Remote Sensing Extraction Method of Illicium verum Based on Functional Characteristics of Vegetation Canopy
Abstract
:1. Introduction
- (1)
- Compare the performance of SVM and RF in tree species classification.
- (2)
- Compare the influence of different feature combination schemes on image classification.
- (3)
- Find the best feature combination scheme to complete the extraction of Illicium verum in Leye County.
2. Materials and Methods
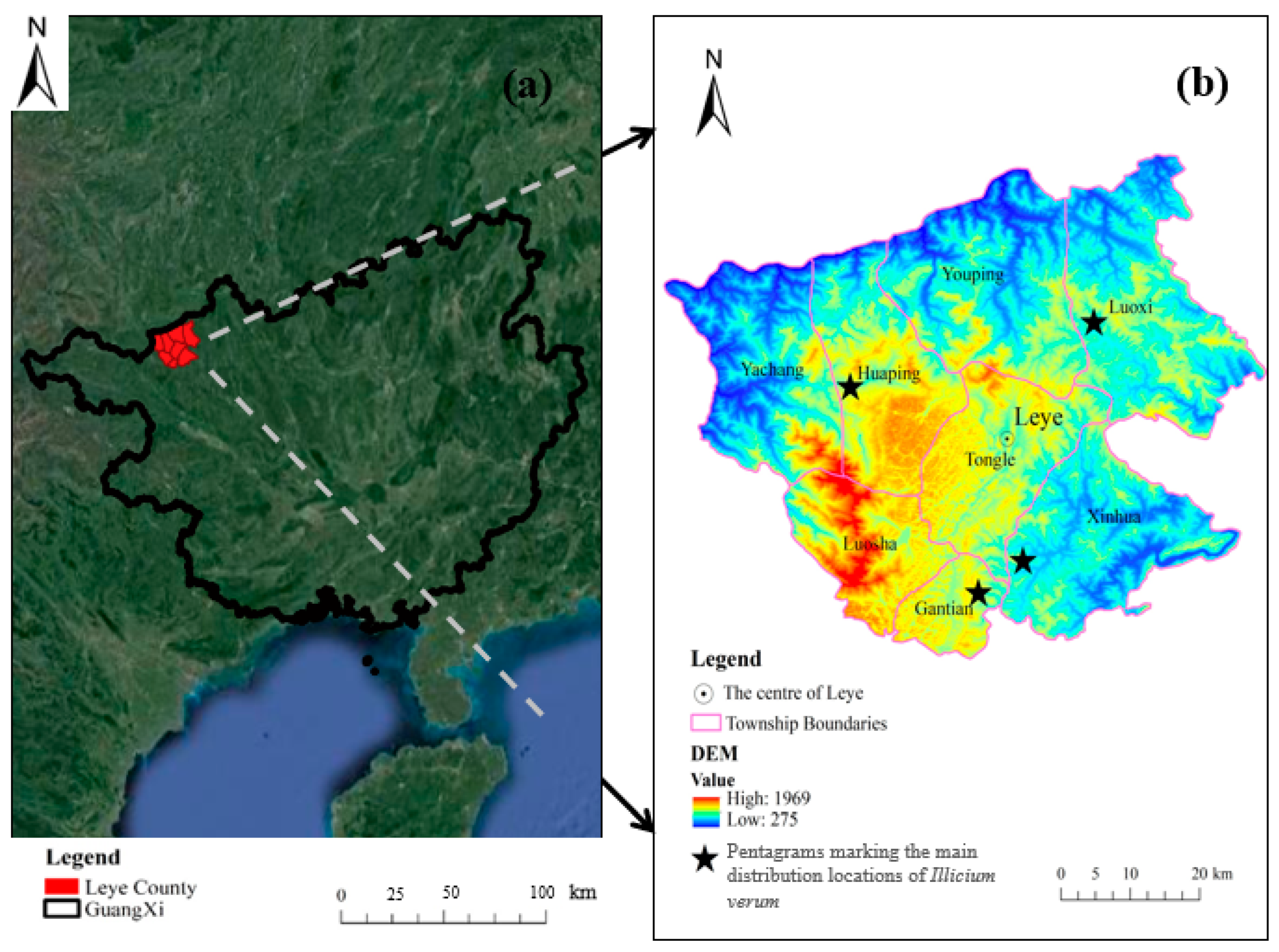
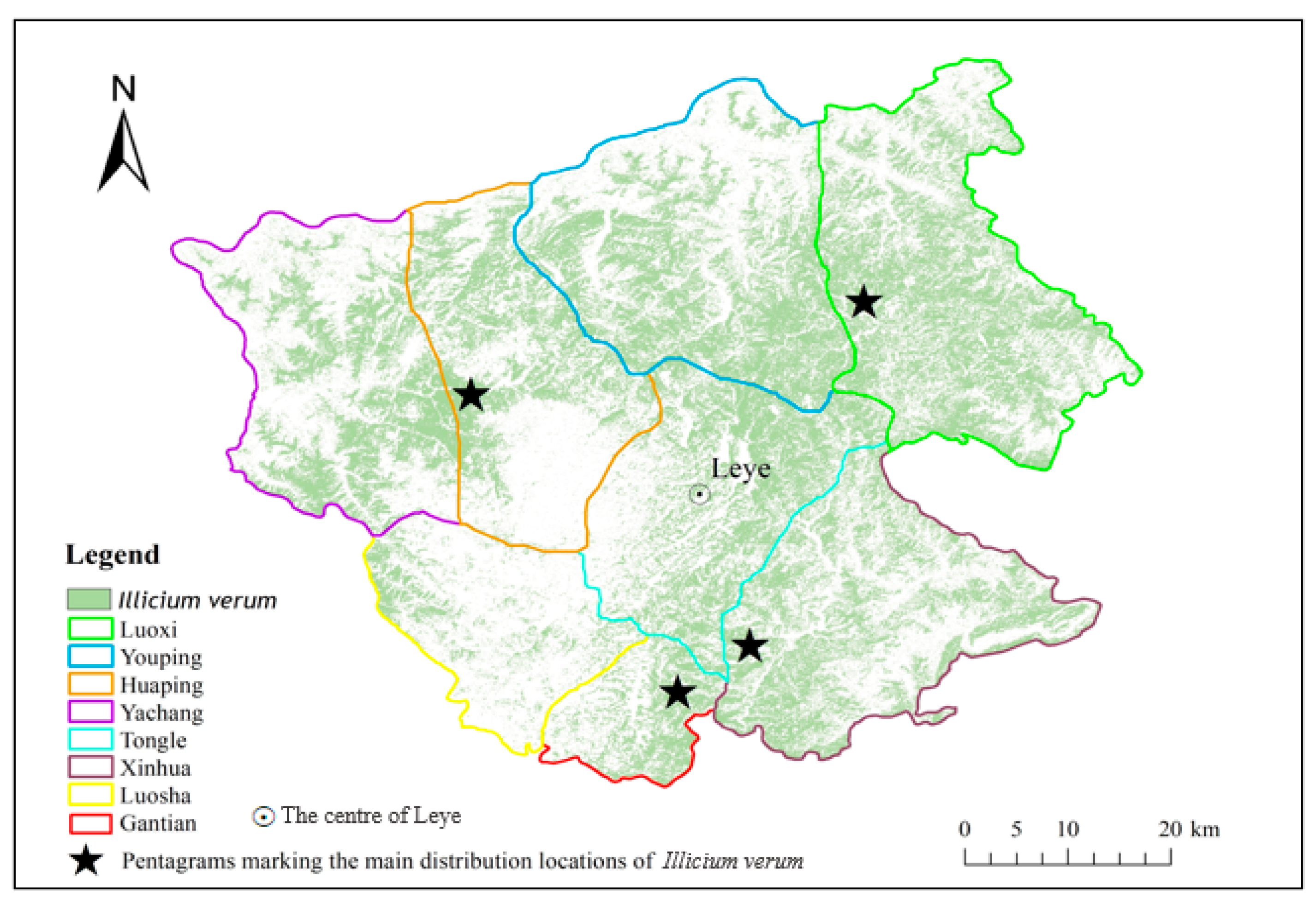
2.1. Study Area
2.2. Experimental Data
2.2.1. Sentinel-1 Data and Sentinel-2 Data
2.2.2. Field Data
2.3. Methods
2.3.1. Vegetation Index Calculation
2.3.2. Environmental Datasets
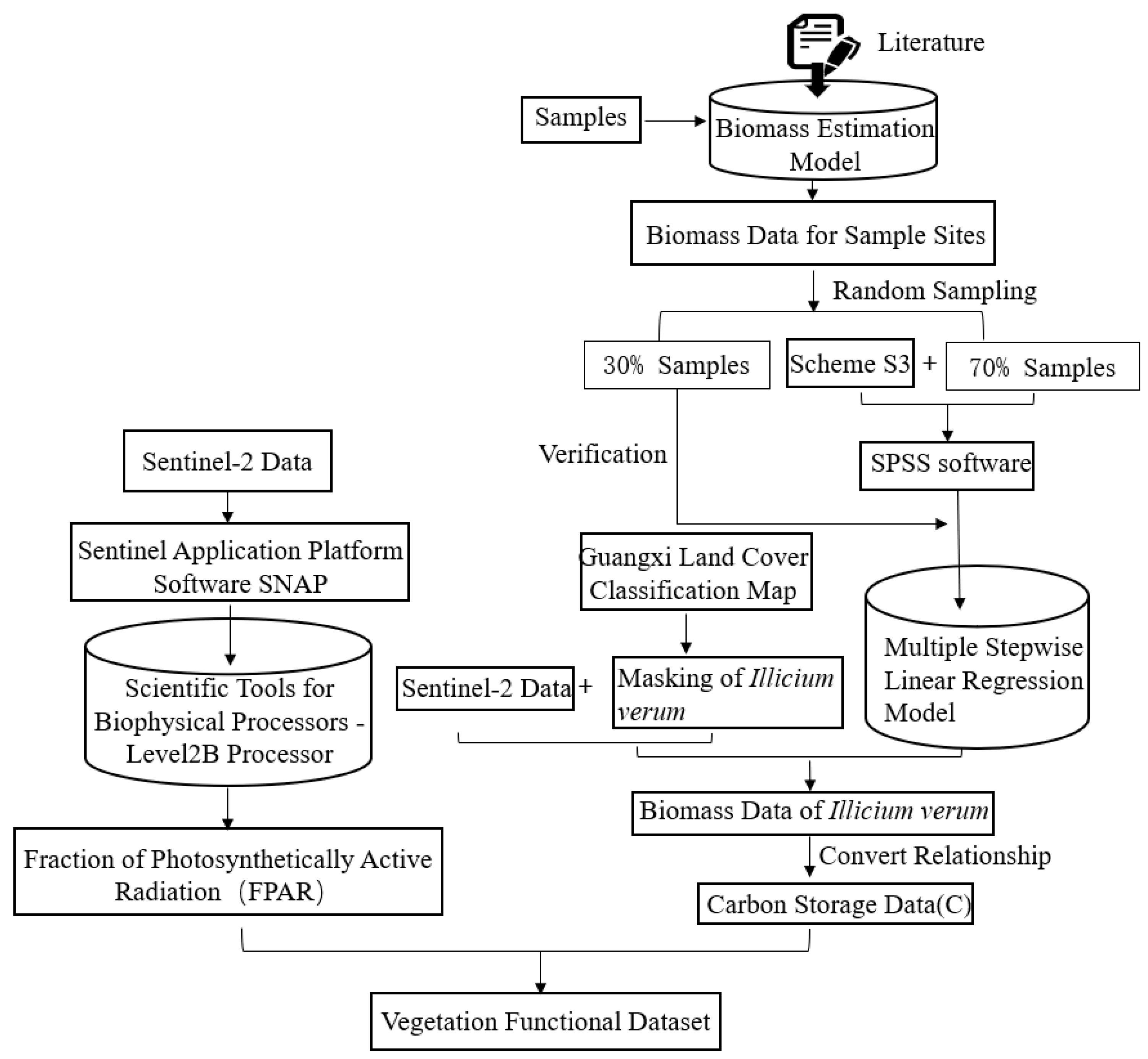
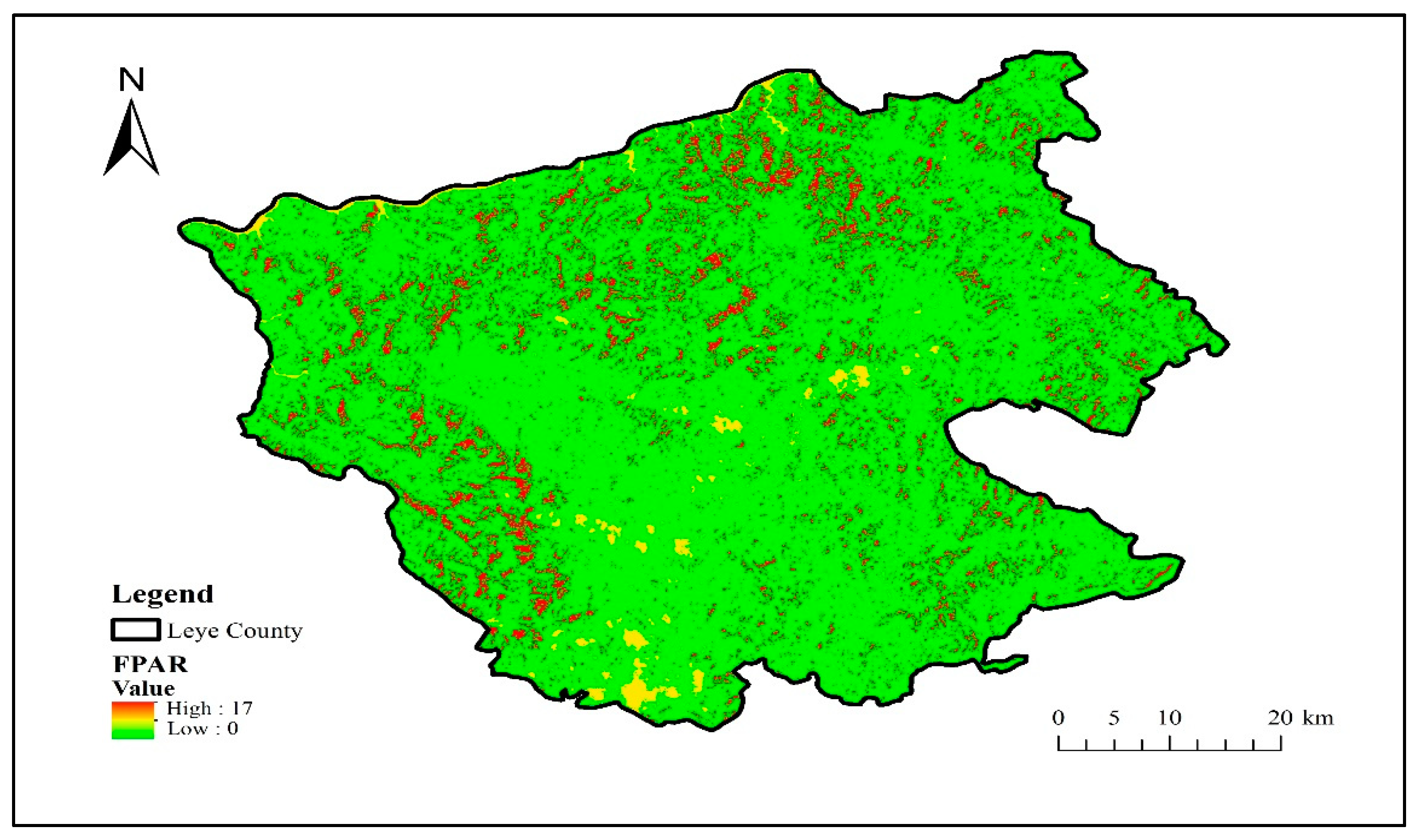
2.3.3. Vegetation Functional Datasets
2.3.4. Random Forest Classifier and Support Vector Machine Classifier
2.3.5. Accuracy Assessment
3. Results
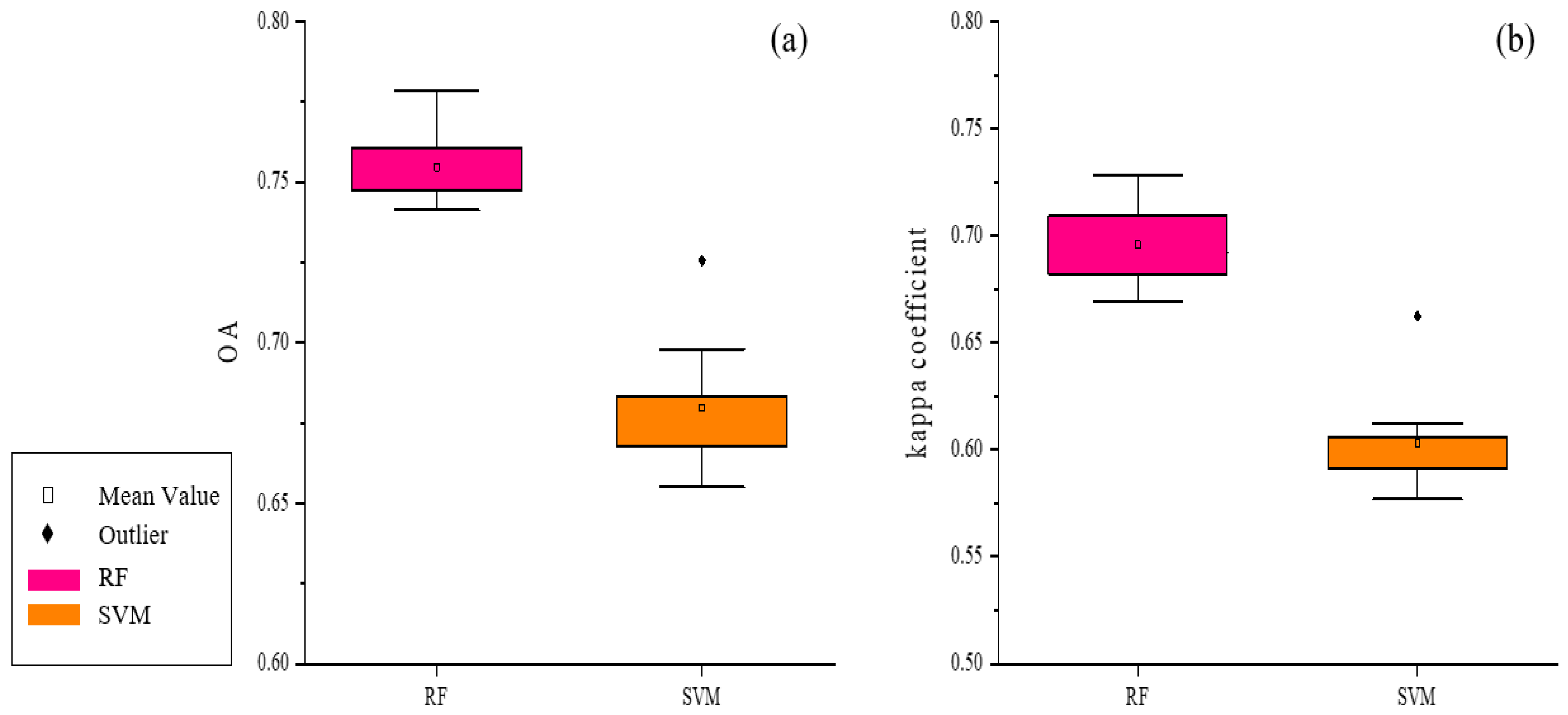
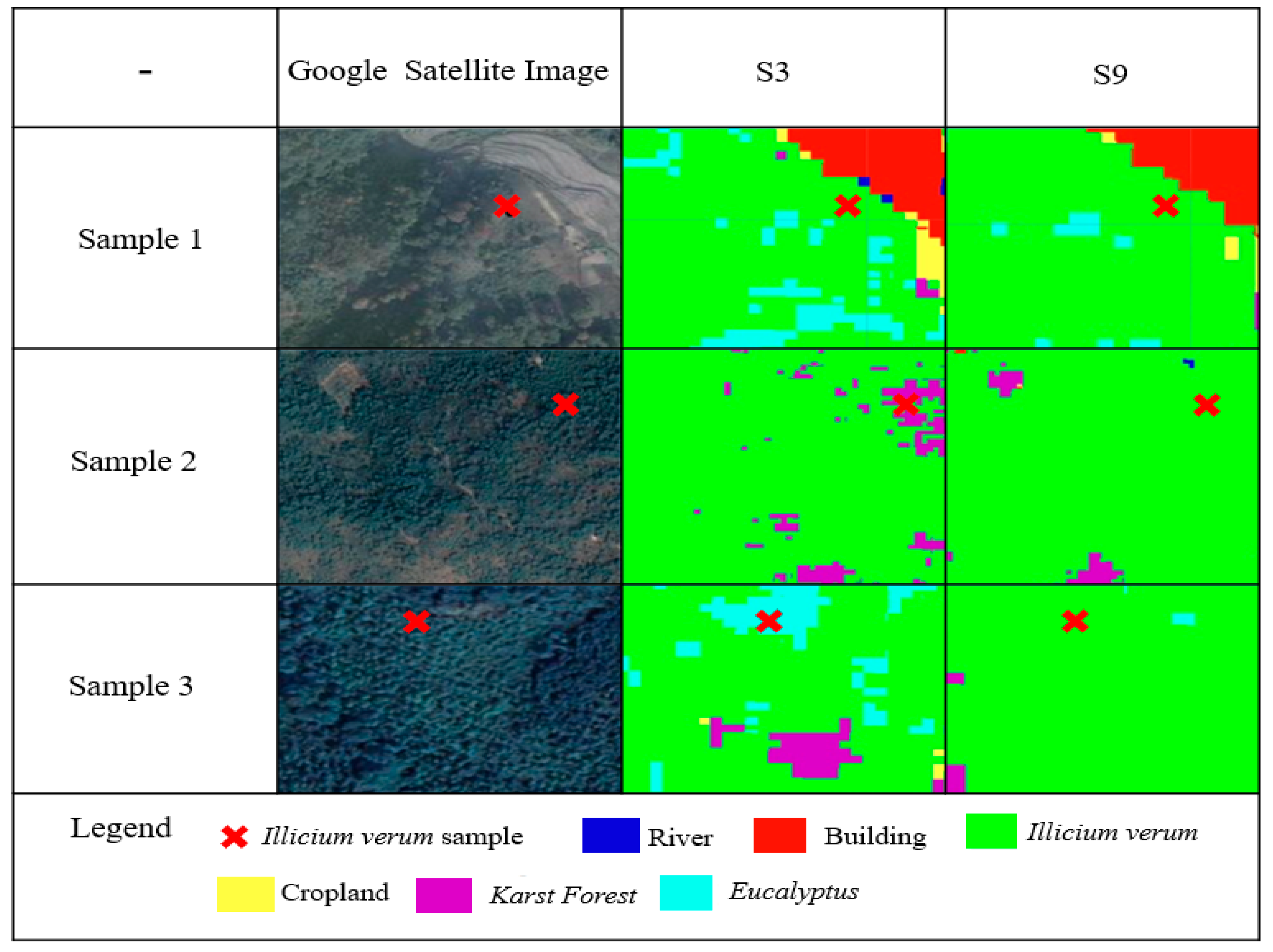
3.1. Comparison of RF and SVM Results
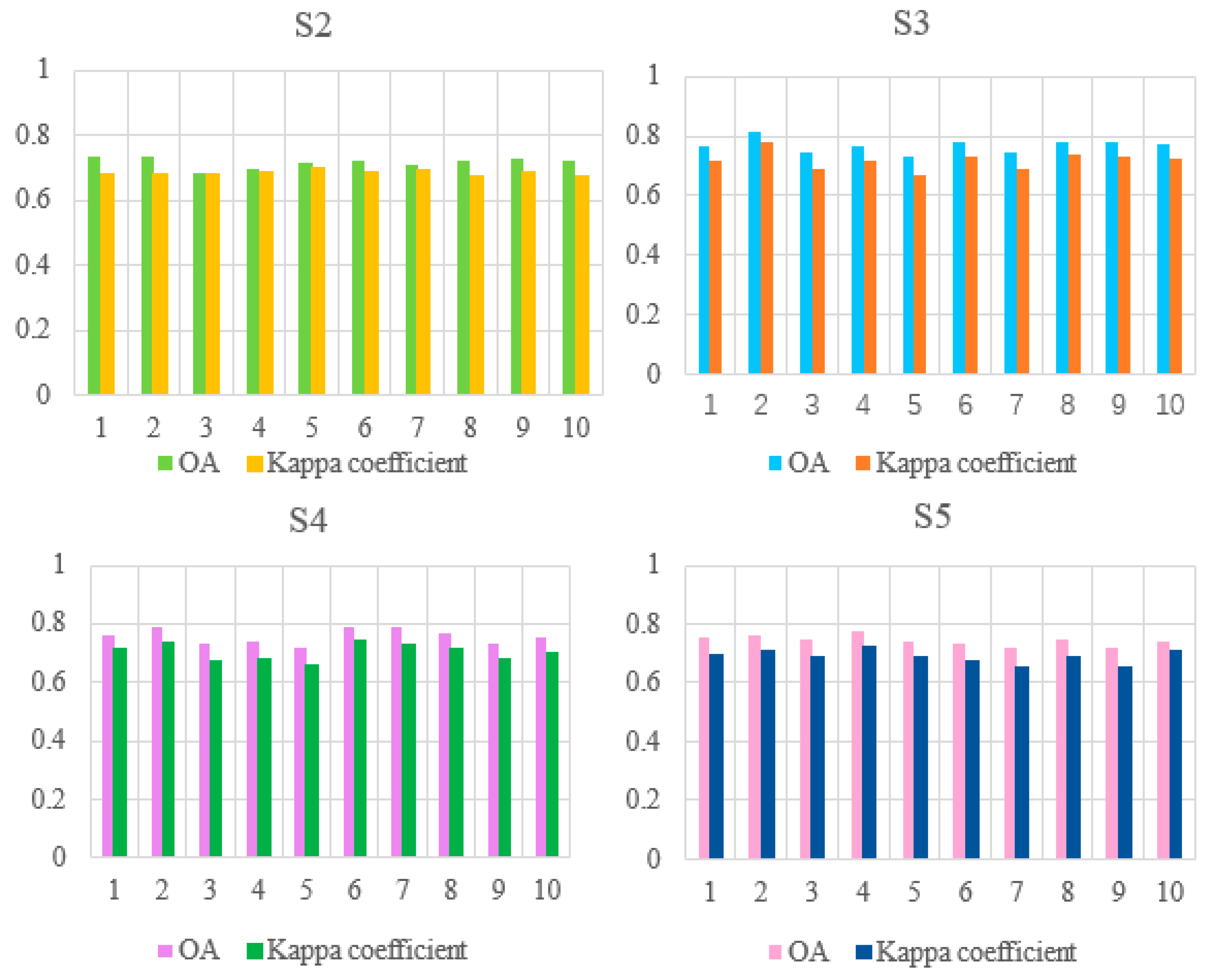
3.2. Impact of Environmental Datasets and Sentinel-1 Data on Classification Results
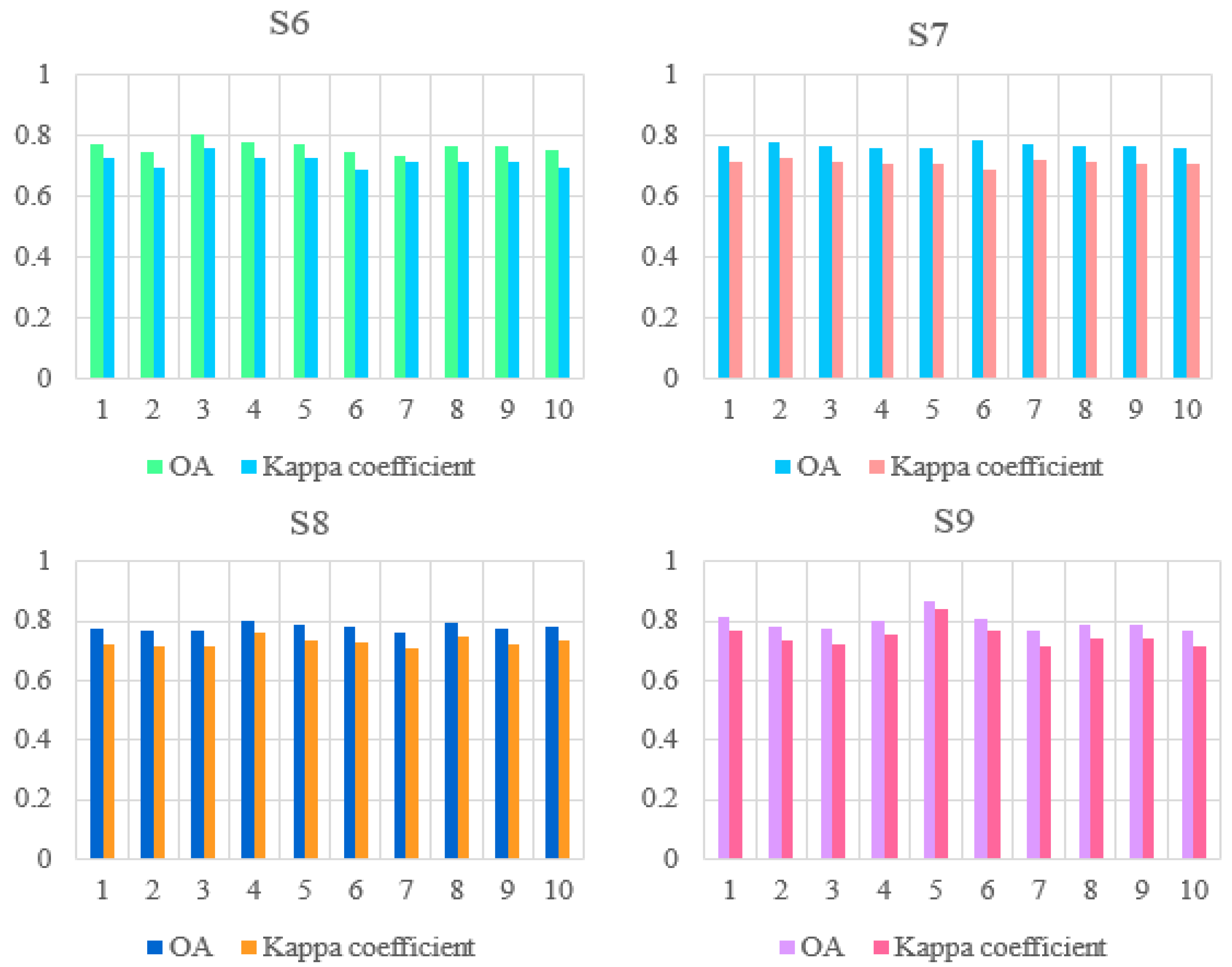
3.3. Effects of Vegetation Functional Datasets on Classification Results
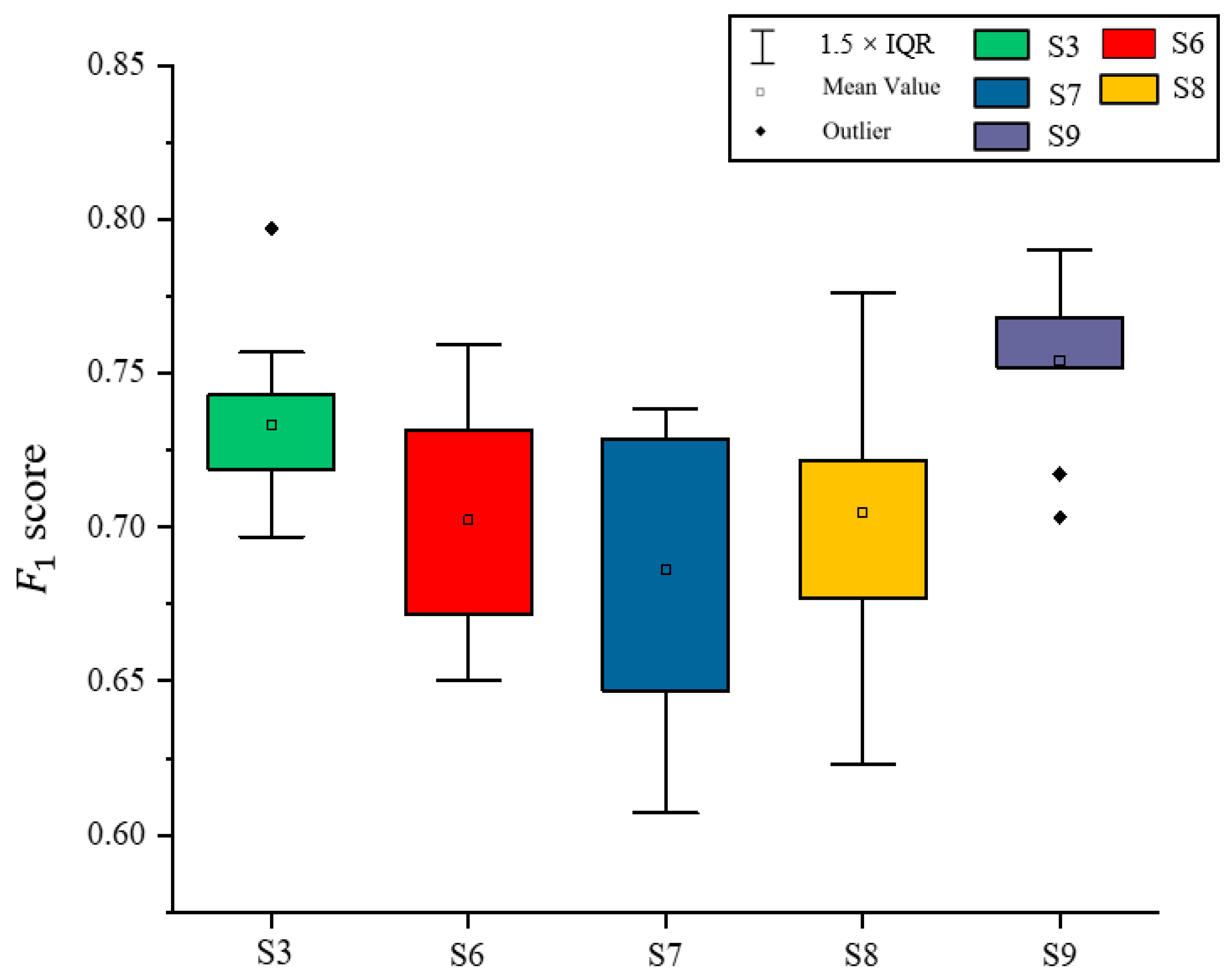
3.4. Effects of Environmental Datasets and the Vegetation Functional Datasets on Classification Results of Illicium verum
