prediction Set | ||
| PLSDA | None | 0.89 | 0.82 | 0.90 |
| SG + SNV | 0.92 | 0.84 | 0.91 | |
| KNN | None | 0.77 | 0.77 | 0.86 |
| SG + SNV | 0.76 | 0.77 | 0.81 | |
| SVM | None | 0.99 | 0.83 | 0.82 |
| SG + SNV | 1.00 | 0.88 | 0.86 | |
| BPNN | None | 1.00 | 0.88 | 0.79 |
| SG + SNV | 1.00 | 0.89 | 0.85 | |
| Variable Selection Method | Wavelength (nm) |
|---|---|
| CARS | 466, 532, 662, 695, 820, 982, 983, 993, 1088, 1176, 1193, 1194, 2034, 2035, 2036, 2037, 2128, 2129, 2130, 2316 |
| SPA | 822, 695, 1232, 1204, 701, 1185, 839, 733, 1176, 697, 719, 837, 702, 693, 747, 827, 2033, 870, 1249, 760 |
| RF | 466, 983, 2129, 2034, 465, 2128, 2130, 2127, 2031, 824, 2042, 2126, 2123, 979, 2039, 2035, 2137, 2033, 1176, 2038 |
| Modelling Method | Accuracy | ||
|---|---|---|---|
| Calibration Set | Validation Set | Prediction Set | |
| CARS + PLSDA | 0.87 | 0.86 | 0.82 |
| CARS + KNN | 0.83 | 0.86 | 0.86 |
| CARS + SVM | 0.84 | 0.84 | 0.86 |
| CARS + BPNN | 0.99 | 0.87 | 0.86 |
| SPA + PLSDA | 0.83 | 0.79 | 0.82 |
| SPA + KNN | 0.81 | 0.79 | 0.82 |
| SPA + SVM | 0.84 | 0.79 | 0.86 |
| SPA + BPNN | 1.00 | 0.70 | 0.73 |
| RF + PLSDA | 0.80 | 0.72 | 0.68 |
| RF + KNN | 0.72 | 0.68 | 0.60 |
| RF + SVM | 0.70 | 0.67 | 0.68 |
| RF + BPNN | 1.00 | 0.72 | 0.71 |
| Vegetation Index | Significance (p-Value) | Accuracy |
|---|---|---|
| NDVI | <0.01 | 0.79 |
| GNDVI | <0.01 | 0.82 |
| REGNDVI | <0.01 | 0.80 |
| NRENDVI | <0.01 | 0.79 |
| GRVI | >0.05 | 0.52 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Aeberli, A.; Robson, A.; Phinn, S.; Lamb, D.W.; Johansen, K. A Comparison of Analytical Approaches for the Spectral Discrimination and Characterisation of Mite Infestations on Banana Plants. Remote Sens. 2022, 14, 5467. https://doi.org/10.3390/rs14215467
Aeberli A, Robson A, Phinn S, Lamb DW, Johansen K. A Comparison of Analytical Approaches for the Spectral Discrimination and Characterisation of Mite Infestations on Banana Plants. Remote Sensing. 2022; 14(21):5467. https://doi.org/10.3390/rs14215467
Chicago/Turabian StyleAeberli, Aaron, Andrew Robson, Stuart Phinn, David W. Lamb, and Kasper Johansen. 2022. "A Comparison of Analytical Approaches for the Spectral Discrimination and Characterisation of Mite Infestations on Banana Plants" Remote Sensing 14, no. 21: 5467. https://doi.org/10.3390/rs14215467
APA StyleAeberli, A., Robson, A., Phinn, S., Lamb, D. W., & Johansen, K. (2022). A Comparison of Analytical Approaches for the Spectral Discrimination and Characterisation of Mite Infestations on Banana Plants. Remote Sensing, 14(21), 5467. https://doi.org/10.3390/rs14215467






