Predicting Leaf Phenology in Forest Tree Species Using UAVs and Satellite Images: A Case Study for European Beech (Fagus sylvatica L.)
Abstract
:1. Introduction
2. Materials and Methods
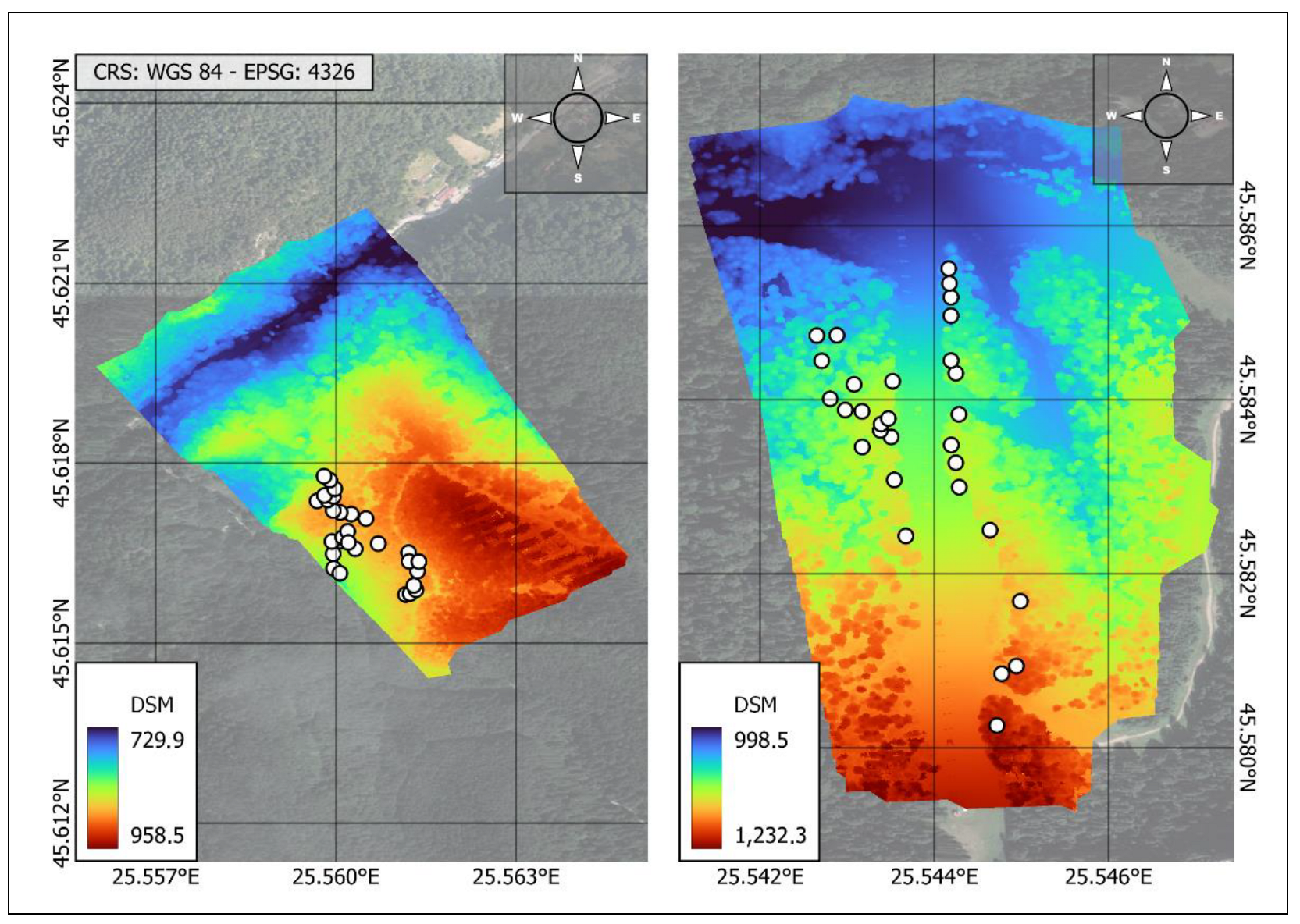
2.1. Study Sites
2.2. Ground Phenological Observations
2.2.1. Leaf Unfolding
2.2.2. Senescence
2.3. Time Series Data Collection of Phenological Observations and Image Processing Using UAVs
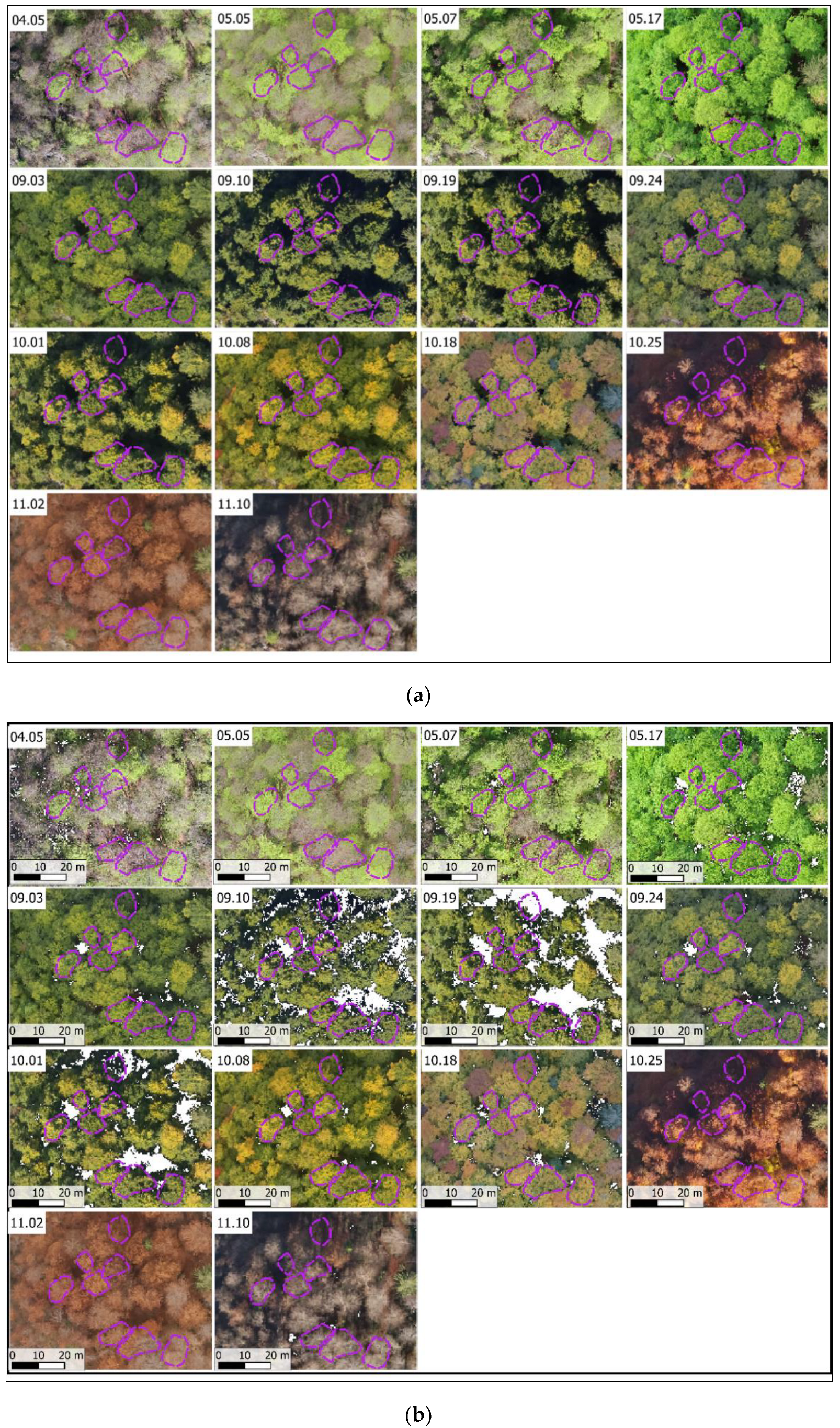
2.3.1. Image Acquisition
2.3.2. UAV Image Processing
2.4. Time Series Data Collection of Phenological Observations and Image Processing Using Copernicus Biophysical Parameters
2.4.1. Data Collection
2.4.2. Data Processing Analysis
2.5. Statistical Analysis
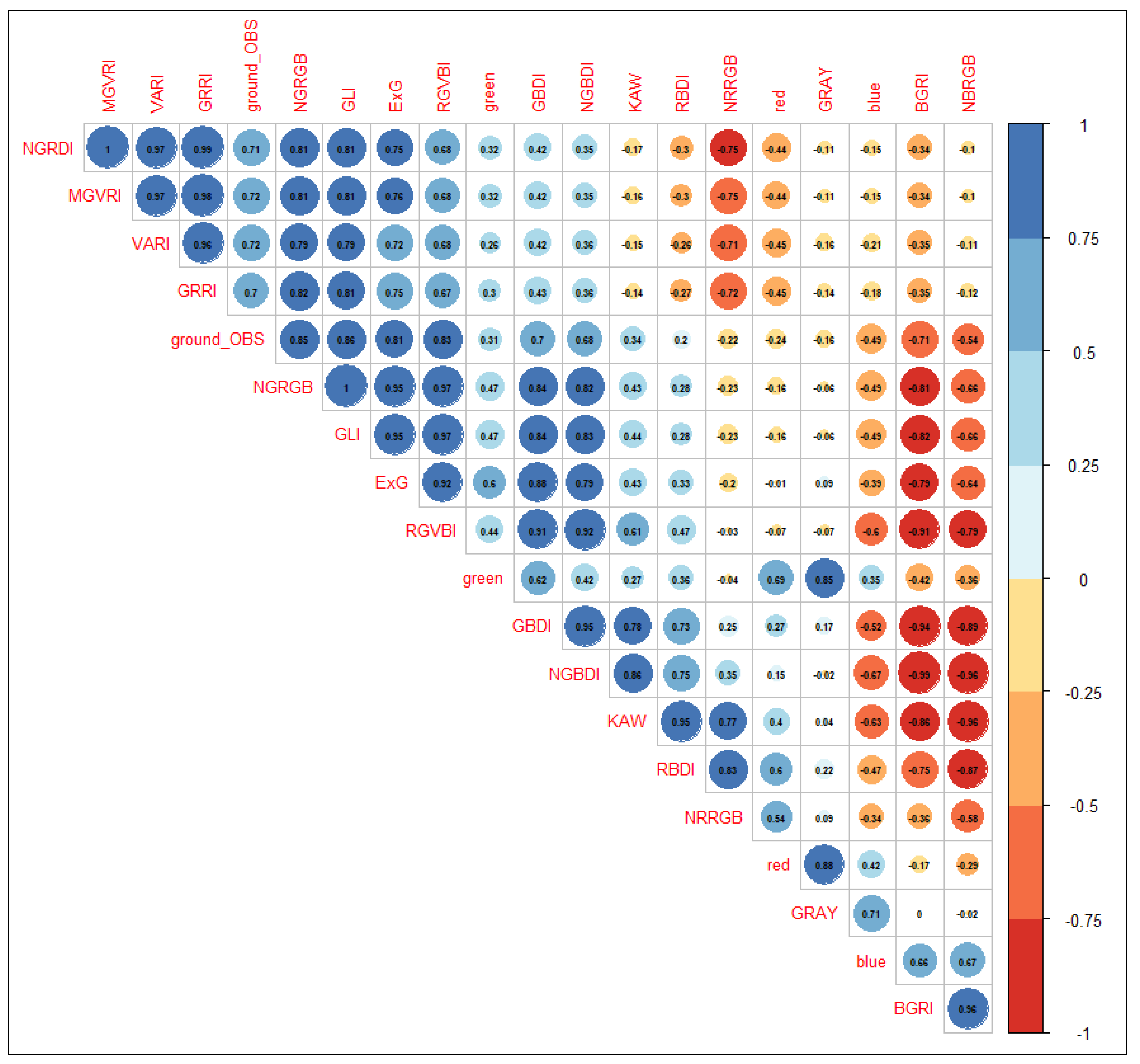
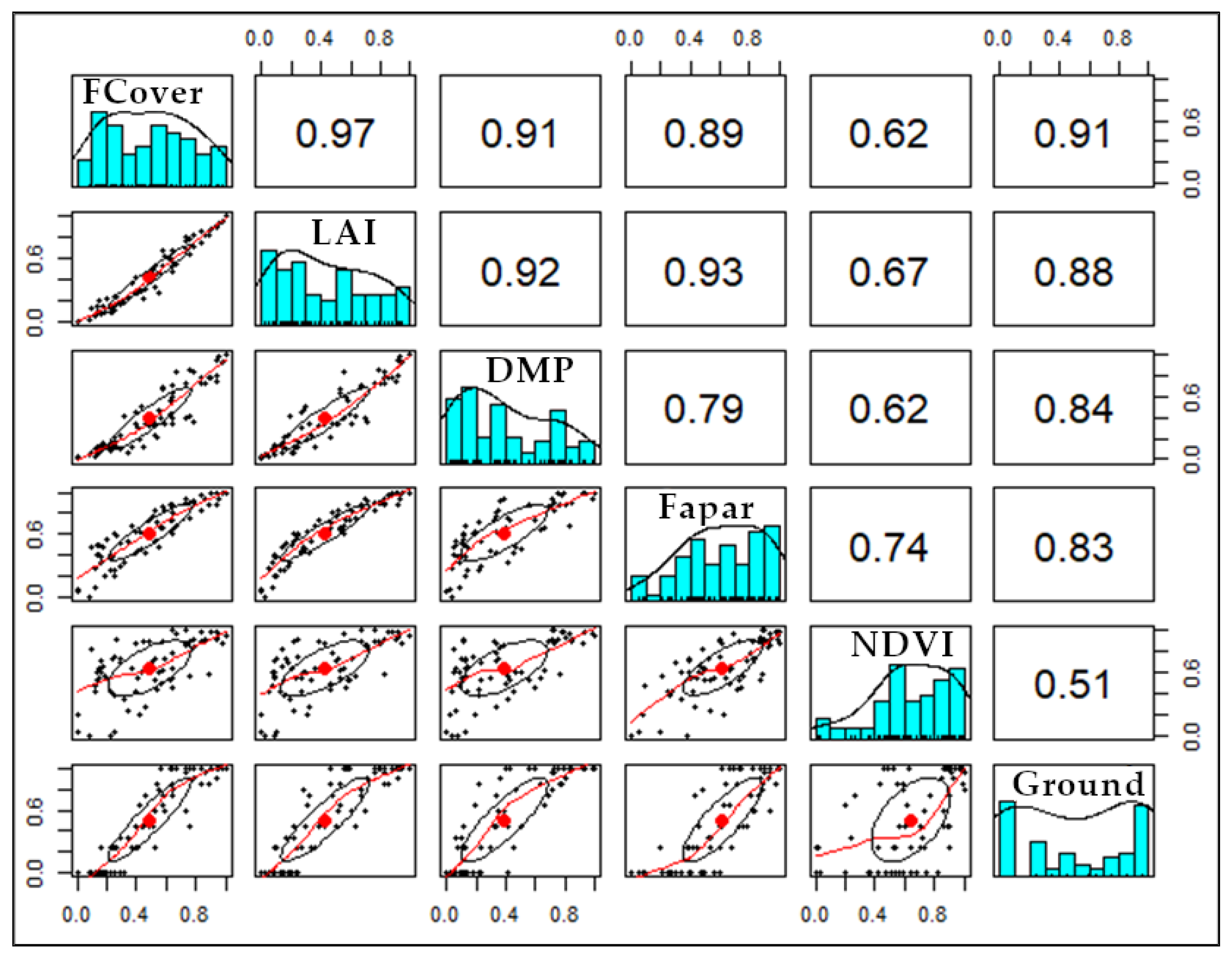
2.5.1. Correlation Analysis
2.5.2. Regression Analysis
Prediction of the Phenology Using Linear Regression Analysis
Prediction of the Phenology Using Non-Linear Regression Analysis
3. Results
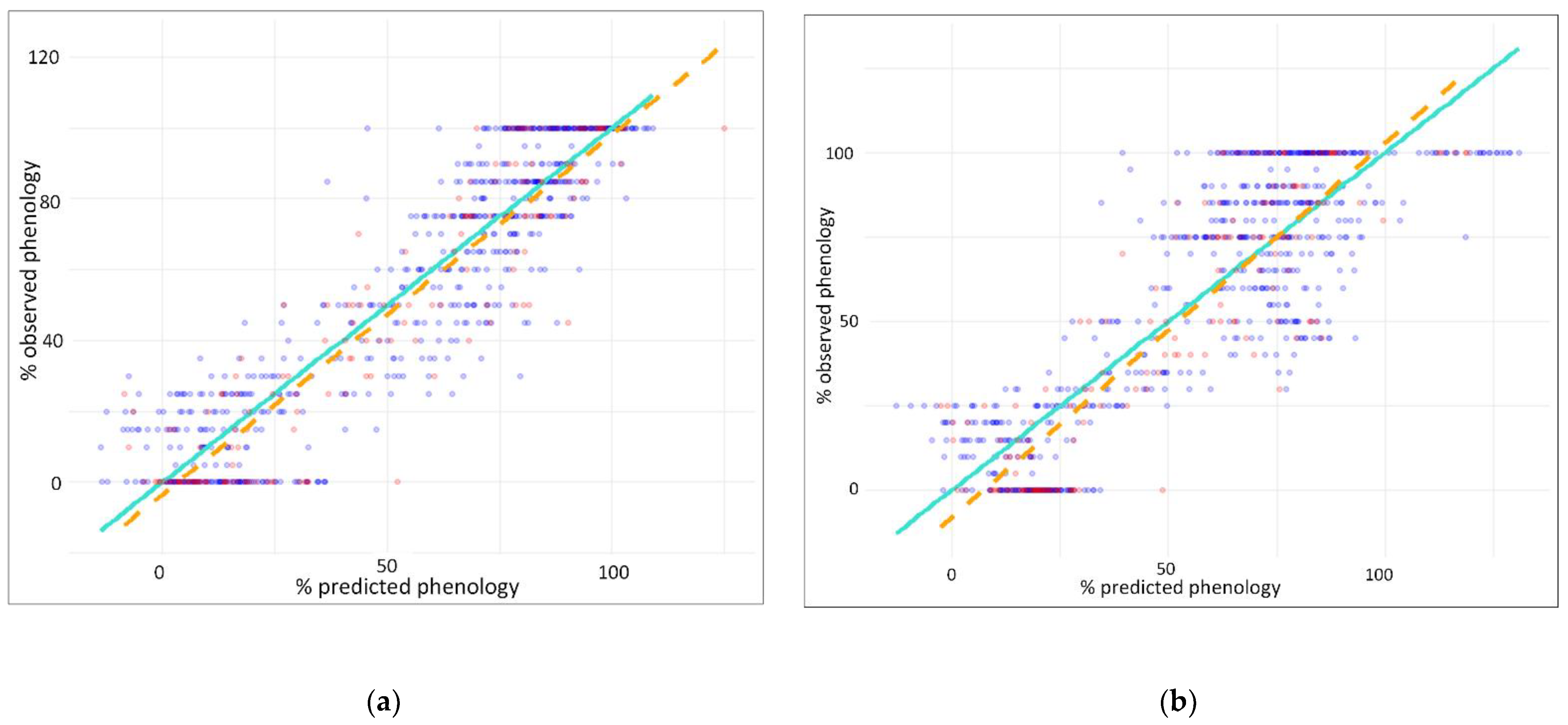
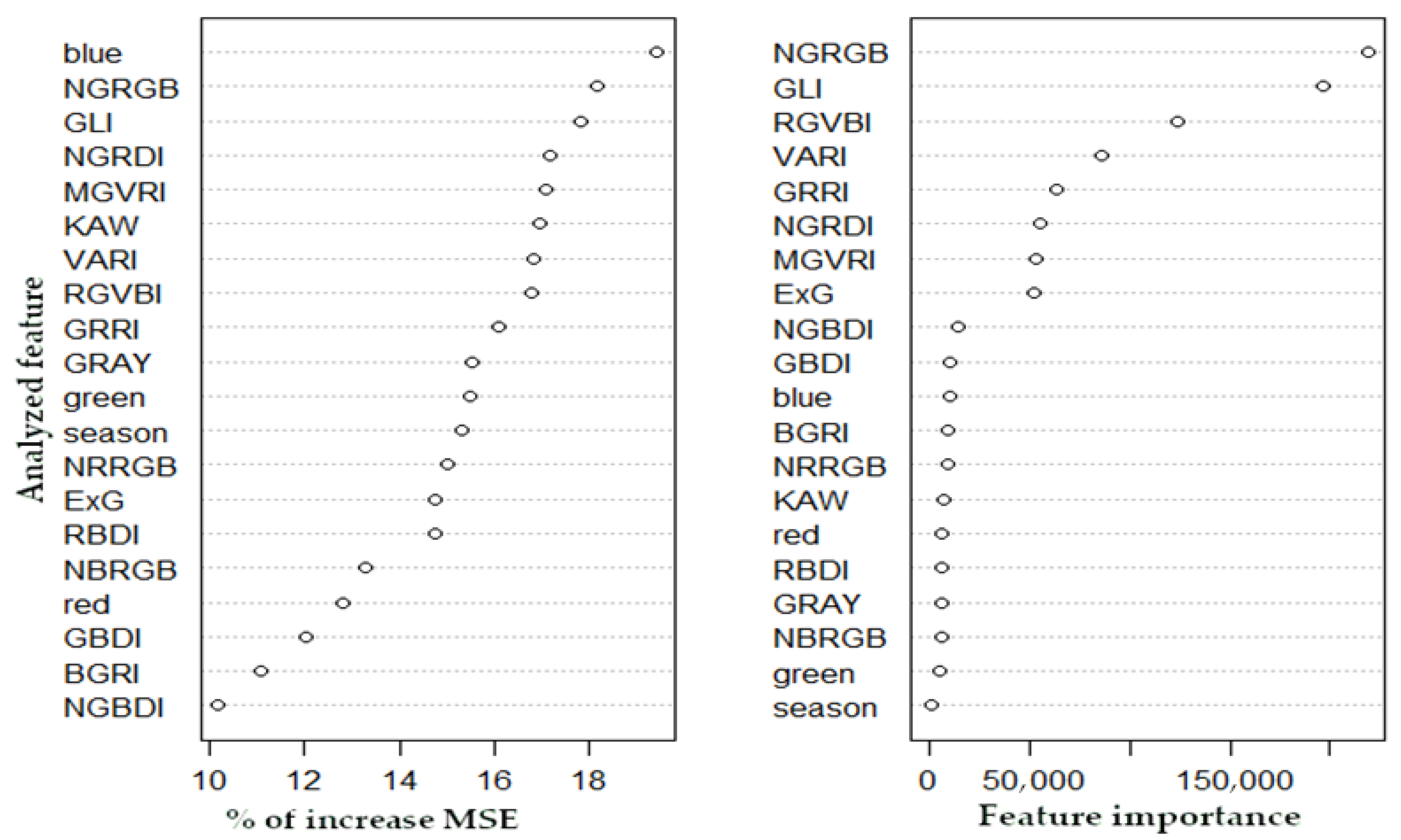
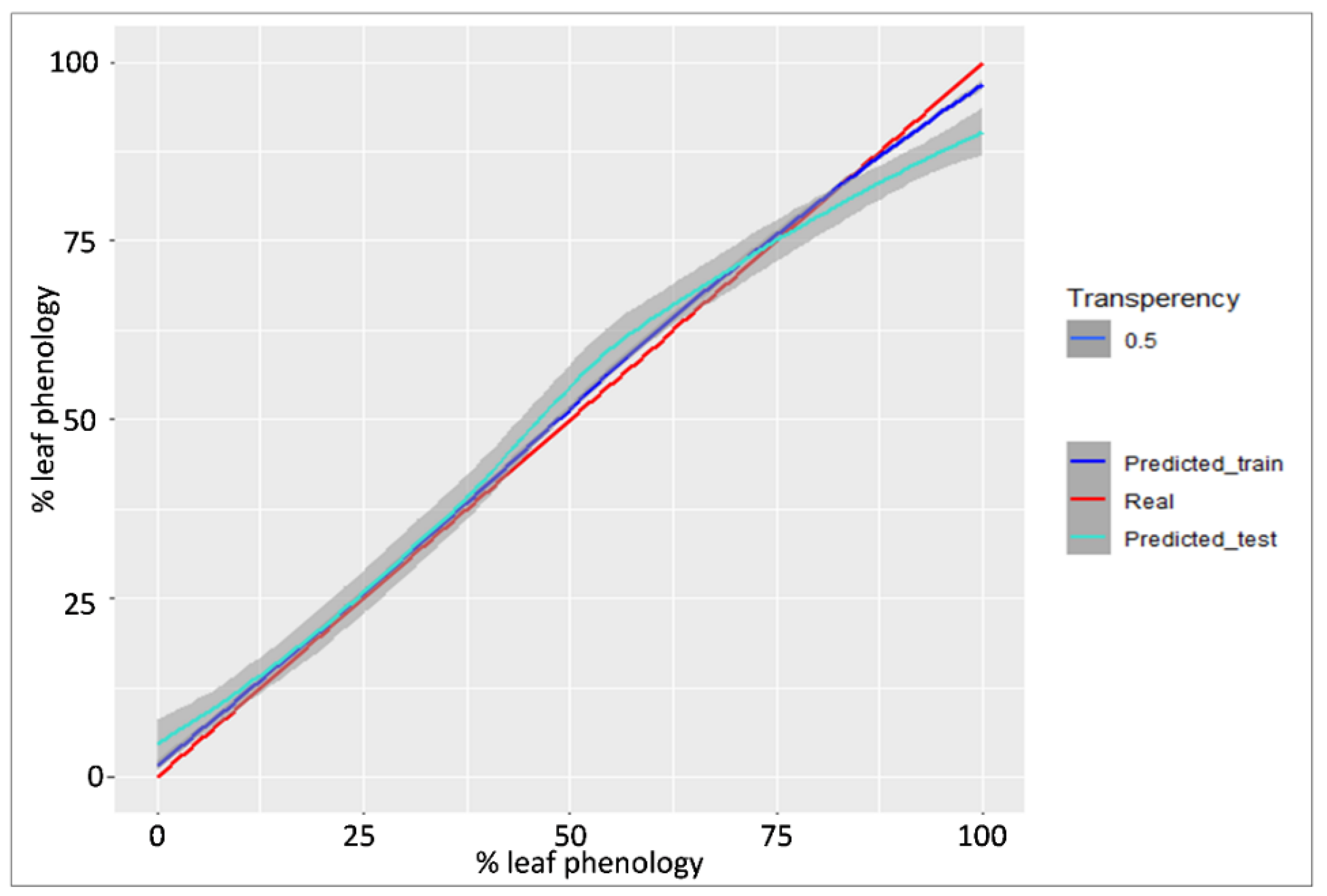
3.1. Predicting Leaf Phenology Using Aerial Images Collected by UAV
3.2. Predicting Leaf Phenology Using Copernicus Biophysical Parameters
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
Appendix A.1. Image Collecting and Processing
Appendix A.2. Correlation Analysis
References
- Klosterman, S.; Melaas, E.; Wang, J.A.; Martinez, A.; Frederick, S.; O’Keefe, J.; Orwig, D.A.; Wang, Z.; Sun, Q.; Schaaf, C.; et al. Fine-Scale Perspectives on Landscape Phenology from Unmanned Aerial Vehicle (UAV) Photography. Agric. For. Meteorol. 2018, 248, 397–407. [Google Scholar] [CrossRef]
- Xie, Y.; Civco, D.L.; Silander, J.A., Jr. Species-Specific Spring and Autumn Leaf Phenology Captured by Time-Lapse Digital Cameras. Ecosphere 2018, 9, e02089. [Google Scholar] [CrossRef] [Green Version]
- Klosterman, S.; Richardson, A.D. Observing Spring and Fall Phenology in a Deciduous Forest with Aerial Drone Imagery. Sensors 2017, 17, 2852. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atkins, J.W.; Stovall, A.E.L.; Yang, X. Mapping Temperate Forest Phenology Using Tower, UAV, and Ground-Based Sensors. Drones 2020, 4, 56. [Google Scholar] [CrossRef]
