Detection of White Leaf Disease in Sugarcane Crops Using UAV-Derived RGB Imagery with Existing Deep Learning Models
Abstract
:1. Introduction
2. Methodology
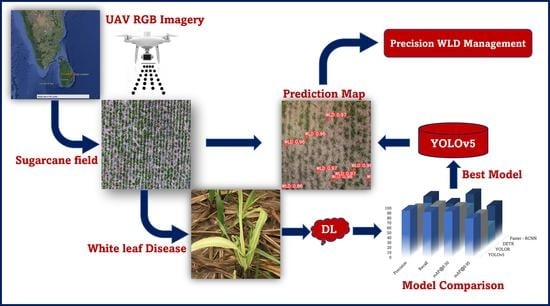
2.1. Process Pipeline
2.2. Study Area
2.3. UAV Image Acquisition
2.4. Ground Truth Data Collection
2.5. Image Orthomosaics
2.6. Image Tiles
2.7. Image Augmentation
2.8. Image Labelling
2.9. Steps in Different DL Models
2.9.1. YOLOv5
2.9.2. YOLOR
2.9.3. DETR
2.9.4. Faster R-CNN
2.10. Evaluation Metrics
3. Results
3.1. Visual Analysis of Evaluation Indicators during Training
3.2. Comparison of DL Model Performances
3.3. Training Duration
3.4. Bounding Box Detection Results from the Different DL Models
3.5. Model Comparison with Previous Work
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Sumesh, K.C.; Ninsawat, S.; Som-ard, J. Integration of RGB-based vegetation index, crop surface model and object-based image analysis approach for sugarcane yield estimation using unmanned aerial vehicle. Comput. Electron. Agric. 2021, 180, 105903. [Google Scholar] [CrossRef]
- Chen, J.; Wu, J.; Qiang, H.; Zhou, B.; Xu, G.; Wang, Z. Sugarcane nodes identification algorithm based on sum of local pixel of minimum points of vertical projection function. Comput. Electron. Agric. 2021, 182, 105994. [Google Scholar] [CrossRef]
- Huang, Y.-K.; Li, W.-F.; Zhang, R.-Y.; Wang, X.-Y. Color Illustration of Diagnosis and Control for Modern Sugarcane Diseases, Pests, and Weeds; Springer: Berlin/Heidelberg, Germany, 2018. [Google Scholar] [CrossRef]
- Braithwaite, K.S.; Croft, B.J.; Magarey, R.C. Progress in Identifying the Cause of Ramu Stunt Disease of Sugarcane. Proc. Aust. Soc. Sugar Cane Technol. 2007, 29, 235–241. [Google Scholar]
- Wang, X.; Zhang, R.; Shan, H.; Fan, Y.; Xu, H.; Huang, P.; Li, Z.; Duan, T.; Kang, N.; Li, W.; et al. Unmanned Aerial Vehicle Control of Major Sugarcane Diseases and Pests in Low Latitude Plateau. Agric. Biotechnol. 2019, 8, 48–51. [Google Scholar]
- Amarasingam, N.; Salgadoe, A.S.A.; Powell, K.; Gonzalez, L.F.; Natarajan, S. A review of UAV platforms, sensors, and applications for monitoring of sugarcane crops. Remote Sens. Appl. 2022, 26, 100712. [Google Scholar] [CrossRef]
- Wickramasinghe, K.P.; Wijesuriya, A.; Ariyawansha, B.D.S.K.; Perera, A.M.M.S.; Chanchala, K.M.G.; Manel, D.; Chandana, R.A.M. Performance of Sugarcane Varieties in a White Leaf Disease (WLD)-Prone Environment at Pelwatte. November 2019. Available online: http://sugarres.lk/wp-content/uploads/2020/05/Best-Paper-Award-–-Seventh-Symposium-on-Plantation-Crop-Research-2019.pdf (accessed on 5 May 2022).