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Dalponte, M.; Bruzzone, L.; Gianelle, D. Tree species classification in the Southern Alps with very high geometrical resolution multispectral and hyperspectral data. In Proceedings of the 2011 3rd Workshop on Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Lisbon, Portugal, 6–9 June 2011. [Google Scholar]
- Waser, L.T.; Ginzler, C.; Kuechler, M.; Baltsavias, E.; Hurni, L. Semi-automatic classification of tree species in different forest ecosystems by spectral and geometric variables derived from Airborne Digital Sensor (ADS40) and RC30 data. Remote Sens. Environ. 2011, 115, 76–85. [Google Scholar] [CrossRef]
- Schimel, D.S.; Asner, G.P.; Moorcroft, P. Observing changing ecological diversity in the Anthropocene. Front. Ecol. Environ. 2013, 11, 129–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, J.; Wang, X.; Wang, T. Classification of tree species and stock volume estimation in ground forest images using Deep Learning. Comput. Electron. Agric. 2019, 166, 105012. [Google Scholar] [CrossRef]
- Turner, W.; Spector, S.; Gardiner, N.; Fladeland, M.; Sterling, E.; Steininger, M. Remote sensing for biodiversity science and conservation. Trends Ecol. Evol. 2003, 18, 306–314. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Asner, G.P.; Martin, R.E.; Anderson, C.; Knapp, D.E.; Kellner, J.R.; Joseph, W.S.; Lalit, K. Operational Tree Species Mapping in a Diverse Tropical Forest with Airborne Imaging Spectroscopy. PLoS ONE 2015, 10, e0118403. [Google Scholar] [CrossRef]
- Ghosh, A.; Fassnacht, F.E.; Joshi, P.K.; Koch, B. A framework for mapping tree species combining hyperspectral and LiDAR data: Role of selected classifiers and sensor across three spatial scales. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 49–63. [Google Scholar] [CrossRef]