- Fawcett, D.; Bennie, J.; Anderson, K. Monitoring Spring Phenology of Individual Tree Crowns Using Drone-Acquired NDVI Data. Remote Sens. Ecol. Conserv. 2021, 7, 227–244. [Google Scholar] [CrossRef]
- Twardosz, R.; Walanus, A.; Guzik, I. Warming in Europe: Recent Trends in Annual and Seasonal Temperatures. Pure Appl. Geophys. 2021, 178, 4021–4032. [Google Scholar] [CrossRef]
- Aitken, S.N.; Yeaman, S.; Holliday, J.A.; Wang, T.; Curtis-McLane, S. Adaptation, Migration or Extirpation: Climate Change Outcomes for Tree Populations. Evol. Appl. 2008, 1, 95–111. [Google Scholar] [CrossRef]
- Scranton, K.; Amarasekare, P. Predicting Phenological Shifts in a Changing Climate. Proc. Natl. Acad. Sci. USA 2017, 114, 13212–13217. [Google Scholar] [CrossRef] [Green Version]
- Weisberg, P.J.; Dilts, T.E.; Greenberg, J.A.; Johnson, K.N.; Pai, H.; Sladek, C.; Kratt, C.; Tyler, S.W.; Ready, A. Phenology-Based Classification of Invasive Annual Grasses to the Species Level. Remote Sens. Environ. 2021, 263, 112568. [Google Scholar] [CrossRef]
- Park, J.Y.; Muller-Landau, H.C.; Lichstein, J.W.; Rifai, S.W.; Dandois, J.P.; Bohlman, S.A. Quantifying Leaf Phenology of Individual Trees and Species in a Tropical Forest Using Unmanned Aerial Vehicle (UAV) Images. Remote Sens. 2019, 11, 1534. [Google Scholar] [CrossRef]
- Budeanu, M.; Petritan, A.M.; Popescu, F.; Vasile, D.; Tudose, N.C. The Resistance of European Beech (Fagus Sylvatica) From the Eastern Natural Limit of Species to Climate Change. Not. Bot. Horti Agrobot. Cluj-Napoca 2016, 44, 625–633. [Google Scholar] [CrossRef] [Green Version]
- Chesnoiu, E.N.; Șofletea, N.; Curtu, A.L.; Toader, A.; Radu, R.; Enescu, M. Bud Burst and Flowering Phenology in a Mixed Oak Forest from Eastern Romania. Ann. For. Res. 2009, 52, 199–206. [Google Scholar] [CrossRef]
- Thapa, S.; Garcia Millan, V.E.; Eklundh, L. Assessing Forest Phenology: A Multi-Scale Comparison of Near-Surface (UAV, Spectral Reflectance Sensor, PhenoCam) and Satellite (MODIS, Sentinel-2) Remote Sensing. Remote Sens. 2021, 13, 1597. [Google Scholar] [CrossRef]