- Sanseechan, P.; Saengprachathanarug, K.; Posom, J.; Wongpichet, S.; Chea, C.; Wongphati, M. Use of vegetation indices in monitoring sugarcane white leaf diseasesymptoms in sugarcane field using multispectral UAV aerial imagery. IOP Conf. Ser. Earth Environ. Sci. 2019, 301, 12025. [Google Scholar] [CrossRef]
- Cherry, R.H.; Nuessly, G.S.; Sandhu, H.S.; Insect Management in Sugarcane. Florida. 2011. Available online: http://edis.ifas.ufl.edu/pdffiles/IG/IG06500.pdf (accessed on 11 May 2022).
- Wilson, B.E. Successful Integrated Pest Management Minimizes the Economic Impact of Diatraea saccharalis (Lepidoptera: Crambidae) on the Louisiana Sugarcane Industry. J. Econ. Entomol. 2021, 114, 468–471. [Google Scholar] [CrossRef]
- Huang, W.; Lu, Y.; Chen, L.; Sun, D.; An, Y. Impact of pesticide/fertilizer mixtures on the rhizosphere microbial community of field-grown sugarcane. 3 Biotech 2021, 11, 210. [Google Scholar] [CrossRef]
- Vennila, A.; Palaniswami, C.; Durai, A.A.; Shanthi, R.M.; Radhika, K. Partitioning of Major Nutrients and Nutrient Use Efficiency of Sugarcane Genotypes. Sugar Tech 2021, 23, 741–746. [Google Scholar] [CrossRef]
- He, S.S.; Zeng, Y.; Liang, Z.X.; Jing, Y.; Tang, S.; Zhang, B.; Li, M. Economic Evaluation of Water-Saving Irrigation Practices for Sustainable Sugarcane Production in Guangxi Province, China. Sugar Tech 2021, 23, 1325–1331. [Google Scholar] [CrossRef]
- Verma, K.; Garg, P.K.; Prasad, K.S.H.; Dadhwal, V.K.; Dubey, S.K.; Kumar, A. Sugarcane Yield Forecasting Model Based on Weather Parameters. Sugar Tech 2021, 23, 158–166. [Google Scholar] [CrossRef]
- Wang, H.; Shang, S.; Wang, D.; He, X.; Feng, K.; Zhu, H. Plant Disease Detection and Classification Method Based on the Optimized Lightweight YOLOv5 Model. Agriculture 2022, 12, 931. [Google Scholar] [CrossRef]
- Narmilan, G.N.; Sumangala, K. Assessment on Consequences and Benefits of the Smart Farming Techniques in Batticaloa District, Sri Lanka. Int. J. Res. Publ. 2020, 61, 14–20. [Google Scholar] [CrossRef]
- Narmilan, A.; Puvanitha, N. Mitigation Techniques for Agricultural Pollution by Precision Technologies with a Focus on the Internet of Things (IoTs): A Review. Agric. Rev. 2020, 41, 279–284. [Google Scholar] [CrossRef]
- Narmilan, A.; Niroash, G. Reduction Techniques for Consequences of Climate Change by Internet of Things (IoT) with an Emphasis on the Agricultural Production: A Review. Int. J. Sci. Technol. Eng. Manag. 2020, 5844, 6–13. [Google Scholar]
- Suresh, K.; Narmilan, A.; Ahmadh, R.K.; Kariapper, R.; Nawaz, S.S.; Suresh, J. Farmers’ Perception on Precision Farming Technologies: A Novel Approach. Indian J. Agric. Econ. 2022, 77, 264–276. [Google Scholar]
- Biffi, L.J.; Mitishita, E.; Liesenberg, V.; Santos, A.A.d.; Gonçalves, D.N.; Estrabis, N.V.; Silva, J.d.A.; Osco, L.P.; Ramos, A.P.M.; Centeno, J.A.S.; et al. Article atss deep learning-based approach to detect apple fruits. Remote Sens. 2021, 13, 54. [Google Scholar] [CrossRef]
- Parvathi, S.; Selvi, S.T. Detection of maturity stages of coconuts in complex background using Faster R-CNN model. Biosyst. Eng. 2021, 202, 119–132. [Google Scholar] [CrossRef]
- Narmilan, A. E-Agricultural Concepts for Improving Productivity: A Review. Sch. J. Eng. Technol. (SJET) 2017, 5, 10–17. [Google Scholar] [CrossRef]