- Piiroinen, R.; Heiskanen, J.; Maeda, E.; Viinikka, A.; Pellikka, P. Classification of Tree Species in a Diverse African Agroforestry Landscape Using Imaging Spectroscopy and Laser Scanning. Remote Sens. 2017, 9, 875. [Google Scholar] [CrossRef] [Green Version]
- Ballanti, L.; Blesius, L.; Hines, E.; Kruse, B. Tree Species Classification Using Hyperspectral Imagery: A Comparison of Two Classifiers. Remote Sens. 2016, 8, 445. [Google Scholar] [CrossRef] [Green Version]
- Ferreira, M.P.; Zortea, M.; Zanotta, D.C.; Shimabukuro, Y.E.; Filho, C. Mapping tree species in tropical seasonal semi-deciduous forests with hyperspectral and multispectral data. Remote Sens. Environ. 2016, 179, 66–78. [Google Scholar] [CrossRef]
- Franklin, S.E.; Ahmed, O.S. Deciduous tree species classification using object-based analysis and machine learning with unmanned aerial vehicle multispectral data. Int. J. Remote Sens. 2018, 39, 5236–5245. [Google Scholar] [CrossRef]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data. Remote Sens. 2018, 10, 1218. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Ferreira, M.P.; Wagner, F.H.; Aragão, L.E.; Shimabukuro, Y.E.; de Souza Filho, C.R. Tree species classification in tropical forests using visible to shortwave infrared WorldView-3 images and texture analysis. ISPRS J. Photogramm. Remote Sens. 2019, 149, 119–131. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Du, P.; Samat, A.; Waske, B.; Liu, S.; Li, Z. Random forest and rotation forest for fully polarized SAR image classification using polarimetric and spatial features. ISPRS J. Photogramm. Remote Sens. 2015, 105, 38–53. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- You, N.; Dong, J. Examining earliest identifiable timing of crops using all available Sentinel 1/2 imagery and Google Earth Engine. ISPRS J. Photogramm. Remote Sens. 2020, 161, 109–123. [Google Scholar] [CrossRef]
- Jafarian, Z.; Kargar, M.; Bahreini, Z. Which spatial distribution model best predicts the occurrence of dominant species in semi-arid rangeland of northern Iran? Ecol. Inform. 2019, 50, 33–42. [Google Scholar] [CrossRef]
- Guo, Q.; Zhang, J.; Guo, S.; Ye, Z.; Deng, H.; Hou, X.; Zhang, H. Urban Tree Classification Based on Object-Oriented Approach and Random Forest Algorithm Using Unmanned Aerial Vehicle (UAV) Multispectral Imagery. Remote Sens. 2022, 14, 3885. [Google Scholar] [CrossRef]
- Teluguntla, P.; Thenkabail, P.S.; Oliphant, A.; Xiong, J.; Gumma, M.K.; Congalton, R.G.; Yadav, K.; Huete, A. A 30-m landsat-derived cropland extent product of Australia and China using random forest machine learning algorithm on Google Earth Engine cloud computing platform. ISPRS J. Photogramm. Remote Sens. 2018, 144, 325–340. [Google Scholar] [CrossRef]
- Azzari, G.; Lobell, D. Landsat-based classification in the cloud: An opportunity for a paradigm shift in land cover monitoring. Remote Sens. Environ. 2017, 202, 64–74. [Google Scholar] [CrossRef]