- Sánchez-Azofeifa, A.; Rivard, B.; Wright, J.; Feng, J.-L.; Li, P.; Chong, M.M.; Bohlman, S.A. Estimation of the Distribution of Tabebuia Guayacan (Bignoniaceae) Using High-Resolution Remote Sensing Imagery. Sensors 2011, 11, 3831–3851. [Google Scholar] [CrossRef]
- Lopes, A.P.; Nelson, B.W.; Wu, J.; de Graça, P.M.L.A.; Tavares, J.V.; Prohaska, N.; Martins, G.A.; Saleska, S.R. Leaf Flush Drives Dry Season Green-up of the Central Amazon. Remote Sens. Environ. 2016, 182, 90–98. [Google Scholar] [CrossRef]
- Berra, E.F.; Gaulton, R.; Barr, S. Assessing Spring Phenology of a Temperate Woodland: A Multiscale Comparison of Ground, Unmanned Aerial Vehicle and Landsat Satellite Observations. Remote Sens. Environ. 2019, 223, 229–242. [Google Scholar] [CrossRef]
- Budianti, N.; Mizunaga, H.; Iio, A. Crown Structure Explains the Discrepancy in Leaf Phenology Metrics Derived from Ground- and UAV-Based Observations in a Japanese Cool Temperate Deciduous Forest. Forests 2021, 12, 425. [Google Scholar] [CrossRef]
- Gray, R.E.J.; Ewers, R.M. Monitoring Forest Phenology in a Changing World. Forests 2021, 12, 297. [Google Scholar] [CrossRef]
- Fuster, B.; Sánchez-Zapero, J.; Camacho, F.; García-Santos, V.; Verger, A.; Lacaze, R.; Weiss, M.; Baret, F.; Smets, B. Quality Assessment of PROBA-V LAI, FAPAR and FCOVER Collection 300 m Products of Copernicus Global Land Service. Remote Sens. 2020, 12, 1017. [Google Scholar] [CrossRef] [Green Version]
- Wu, S.; Wang, J.; Yan, Z.; Song, G.; Chen, Y.; Ma, Q.; Deng, M.; Wu, Y.; Zhao, Y.; Guo, Z.; et al. Monitoring Tree-Crown Scale Autumn Leaf Phenology in a Temperate Forest with an Integration of PlanetScope and Drone Remote Sensing Observations. ISPRS J. Photogramm. Remote Sens. 2021, 171, 36–48. [Google Scholar] [CrossRef]
- Liu, Y.; Hatou, K.; Aihara, T.; Kurose, S.; Akiyama, T.; Kohno, Y.; Lu, S.; Omasa, K. A Robust Vegetation Index Based on Different UAV RGB Images to Estimate SPAD Values of Naked Barley Leaves. Remote Sens. 2021, 13, 686. [Google Scholar] [CrossRef]
- Barbosa, B.D.S.; Ferraz, G.A.S.; Gonçalves, L.M.; Marin, D.B.; Maciel, D.T.; Ferraz, P.F.P.; Rossi, G. RGB Vegetation Indices Applied to Grass Monitoring: A Qualitative Analysis. Agron. Res. 2019, 17, 349–357. [Google Scholar] [CrossRef]