- Chandra, L.; Desai, S.V.; Guo, W.; Balasubramanian, V.N. Computer Vision with Deep Learning for Plant Phenotyping in Agriculture: A Survey. arXiv 2020, arXiv:2006.11391. [Google Scholar] [CrossRef]
- Seyyedhasani, H.; Digman, M.; Luck, B.D. Utility of a commercial unmanned aerial vehicle for in-field localization of biomass bales. Comput. Electron. Agric. 2021, 180, 105898. [Google Scholar] [CrossRef]
- Nebiker, S.; Annen, A.; Scherrer, M.; Oesch, D. A lightweight multispectral sensor for micro-UAV—Opportunities for very high resolution airborne remote sensing. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2008, 37, 1193–1200. [Google Scholar]
- Yue, J.; Lei, T.; Li, C.; Zhu, J. The Application of Unmanned Aerial Vehicle Remote Sensing in Quickly Monitoring Crop Pests. Intell. Autom. Soft Comput. 2012, 18, 1043–1052. [Google Scholar] [CrossRef]
- Aasen, H.; Honkavaara, E.; Lucieer, A.; Zarco-Tejada, P.J. Quantitative remote sensing at ultra-high resolution with UAV spectroscopy: A review of sensor technology, measurement procedures, and data correctionworkflows. Remote Sens. 2018, 10, 1091. [Google Scholar] [CrossRef]
- Casagli, N.; Frodella, W.; Morelli, S.; Tofani, V.; Ciampalini, A.; Intrieri, E.; Lu, P. Spaceborne, UAV and ground-based remote sensing techniques for landslide mapping, monitoring and early warning. Geoenvironmental Disasters 2017, 4, 9. [Google Scholar] [CrossRef]
- Xiang, H.; Tian, L. Development of a low-cost agricultural remote sensing system based on an autonomous unmanned aerial vehicle (UAV). Biosyst. Eng. 2011, 108, 174–190. [Google Scholar] [CrossRef]
- Chivasa, W.; Mutanga, O.; Burgueño, J. UAV-based high-throughput phenotyping to increase prediction and selection accuracy in maize varieties under artificial MSV inoculation. Comput. Electron. Agric. 2021, 184. [Google Scholar] [CrossRef]
- Aboutalebi, M.; Torres-Rua, A.F.; Kustas, W.P.; Nieto, H.; Coopmans, C.; McKee, M. Assessment of different methods for shadow detection in high-resolution optical imagery and evaluation of shadow impact on calculation of NDVI, and evapotranspiration. Irrig. Sci. 2019, 37, 407–429. [Google Scholar] [CrossRef]
- Sandino, J.; Gonzalez, F.; Mengersen, K.; Gaston, K.J. UAVs and machine learning revolutionizing invasive grass and vegetation surveys in remote arid lands. Sensors 2018, 18, 605. [Google Scholar] [CrossRef] [Green Version]
- Sandino, J.; Gonzalez, F. A Novel Approach for Invasive Weeds and Vegetation Surveys Using UAS and Artificial Intelligence. In Proceedings of the 2018 23rd International Conference on Methods and Models in Automation and Robotics, MMAR 2018, Miedzyzdroje, Poland, 27–30 August 2018; pp. 515–520. [Google Scholar] [CrossRef]
- Sandino, J.; Pegg, G.; Gonzalez, F.; Smith, G. Aerial Mapping of Forests Affected by Pathogens Using UAVs, Hyperspectral Sensors, and Artificial Intelligence. Sensors 2018, 18, 944. [Google Scholar] [CrossRef] [Green Version]
- Ampatzidis, Y.; Partel, V. UAV-based high throughput phenotyping in citrus utilizing multispectral imaging and artificial intelligence. Remote Sens. 2019, 11, 410. [Google Scholar] [CrossRef] [Green Version]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Yang, H. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vergouw, B.; Nagel, H.; Bondt, G.; Custers, B. Drone Technology: Types, Payloads, Applications, Frequency Spectrum Issues and Future Developments; Springer: Berlin/Heidelberg, Germany, 2016. [Google Scholar] [CrossRef]