- Zhang, X.; Wu, B.; Ponce-Campos, G.E.; Zhang, M.; Chang, S.; Tian, F. Mapping up-to-date paddy rice extent at 10 m resolution in china through the integration of optical and synthetic aperture radar images. Remote Sens. 2018, 10, 1200. [Google Scholar] [CrossRef] [Green Version]
- Qin, H.; Zhou, W.; Yao, Y.; Wang, W. Individual tree segmentation and tree species classification in subtropical broadleaf forests using UAV-based LiDAR, hyperspectral, and ultrahigh-resolution RGB data. Remote Sens. Environ. 2022, 280, 113143. [Google Scholar] [CrossRef]
- Plakman, V.; Janssen, T.; Brouwer, N.; Veraverbeke, S. Mapping species at an individual-tree scale in a temperate Forest, using Sentinel-2 images, airborne laser scanning data, and random Forest classification. Remote Sens. 2020, 12, 3710. [Google Scholar] [CrossRef]
- Raczko, E.; Zagajewski, B. Comparison of support vector machine, random forest and neural network classifiers for tree species classification on airborne hyperspectral APEX images. Eur. J. Remote Sens. 2017, 50, 144–154. [Google Scholar] [CrossRef] [Green Version]
- Zhang, B.; Zhao, L.; Zhang, X. Three-dimensional convolutional neural network model for tree species classification using airborne hyperspectral images. Remote Sens. Environ. 2020, 247, 111938. [Google Scholar] [CrossRef]
- Axelsson, A.; Lindberg, E.; Reese, H.; Olsson, H. Tree species classification using Sentinel-2 imagery and Bayesian inference. Int. J. Appl. Earth Obs. Geoinf. 2021, 100, 102318. [Google Scholar] [CrossRef]
- Wang, K.; Wang, T.; Liu, X. A Review: Individual Tree Species Classification Using Integrated Airborne LiDAR and Optical Imagery with a Focus on the Urban Environment. Forests 2019, 10, 1. [Google Scholar] [CrossRef] [Green Version]