- Kawashima, S.; Nakatani, M. An Algorithm for Estimating Chlorophyll Content in Leaves Using a Video Camera. Ann. Bot. 1998, 81, 49–54. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Wang, D.; Shi, P.; Omasa, K. Estimating Rice Chlorophyll Content and Leaf Nitrogen Concentration with a Digital Still Color Camera under Natural Light. Plant Methods 2014, 10, 36. [Google Scholar] [CrossRef] [Green Version]
- Morley, P.J.; Jump, A.S.; West, M.D.; Donoghue, D.N.M. Spectral Response of Chlorophyll Content during Leaf Senescence in European Beech Trees. Environ. Res. Commun. 2020, 2, 071002. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel Algorithms for Remote Estimation of Vegetation Fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Geßler, A.; Keitel, C.; Kreuzwieser, J.; Matyssek, R.; Seiler, W.; Rennenberg, H. Potential Risks for European Beech (Fagus Sylvatica L.) in a Changing Climate. Trees 2007, 21, 1–11. [Google Scholar] [CrossRef]
- Zlatník: Lesnická Fytocenologie: Příručka Pro...—Google Scholar. Available online: https://scholar.google.com/scholar_lookup?title=Lesnick%C3%A1%20fytocenologie&publication_year=1978&author=Zlatn%C3%ADk%2CA. (accessed on 13 October 2022).
- Schieber, B. Spring Phenology of European Beech (Fagus Sylvatica L.) in a Submountain Beech Stand with Different Stocking in 1995–2004. J. For. Sci. 2006, 52, 208–216. [Google Scholar] [CrossRef] [Green Version]
- Leuschner, C. Drought Response of European Beech (Fagus Sylvatica L.)—A Review. Perspect. Plant Ecol. Evol. Syst. 2020, 47, 125576. [Google Scholar] [CrossRef]
- Vitasse, Y.; Delzon, S.; Dufrêne, E.; Pontailler, J.-Y.; Louvet, J.-M.; Kremer, A.; Michalet, R. Leaf Phenology Sensitivity to Temperature in European Trees: Do within-Species Populations Exhibit Similar Responses? Agric. For. Meteorol. 2009, 149, 735–744. [Google Scholar] [CrossRef]
- Alberto, F.; Bouffier, L.; Louvet, J.-M.; Lamy, J.-B.; Delzon, S.; Kremer, A. Adaptive Responses for Seed and Leaf Phenology in Natural Populations of Sessile Oak along an Altitudinal Gradient. J. Evol. Biol. 2011, 24, 1442–1454. [Google Scholar] [CrossRef]
- PIX4Dmapper: Professional Photogrammetry Software for Drone Mapping. Available online: https://www.pix4d.com/product/pix4dmapper-photogrammetry-software (accessed on 26 November 2022).