- Olson, D.; Anderson, J. Review on unmanned aerial vehicles, remote sensors, imagery processing, and their applications in agriculture. Agron. J. 2021, 113, 971–992. [Google Scholar] [CrossRef]
- Anagnostis, A.; Tagarakis, A.C.; Asiminari, G.; Papageorgiou, E.; Kateris, D.; Moshou, D.; Bochtis, D. A deep learning approach for anthracnose infected trees classification in walnut orchards. Comput. Electron. Agric. 2021, 182, 105998. [Google Scholar] [CrossRef]
- Gonzalo-Martín, C.; García-Pedrero, A.; Lillo-Saavedra, M. Improving deep learning sorghum head detection through test time augmentation. Comput. Electron. Agric. 2021, 186, 106179. [Google Scholar] [CrossRef]
- Hasan, S.M.M.; Sohel, F.; Diepeveen, D.; Laga, H.; Jones, M.G.K. A survey of deep learning techniques for weed detection from images. Comput. Electron. Agric. 2021, 184, 106067. [Google Scholar] [CrossRef]
- Shin, J.; Chang, Y.K.; Heung, B.; Nguyen-Quang, T.; Price, G.W.; Al-Mallahi, A. A deep learning approach for RGB image-based powdery mildew disease detection on strawberry leaves. Comput. Electron. Agric. 2020, 183, 106042. [Google Scholar] [CrossRef]
- Ahmad, A.; Saraswat, D.; Aggarwal, V.; Etienne, A.; Hancock, B. Performance of deep learning models for classifying and detecting common weeds in corn and soybean production systems. Comput. Electron. Agric. 2021, 184, 106081. [Google Scholar] [CrossRef]
- Vong, N.; Conway, L.S.; Zhou, J.; Kitchen, N.R.; Sudduth, K.A. Early corn stand count of different cropping systems using UAV-imagery and deep learning. Comput. Electron. Agric. 2021, 186, 106214. [Google Scholar] [CrossRef]
- Hong, H.; Lin, J.; Huang, F. Tomato Disease Detection and Classification by Deep Learning. In Proceedings of the 2020 International Conference on Big Data, Artificial Intelligence and Internet of Things Engineering, ICBAIE 2020, Fuzhou, China, 12–14 June 2020; pp. 25–29. [Google Scholar] [CrossRef]
- Chen, Z.; Wu, R.; Lin, Y.; Li, C.; Chen, S.; Yuan, Z.; Zou, X. Plant Disease Recognition Model Based on Improved YOLOv5. Agronomy 2022, 12, 365. [Google Scholar] [CrossRef]
- Cao, J.; Zhang, Z.; Tao, F.; Zhang, L.; Luo, Y.; Zhang, J.; Xie, J. Integrating Multi-Source Data for Rice Yield Prediction across China using Machine Learning and Deep Learning Approaches. Agric. For. Meteorol. 2021, 297, 108275. [Google Scholar] [CrossRef]
- Osorio, K.; Puerto, A.; Pedraza, C.; Jamaica, D.; Rodríguez, L. A Deep Learning Approach for Weed Detection in Lettuce Crops Using Multispectral Images. AgriEngineering 2020, 2, 471–488. [Google Scholar] [CrossRef]
- Zhang, P.; Li, D. EPSA-YOLO-V5s: A novel method for detecting the survival rate of rapeseed in a plant factory based on multiple guarantee mechanisms. Comput. Electron. Agric. 2022, 193, 106714. [Google Scholar] [CrossRef]
- Santos, T.T.; de Souza, L.L.; Santos, A.A.d.; Avila, S. Grape detection, segmentation, and tracking using deep neural networks and three-dimensional association. Comput. Electron. Agric. 2020, 170, 105247. [Google Scholar] [CrossRef] [Green Version]