- Lechner, M.; Dostálová, A.; Hollaus, M.; Atzberger, C.; Immitzer, M. Combination of Sentinel-1 and Sentinel-2 Data for Tree Species Classification in a Central European Biosphere Reserve. Remote Sens. 2022, 14, 2687. [Google Scholar] [CrossRef]
- Ewa, G.; David, F.; Katarzyna, O. Evaluation of machine learning algorithms for forest stand species mapping using Sentinel-2 imagery and environmental data in the Polish Carpathians. Remote Sens. Environ. 2020, 251, 7. [Google Scholar]
- Deur, M.; Gašparović, M.; Balenović, I. Tree species classification in mixed deciduous forests using very high spatial resolution satellite imagery and machine learning methods. Remote Sens. 2020, 12, 3926. [Google Scholar] [CrossRef]
- Götze, C.; Gerstmann, H.; Gläßer, C.; Jung, A. An approach for the classification of pioneer vegetation based on species-specific phenological patterns using laboratory spectrometric measurements. Phys. Geogr. 2017, 38, 524–540. [Google Scholar] [CrossRef]
- Violle, C.; Navas, M.L.; Vile, D.; Kazakou, E.; Fortunel, C.; Hummel, I.; Garnier, E. Let the concept of trait be functional! Oikos 2007, 116, 882–892. [Google Scholar] [CrossRef]
- Violle, C.; Reich, P.B.; Pacala, S.W.; Enquist, B.J.; Kattge, J. The emergence and promise of functional biogeography. Proc. Natl. Acad. Sci. USA 2014, 111, 13690–13696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.; Huang, Z.; Sun, H.; Wang, G. Mapping Forest Ecosystem Biomass Density for Xiangjiang River Basin by Combining Plot and Remote Sensing Data and Comparing Spatial Extrapolation Methods. Remote Sens. 2017, 9, 241. [Google Scholar] [CrossRef] [Green Version]