- OpenDroneMap. WebODM: Drone Mapping Software (Version 1.1.0). Available online: https://www.opendronemap.org/webodm/ (accessed on 1 March 2021).
- QGIS. QGIS Project 3.26.3. 2017. Available online: https://www.qgis.org/nl/site/ (accessed on 1 March 2021).
- Pearson, R.L.; Miller, L.D.; Tucker, C.J. Hand-Held Spectral Radiometer to Estimate Gramineous Biomass. Appl. Opt. 1976, 15, 416–418. [Google Scholar] [CrossRef]
- Fuentes, D.A.; Gamon, J.A.; Qiu, H.; Sims, D.A.; Roberts, D.A. Mapping Canadian Boreal Forest Vegetation Using Pigment and Water Absorption Features Derived from the AVIRIS Sensor. J. Geophys. Res. Atmos. 2001, 106, 33565–33577. [Google Scholar] [CrossRef] [Green Version]
- Zarco-Tejada, P.J.; Berjón, A.; López-Lozano, R.; Miller, J.R.; Martín, P.; Cachorro, V.; González, M.R.; de Frutos, A. Assessing Vineyard Condition with Hyperspectral Indices: Leaf and Canopy Reflectance Simulation in a Row-Structured Discontinuous Canopy. Remote Sens. Environ. 2005, 99, 271–287. [Google Scholar] [CrossRef]
- Woebbecke, D.M.; Meyer, G.E.; Von Bargen, K.; Mortensen, D.A. Color Indices for Weed Identification Under Various Soil, Residue, and Lighting Conditions. Trans. ASAE 1995, 38, 259–269. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. Nasa Spec. Publ. 1974, 351, 309. [Google Scholar]
- Hunt, E.R.; Cavigelli, M.; Daughtry, C.S.T.; Mcmurtrey, J.E.; Walthall, C.L. Evaluation of Digital Photography from Model Aircraft for Remote Sensing of Crop Biomass and Nitrogen Status. Precis. Agric. 2005, 6, 359–378. [Google Scholar] [CrossRef]
- Louhaichi, M.; Borman, M.M.; Johnson, D.E. Spatially Located Platform and Aerial Photography for Documentation of Grazing Impacts on Wheat. Geocarto Int. 2001, 16, 65–70. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-Based Plant Height from Crop Surface Models, Visible, and near Infrared Vegetation Indices for Biomass Monitoring in Barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Copernicus Land Monitoring Service. Available online: https://land.copernicus.eu/ (accessed on 13 March 2022).
- R Project for Statistical Computing. R Version 4.1.3, Released on 10.03.2022. Available online: https://www.r-project.org/ (accessed on 31 March 2022).