- Wu, B.; Liang, A.; Zhang, H.; Zhu, T.; Zou, Z.; Yang, D.; Su, J. Application of conventional UAV-based high-throughput object detection to the early diagnosis of pine wilt disease by deep learning. For. Ecol. Manag. 2021, 486, 118986. [Google Scholar] [CrossRef]
- Tan, L.; Lu, J.; Jiang, H. Tomato Leaf Diseases Classification Based on Leaf Images: A Comparison between Classical Machine Learning and Deep Learning Methods. AgriEngineering 2021, 3, 542–558. [Google Scholar] [CrossRef]
- Dananjayan, S.; Tang, Y.; Zhuang, J.; Hou, C.; Luo, S. Assessment of state-of-the-art deep learning based citrus disease detection techniques using annotated optical leaf images. Comput. Electron. Agric. 2022, 193, 106658. [Google Scholar] [CrossRef]
- Qi, J.; Liu, X.; Liu, K.; Xu, F.; Guo, H.; Tian, X.; Li, Y. An improved YOLOv5 model based on visual attention mechanism: Application to recognition of tomato virus disease. Comput. Electron. Agric. 2022, 194, 106780. [Google Scholar] [CrossRef]
- Temniranrat, P.; Kiratiratanapruk, K.; Kitvimonrat, A.; Sinthupinyo, W.; Patarapuwadol, S. A system for automatic rice disease detection from rice paddy images serviced via a Chatbot. Comput. Electron. Agric. 2021, 185, 106156. [Google Scholar] [CrossRef]
- Zhang, Y.; Yu, J.; Chen, Y.; Yang, W.; Zhang, W.; He, Y. Real-time strawberry detection using deep neural networks on embedded system (rtsd-net): An edge AI application. Comput. Electron. Agric. 2022, 192, 106586. [Google Scholar] [CrossRef]
- Kang, H.; Chen, C. Fast implementation of real-time fruit detection in apple orchards using deep learning. Comput. Electron. Agric. 2020, 168, 105108. [Google Scholar] [CrossRef]
- Yu, Y.; Zhang, K.; Yang, L.; Zhang, D. Fruit detection for strawberry harvesting robot in non-structural environment based on Mask-R-CNN. Comput. Electron. Agric. 2019, 163, 104846. [Google Scholar] [CrossRef]
- Wang, X.; Tang, J.; Whitty, M. DeepPhenology: Estimation of apple flower phenology distributions based on deep learning. Comput. Electron. Agric. 2021, 185, 106123. [Google Scholar] [CrossRef]
- Yang, G.F.; Yong, Y.A.N.G.; He, Z.K.; Zhang, X.Y.; Yong, H.E. A rapid, low-cost deep learning system to classify strawberry disease based on cloud service. J. Integr. Agric. 2022, 21, 460–473. [Google Scholar] [CrossRef]
- Kathiresan, G.; Anirudh, M.; Nagharjun, M.; Karthik, R. Disease detection in rice leaves using transfer learning techniques. J. Phys. Conf. Ser. 2021, 1911, 012004. [Google Scholar] [CrossRef]
- Yao, J.; Qi, J.; Zhang, J.; Shao, H.; Yang, J.; Li, X. A real-time detection algorithm for kiwifruit defects based on yolov5. Electronics 2021, 10, 1711. [Google Scholar] [CrossRef]
- Sethy, P.K.; Barpanda, N.K.; Rath, A.K.; Behera, S.K. Rice false smut detection based on faster R-CNN. Indones. J. Electr. Eng. Comput. Sci. 2020, 19, 1590–1595. [Google Scholar] [CrossRef]
- Ieamsaard, J.; Charoensook, S.N.; Yammen, S. Deep Learning-based Face Mask Detection Using YoloV5. In Proceedings of the 2021 9th International Electrical Engineering Congress, iEECON 2021, Pattaya, Thailand, 10–12 March 2021; pp. 428–431. [Google Scholar] [CrossRef]
- Wang, C.-Y.; Yeh, I.-H.; Liao, H.-Y.M.; You Only Learn One Representation: Unified Network for Multiple Tasks. May 2021. Available online: http://arxiv.org/abs/2105.04206 (accessed on 12 May 2022).