- Bilous, A.; Myroniuk, V.; Holiaka, D.; Bilous, S.; See, L.; Schepaschenko, D. Mapping growing stock volume and forest live biomass: A case study of the Polissya region of Ukraine. Environ. Res. Lett. 2017, 12, 105001. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Ma, Y.; Chen, P.; Yang, Y.; Fu, H.; Yang, F.; Raza, M.A.; Guo, C.; Shu, C.; Sun, Y.; et al. Estimation of Rice Aboveground Biomass by Combining Canopy Spectral Reflectance and Unmanned Aerial Vehicle-Based Red Green Blue Imagery Data. Front. Plant Sci. 2022, 13, 903643. [Google Scholar] [CrossRef]
- Santi, E.; Paloscia, S.; Pettinato, S.; Fontanelli, G.; Mura, M.; Zolli, C.; Maselli, F.; Chiesi, M.; Bottai, L.; Chirici, G. The potential of multifrequency SAR images for estimating forest biomass in Mediterranean areas. Remote Sens. Environ. 2017, 200, 63–73. [Google Scholar] [CrossRef]
- Hongwei, M.; Hai, L.; Shunbin, Y.; Wei, Z. Analysis and Prospect on the Application of Tree Species Classification Based on Forestry Remote Sensing. For. Resour. Manag. 2020, 3, 118. [Google Scholar] [CrossRef]
- Udali, A.; Lingua, E.; Persson, H.J. Assessing Forest Type and Tree Species Classification Using Sentinel-1 C-Band SAR Data in Southern Sweden. Remote Sens. 2021, 13, 3237. [Google Scholar] [CrossRef]
- Hemmerling, J.; Pflugmacher, D.; Hostert, P. Mapping temperate forest tree species using dense Sentinel-2 time series. Remote Sens. Environ. 2021, 267, 112743. [Google Scholar] [CrossRef]