- Berra, E.F.; Gaulton, R.; Barr, S. Use of a Digital Camera Onboard a UAV to Monitor Spring Phenology at Individual Tree Level. In Proceedings of the 2016 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Beijing, China, 10–15 July 2016; pp. 3496–3499. [Google Scholar]
- Yengoh, G.T.; Dent, D.; Olsson, L.; Tengberg, A.E.; Tucker, C.J., III. Use of the Normalized Difference Vegetation Index (NDVI) to Assess Land Degradation at Multiple Scales: Current Status, Future Trends, and Practical Considerations; SpringerBriefs in Environmental Science; Springer International Publishing: Cham, Switzerland, 2016; ISBN 978-3-319-24110-4. [Google Scholar]
- Zhao, B.; Zhang, J.; Yang, C.; Zhou, G.; Ding, Y.; Shi, Y.; Zhang, D.; Xie, J.; Liao, Q. Rapeseed Seedling Stand Counting and Seeding Performance Evaluation at Two Early Growth Stages Based on Unmanned Aerial Vehicle Imagery. Front. Plant Sci. 2018, 9, 1362. [Google Scholar] [CrossRef]
- Bannari, A.; Guedon, A.M.; El Harti, A.; Cherkaoui, F.Z.; El-Ghmari, A.; Saquaque, A. Slight and Moderate Saline and Sodic Soils Characterization in Irrigated Agricultural Land Using Multispectral Remote Sensing. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2013, 34, 1–6. [Google Scholar]
- Dixon, D.J.; Callow, J.N.; Duncan, J.M.A.; Setterfield, S.A.; Pauli, N. Satellite Prediction of Forest Flowering Phenology. Remote Sens. Environ. 2021, 255, 112197. [Google Scholar] [CrossRef]
- D’Odorico, P.; Besik, A.; Wong, C.Y.S.; Isabel, N.; Ensminger, I. High-Throughput Drone-Based Remote Sensing Reliably Tracks Phenology in Thousands of Conifer Seedlings. New Phytol. 2020, 226, 1667–1681. [Google Scholar] [CrossRef]
- Lukasová, V.; Bucha, T.; Škvareninová, J.; Škvarenina, J. Validation and Application of European Beech Phenological Metrics Derived from MODIS Data along an Altitudinal Gradient. Forests 2019, 10, 60. [Google Scholar] [CrossRef]
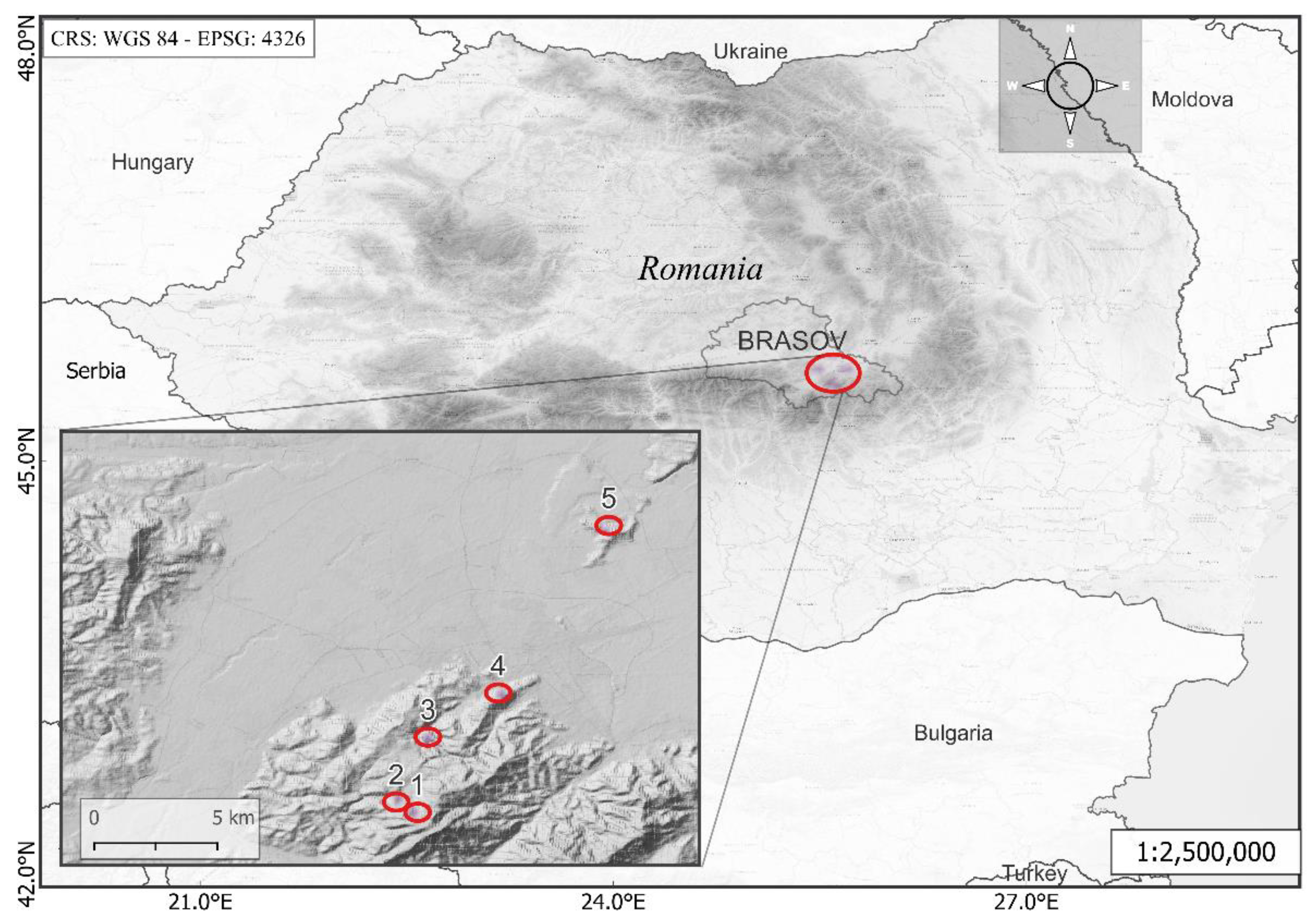
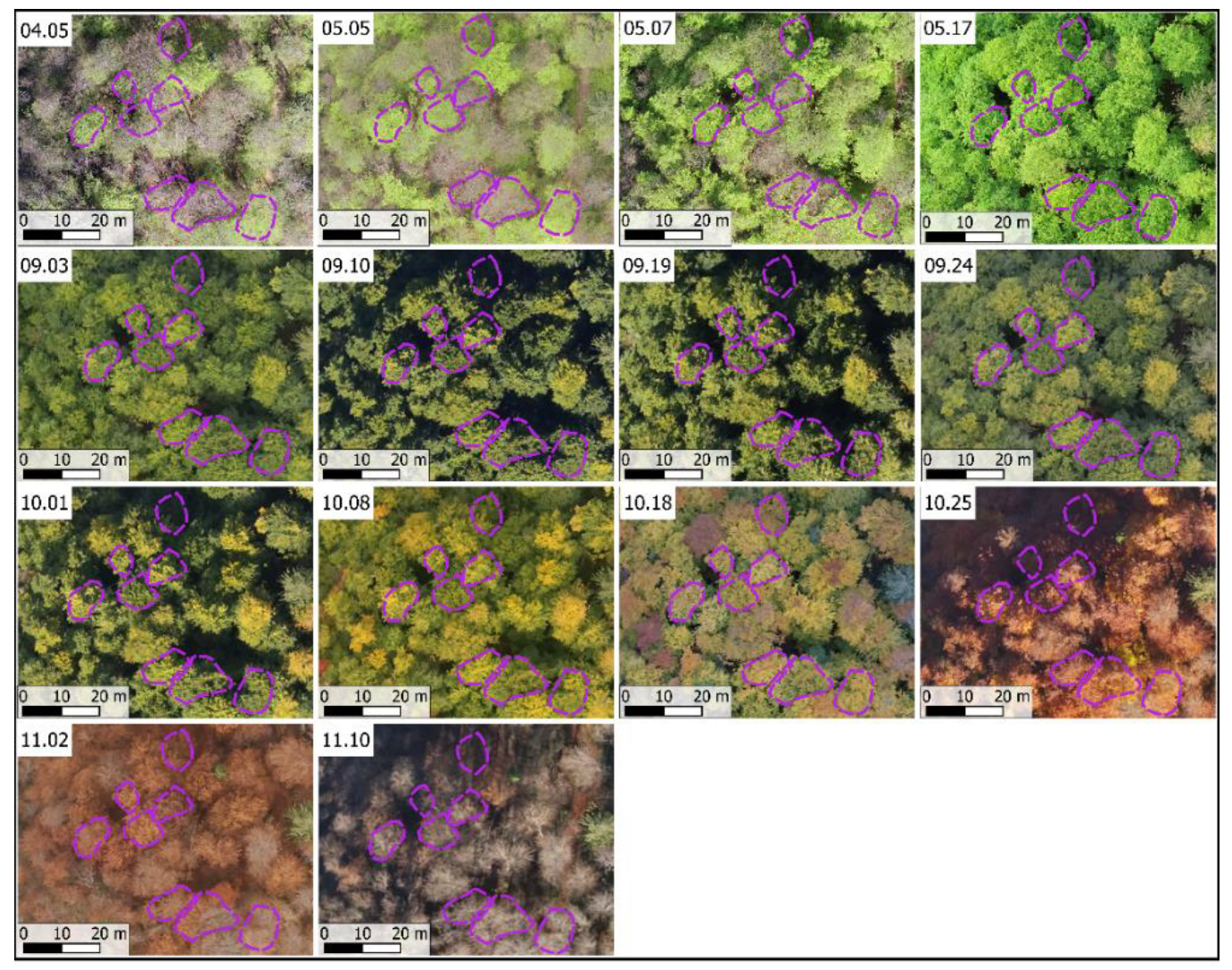
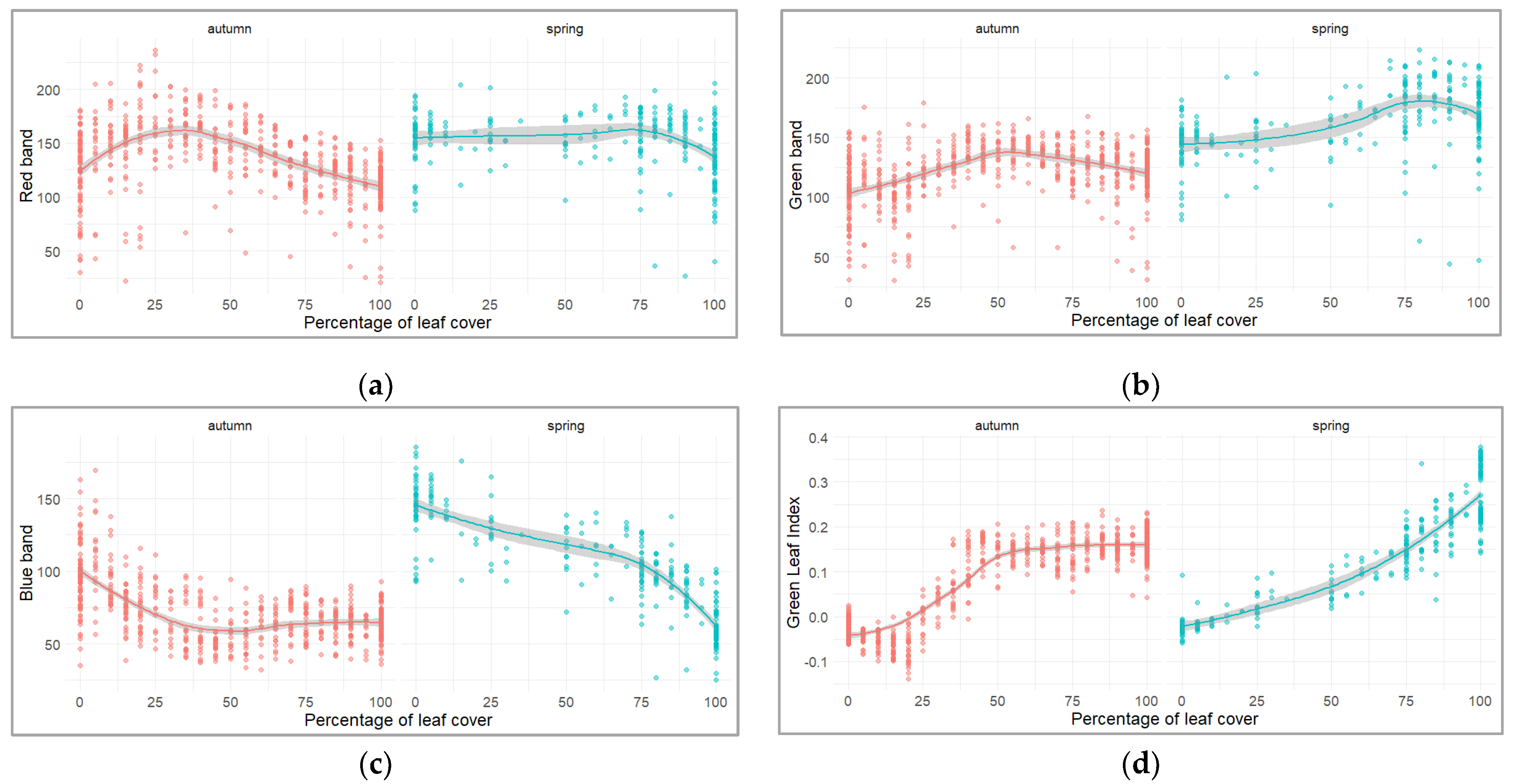
| Number | Name | Geographic Coordinates | Altitude Range (Meters) | Mean Temperature (°C) 1 | Mean Precipitation (Millimeters) 1 |
|---|---|---|---|---|---|
| Site 1 | Ruia | 45°34′25.41″N 25°33′11.67″E | 1300–1450 | 3.5 | 1023 |
| Site 2 | Lupului | 45°34′54.64″N 25°32′36.43″E | 1000–1150 | 5.2 | 957 |
| Site 3 | Solomon | 45°36′59.75″N 25°33′39.87″E | 800–1000 | 6.2 | 855 |
| Site 4 | Tampa | 45°38′18.86″N 25°35′38.56″E | 650–750 | 7.2 | 791 |
| Site 5 | Lempes | 45°43′34.88″N 25°39′30.66″E | 550–610 | 7.5 | 712 |