- Brungel, R.; Friedrich, C.M. DETR and YOLOv5: Exploring performance and self-training for diabetic foot ulcer detection. In Proceedings of the IEEE Symposium on Computer-Based Medical Systems, Aveiro, Portugal, 7–9 June 2021; Volume 2021, pp. 148–153. [Google Scholar] [CrossRef]
- Carion, N.; Massa, F.; Synnaeve, G.; Usunier, N.; Kirillov, A.; Zagoruyko, S. End-to-End Object Detection with Transformers. Available online: https://github.com/facebookresearch/detr (accessed on 15 August 2022).
- Paliyam, M.; Nakalembe, C.; Liu, K.; Nyiawung, R.; Kerner, H. Street2Sat: A Machine Learning Pipeline for Generating Ground-truth Geo-Referenced Labeled Datasets from Street-Level Images. 2021. Available online: https://github.com/ultralytics/yolov5 (accessed on 23 June 2022).
- Murugeswari, R.; Anwar, Z.S.; Dhananjeyan, V.R.; Karthik, C.N. Automated Sugarcane Disease Detection Using Faster R-CNN with an Android Application. In Proceedings of the 2022 6th International Conference on Trends in Electronics and Informatics, ICOEI 2022—Proceedings, Tirunelveli, India, 28–30 April 2022; pp. 1–7. [Google Scholar] [CrossRef]
- Chen, W.; Ju, C.; Li, Y.; Hu, S.; Qiao, X. Sugarcane stem node recognition in field by deep learning combining data expansion. Appl. Sci. 2021, 11, 8663. [Google Scholar] [CrossRef]
- Zhu, C.; Wu, C.; Li, Y.; Hu, S.; Gong, H. Spatial Location of Sugarcane Node for Binocular Vision-Based Harvesting Robots Based on Improved YOLOv4. Appl. Sci. 2022, 12, 3088. [Google Scholar] [CrossRef]
- Malik, H.S.; Dwivedi, M.; Omkar, S.N.; Javed, T.; Bakey, A.; Pala, M.R.; Chakravarthy, A. Disease Recognition in Sugarcane Crop Using Deep Learning. In Advances in Artificial Intelligence and Data Engineering; Kacprzyk, J., Ed.; Springer: Singapore, 2019; Volume 1133, pp. 189–205. Available online: http://www.springer.com/series/11156 (accessed on 3 May 2022).
- Kumpala, I.; Wichapha, N.; Prasomsab, P. Sugar Cane Red Stripe Disease Detection using YOLO CNN of Deep Learning Technique. Eng. Access 2022, 8, 192–197. [Google Scholar]
- Narmilan, A.; Gonzalez, F.; Salgadoe, A.S.A.; Powell, K. Detection of White Leaf Disease in Sugarcane Using Machine Learning Techniques over UAV Multispectral Images. Drones 2022, 6, 230. [Google Scholar] [CrossRef]
- Sugar Research Australia (SRA). WLD Information Sheet. 2013. Available online: Sugarresearch.com.au (accessed on 13 April 2022).
- Zhou, F.; Zhao, H.; Nie, Z. Safety Helmet Detection Based on YOLOv5. In Proceedings of the 2021 IEEE International Conference on Power Electronics, Computer Applications, ICPECA 2021, Shenyang, China, 22–24 January 2021; pp. 6–11. [Google Scholar] [CrossRef]
- Du, X.; Song, L.; Lv, Y.; Qiu, S. A Lightweight Military Target Detection Algorithm Based on Improved YOLOv5. Electronics 2022, 11, 3263. [Google Scholar] [CrossRef]
- Wang, Q.; Cheng, M.; Huang, S.; Cai, Z.; Zhang, J.; Yuan, H. A deep learning approach incorporating YOLO v5 and attention mechanisms for field real-time detection of the invasive weed Solanum rostratum Dunal seedlings. Comput. Electron. Agric. 2022, 199, 107194. [Google Scholar] [CrossRef]
- Li, X.; Wang, C.; Ju, H.; Li, Z. Surface Defect Detection Model for Aero-Engine Components Based on Improved YOLOv5. Appl. Sci. 2022, 12, 7235. [Google Scholar] [CrossRef]
- Jing, Y.; Ren, Y.; Liu, Y.; Wang, D.; Yu, L. Automatic Extraction of Damaged Houses by Earthquake Based on Improved YOLOv5: A Case Study in Yangbi. Remote Sens. 2022, 14, 382. [Google Scholar] [CrossRef]
- Training, Validation, and Test Datasets—Machine Learning Glossary. Available online: https://machinelearning.wtf/terms/training-validation-test-datasets/ (accessed on 31 October 2022).
- Why No Augmentation Applied to Test or Validation Data and Only to Train Data? | Data Science and Machine Learning | Kaggle. Available online: https://www.kaggle.com/questions-and-answers/291581 (accessed on 31 October 2022).
- Data Augmentation | Baeldung on Computer Science. Available online: https://www.baeldung.com/cs/ml-data-augmentation (accessed on 31 October 2022).
- Abayomi-Alli, O.; Damaševičius, R.; Misra, S.; Maskeliūnas, R. Cassava disease recognition from low-quality images using enhanced data augmentation model and deep learning. Expert Syst. 2021, 38, e12746. [Google Scholar] [CrossRef]
- Li, J.; Zhu, X.; Jia, R.; Liu, B.; Yu, C. Apple-YOLO: A Novel Mobile Terminal Detector Based on YOLOv5 for Early Apple Leaf Diseases. In Proceedings of the 2022 IEEE 46th Annual Computers, Software, and Applications Conference (COMPSAC), Los Alamitos, CA, USA, 27 June–1 July 2022; pp. 352–361. [Google Scholar] [CrossRef]
- Cruz, M.; Mafra, S.; Teixeira, E.; Figueiredo, F. Smart Strawberry Farming Using Edge Computing and IoT. Sensors 2022, 22, 5866. [Google Scholar] [CrossRef]
- Mathew, P.; Mahesh, T.Y. Leaf-based disease detection in bell pepper plant using YOLO v5. Signal Image Video Process 2022, 16, 841–847. [Google Scholar] [CrossRef]
- Wang, Y.; Sun, F.; Wang, Z.; Zhou, Z.; Lan, P. Apple Leaf Disease Identification Method Based on Improved YoloV5; Springer: Singapore, 2022; pp. 1246–1252. [Google Scholar] [CrossRef]
- Jhatial, M.J.; Shaikh, R.A.; Shaikh, N.A.; Rajper, S.; Arain, R.H.; Chandio, G.H.; Shaikh, K.H. Deep Learning-Based Rice Leaf Diseases Detection Using Yolov5. Sukkur IBA J. Comput. Math. Sci. 2022, 6, 49–61. [Google Scholar]
- Yu, R.; Luo, Y.; Zhou, Q.; Zhang, X.; Wu, D.; Ren, L. Early detection of pine wilt disease using deep learning algorithms and UAV-based multispectral imagery. For. Ecol. Manag. 2021, 497, 119493. [Google Scholar] [CrossRef]
- Sun, Z.; Ibrayim, M.; Hamdulla, A. Detection of Pine Wilt Nematode from Drone Images Using UAV. Sensors 2022, 22, 4704. [Google Scholar] [CrossRef] [PubMed]
- Cynthia, S.T.; Hossain, K.M.S.; Hasan, M.N.; Asaduzzaman, M.; Das, A.K. Automated Detection of Plant Diseases Using Image Processing and Faster R-CNN Algorithm. In Proceedings of the 2019 International Conference on Sustainable Technologies for Industry 4.0 (STI), Dhaka, Bangladesh, 24–25 December 2019. [Google Scholar]
- Wang, Q.; Qi, F. Tomato diseases recognition based on faster R-CNN. In Proceedings of the 10th International Conference on Information Technology in Medicine and Education, ITME 2019, Qingdao, China, 23–25 August 2019; pp. 772–776. [Google Scholar] [CrossRef]
- Wu, J.; Wen, C.; Chen, H.; Ma, Z.; Zhang, T.; Su, H.; Yang, C. DS-DETR: A Model for Tomato Leaf Disease Segmentation and Damage Evaluation. Agronomy 2022, 12, 2023. [Google Scholar] [CrossRef]


















| Location | Application | DL Technique | Literature |
|---|---|---|---|
| Brazil | Detection of apple fruits | Adaptive Training Sample Selection (ATSS) Retina Net, Cascade R-CNN, Faster R-CNN, Feature Selective Anchor-Free (FSAF), and High-Resolution Network (HRNet) | [20] |