| Code | Phenological Stage (3) | Range of the Percentage of Leaf Cover (%) (2) |
|---|---|---|
| 0 | Dormant winter bud | <25 |
| 1 | Bud-swollen | 26–50 |
| 2 | Bud-burst | 51–75 |
| 3 | At least one leaf unfolding | >75 |
| No. | Name | Abbreviation | Equation | Reference |
|---|---|---|---|---|
| 1 | Digital number for red band | R | red/255 | [36] |
| 2 | Digital number for green band | G | green/255 | [36] |
| 3 | Digital number for blue band | B | blue/255 | [36] |
| 4 | Green Red Ratio Index | GRRI | G/R | [37] |
| 5 | Blue Green Ratio Index | BGRI | B/G | [38] |
| 6 | Green Blue Difference Index | GBDI | G − B | [23] |
| 7 | Red Blue Difference Index | RBDI | R − B | [23] |
| 8 | Excess of green index | ExG | 2G − R − B | [39] |
| 9 | Grayscale Index | GRAY | (R + G + B)/3 | [24] |
| 10 | Chromatic coordinates for red/ Normalized red of RGB | NRRGB | R/(R + G + B) | [39] |
| 11 | Chromatic coordinates for green/ Normalized green of RGB | NGRGB | G/(R + G + B) | [39] |
| 12 | Chromatic coordinates for blue/ Normalized blue of RGB | NBRGB | B/(R + G + B) | [39] |
| 13 | Normalized Green Red Difference Index | NGRDI | (G − R)/(G + R) | [40] |
| 14 | Kawashima index | KAW | (R − B)/(R + B) | [23] |
| 15 | Normalized Green Blue Difference index | NGBDI | (G − B)/(G + B) | [41] |
| 16 | Green Leaf Index | GLI | (2G − R − B)/(2G + R + B) | [42] |
| 17 | Modified Green Red Vegetation Index | MGVRI | (G2 − R2)/(G2 + R2) | [43] |
| 18 | Red Green Blue Vegetation Index | RGVBI | (G − B × R)/(G2 + B × R) | [43] |
| 19 | Visible Atmospherically Resistant Index | VARI | (G − R)/(G + R − B) | [26] |
| No. | Name | Abbreviation | Description |
|---|---|---|---|
| 1 | Dry Matter Productivity | DMP | the overall growth rate or dry biomass increase of the vegetation (kg/ha/day) [44] |
| 2 | Fraction of Absorbed Photosynthetically Active Radiation | FAPAR | quantifies the fraction of the solar radiation absorbed by live leaves for photosynthesis activity [44] |
| 3 | Fraction of Vegetation Cover | FCover | fraction of ground covered by green vegetation [44] |
| 4 | Leaf Area Index | LAI | half the total area of green elements of the canopy per unit of the horizontal ground area [44] |
| 5 | Normalized Difference Vegetation Index | NDVI | indicator of the greenness of the biomass [44] |
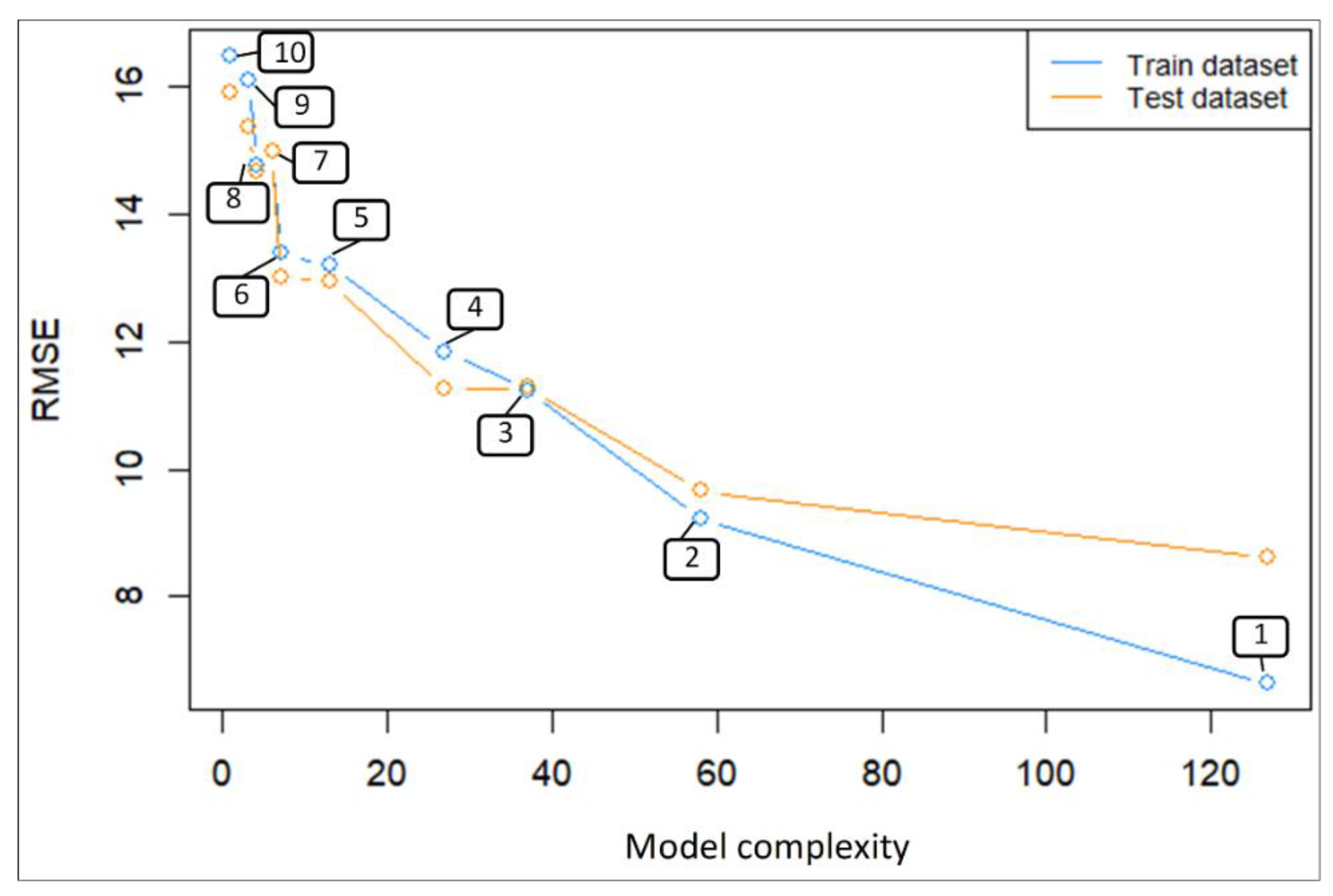
| No. | Train RMSE | Test RMSE | R2 Train Data | R2 Test Data | Model Complexity | Independent Variable Component of the Linear Model Equation |
|---|---|---|---|---|---|---|
| 1 | 6.63 | 8.61 | 0.94 | 0.90 | 127 | 1 F(x) = NGRGB × GLI × ExG × RGVBI × GBDI × NGBDI × season |