| Colombia | Weed detection in a lettuce field | YOLOV3, Mask R-CNN | [48] |
| China | Detection of the survival rate of rape | YOLOV5, Faster R-CNN, YOLOv3, and YOLOv4 | [49] |
| Brazil | Detection of grape | YOLOv2 and YOLOv3 | [50] |
| Florida | Detect, count, and geolocate Citrus trees | YOLOv3 | [35] |
| China | Detection of Pine wilt disease | YOLOv3 and Faster R-CNN | [51] |
| China | Tomato Leaf Diseases Classification | GG16, VGG19, ResNet34, ResNeXt50 (32 × 4 d), EfficientNet-b7, and MobileNetV2 | [52] |
| China | Detection of citrus leaf diseases | CenterNet, YOLOv4, Faster R-CNN, DetectoRS, Cascade R-CNN, Foveabox and Deformabe DETR | [53] |
| China | Detection of tomato virus diseases | YOLOv5 | [54] |
| China | Detection of plant diseases | YOLOv5 | [15] |
| Thailand | Detection of rice disease | LINE Bot System | [55] |
| China | Detection strawberry | RTSD-Net | [56] |
| Australia | real-time fruit detection in apple orchards | LedNet | [57] |
| China | Fruit detection for strawberry harvesting | Mask R-CNN | [58] |
| Australia | Estimation of apple flower phenology | VGG-16, YOLOv5 | [59] |
| China | classify strawberry disease | LFC-Net | [60] |
| India | Disease detection in rice | MobileNet, ResNet 50, ResNet 101, Inception V3, Xception, and RiceDenseNet | [61] |
| China | Plant Disease Recognition | YOLOv5 | [46] |
| China | Detection of Kiwifruit Defects | YOLOv5 | [62] |
| India | Detection of maturity stages of coconuts | Faster R-CNN | [21] |
| India | Rice false smut detection | Faster R-CNN | [63] |
| Model | Precision | Recall | [email protected] | [email protected] | Model Size |
|---|---|---|---|---|---|
| YOLOv5 | 95 | 92 | 93 | 79 | 14 MB |
| YOLOR | 87 | 93 | 90 | 75 | 281 MB |
| DETR | 77 | 69 | 77 | 41 | 473 MB |
| Faster R-CNN | 90 | 76 | 95 | 71 | 158 MB |
| Model | Time (Hours: Minutes: Seconds) |
|---|---|
| YOLOv5 | 06:02:55 |
| YOLOR | 12:10:31 |
| DETR | 30:22:47 |
| Faster R-CNN | 03:03:21 |
| XGB | RF | DT | KNN | |
|---|---|---|---|---|
| Precision (%) | 72 | 71 | 69 | 71 |
| Recall (%) | 72 | 72 | 65 | 67 |
| F1-score (%) | 71 | 71 | 67 | 69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Amarasingam, N.; Gonzalez, F.; Salgadoe, A.S.A.; Sandino, J.; Powell, K. Detection of White Leaf Disease in Sugarcane Crops Using UAV-Derived RGB Imagery with Existing Deep Learning Models. Remote Sens. 2022, 14, 6137. https://doi.org/10.3390/rs14236137
Amarasingam N, Gonzalez F, Salgadoe ASA, Sandino J, Powell K. Detection of White Leaf Disease in Sugarcane Crops Using UAV-Derived RGB Imagery with Existing Deep Learning Models. Remote Sensing. 2022; 14(23):6137. https://doi.org/10.3390/rs14236137
Chicago/Turabian StyleAmarasingam, Narmilan, Felipe Gonzalez, Arachchige Surantha Ashan Salgadoe, Juan Sandino, and Kevin Powell. 2022. "Detection of White Leaf Disease in Sugarcane Crops Using UAV-Derived RGB Imagery with Existing Deep Learning Models" Remote Sensing 14, no. 23: 6137. https://doi.org/10.3390/rs14236137