| 2 | 9.21 | 9.65 | 0.88 | 0.87 | 58 | 1 F(x) = R + G + B + GRRI + BGRI + GBDI + RBDI + ExG + GRAY + NRRGB + NGRGB + NBRGB + NGRDI + KAW + NGBDI + GLI + MGVRI + RGVBI + VARI |
| 3 | 11.24 | 11.27 | 0.84 | 0.84 | 37 | 2 F(x) = (NGRGB_m + NGRGB_me + NGRGB_sd + GLI_m + GLI_me + GLI_sd + ExG_m + ExG_me + ExG_sd + RGVBI_m + RGVBI_me + RGVBI_sd + GBDI_m + GBDI_me + GBDI_sd + NGBDI_m + NGBDI_me + NGBDI_sd) × season |
| 4 | 11.84 | 11.27 | 0.83 | 0.84 | 27 | 1 F(x) = (NGRGB + GLI + ExG + RGVBI + GBDI + NGBDI) × season × location |
| 5 | 13.19 | 12.95 | 0.80 | 0.80 | 13 | 1 F(x) = (NGRGB + GLI + ExG + RGVBI + GBDI + NGBDI) × season |
| 6 | 13.38 | 13.02 | 0.79 | 0.80 | 7 | 1 F(x) = (NGRGB + GLI + RGVBI) × season |
| 7 | 14.99 | 15.00 | 0.76 | 0.76 | 6 | 1 F(x) = NGRGB + GLI + ExG + RGVBI + GBDI + NGBDI |
| 8 | 14.76 | 14.67 | 0.77 | 0.77 | 4 | 1 F(x) = NGRGB + GLI + RGVBI + season |
| 9 | 16.11 | 15.35 | 0.74 | 0.75 | 3 | 1 F(x) = GLI × season |
| 10 | 16.47 | 15.91 | 0.73 | 0.74 | 1 | 1 F(x) = GLI |
| Error Type | Train Data | Test Data |
|---|---|---|
| MSE | 23.11 | 159.39 |
| RMSE | 3.28 | 8.12 |
| No. | RMSE | R2 | Model Complexity | Linear Model |
|---|---|---|---|---|
| 1 | 11.65 | 0.87 | 11 | F(x) = (FCover + LAI + FAPAR + DMP + NDVI) × season |
| 2 | 7.84 | 0.94 | 9 | F(x) = FCover + LAI + FAPAR + DMP + NDVI + location |
| 3 | 11.89 | 0.85 | 9 | F(x) = (FCover + LAI + FAPAR + DMP) × season |
| 4 | 12.32 | 0.85 | 7 | F(x) = (FCover + LAI + FAPAR) × season |
| 5 | 12.57 | 0.85 | 5 | F(x) = FCover + LAI + FAPAR + DMP + NDVI |
| 6 | 12.99 | 0.84 | 5 | F(x) = (FCover + LAI) × season |
| 7 | 13.00 | 0.84 | 3 | F(x) = FCover × season |
| 8 | 13.11 | 0.83 | 1 | F(x) = FCover |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ciocîrlan, M.I.C.; Curtu, A.L.; Radu, G.R. Predicting Leaf Phenology in Forest Tree Species Using UAVs and Satellite Images: A Case Study for European Beech (Fagus sylvatica L.). Remote Sens. 2022, 14, 6198. https://doi.org/10.3390/rs14246198
Ciocîrlan MIC, Curtu AL, Radu GR. Predicting Leaf Phenology in Forest Tree Species Using UAVs and Satellite Images: A Case Study for European Beech (Fagus sylvatica L.). Remote Sensing. 2022; 14(24):6198. https://doi.org/10.3390/rs14246198
Chicago/Turabian StyleCiocîrlan, Mihnea Ioan Cezar, Alexandru Lucian Curtu, and Gheorghe Raul Radu. 2022. "Predicting Leaf Phenology in Forest Tree Species Using UAVs and Satellite Images: A Case Study for European Beech (Fagus sylvatica L.)" Remote Sensing 14, no. 24: 6198. https://doi.org/10.3390/rs14246198