- Louis, J.; Debaecker, V.; Pflug, B.; Main-Knorn, M.; Gascon, F. SENTINEL-2 SEN2COR: L2A Processor for Users. In Proceedings of the Living Planet Symposium, Prague, Czech Republic, 9–13 May 2016. [Google Scholar]
- Wan, H.; Tang, Y.; Jing, L.; Li, H.; Qiu, F.; Wu, W. Tree species classification of forest stands using multisource remote sensing data. Remote Sens. 2021, 13, 144. [Google Scholar] [CrossRef]
- Venkatappa, M.; Sasaki, N.; Anantsuksomsri, S.; Smith, B. Applications of the Google Earth Engine and Phenology-Based Threshold Classification Method for Mapping Forest Cover and Carbon Stock Changes in Siem Reap Province, Cambodia. Remote Sens. 2020, 12, 3110. [Google Scholar] [CrossRef]
- Han, Y.; Meng, J.; Xu, J. Soybean growth assessment method based on NDVI and phenological calibration. Trans. Chin. Soc. Agric. Eng. 2017, 33, 177–182. [Google Scholar] [CrossRef]
- Madhusudhan, M.; Ambujam, N.K. An urban ecology approach to land-cover changes in the Adyar sub-basin: Comparative analysis of NDWI, NDVI and NDBI using remote sensing. Int. J. Environ. Sustain. Dev. 2021, 1, 1. [Google Scholar] [CrossRef]
- Panigrahi, S.; Verma, K.; Tripathi, P. Review of MODIS EVI and NDVI Data for Data Mining Applications. In Data Deduplication Approaches: Concepts, Strategies, and Challenges; Academic Press: Cambridge, MA, USA, 2021; pp. 231–253. [Google Scholar] [CrossRef]
- Zefeng, X.; Ying, L.; Rongxin, D.; Honglei, Z.; Bolin, F. Extracting Farmland Shelterbelt Automatically Based on ZY-3 Remote Sensing Images. Sci. Silvae Sin. 2016, 52, 11–20. [Google Scholar] [CrossRef]
- Xu, D.; Wang, C.; Chen, J.; Shen, M.; Shen, B.; Yan, R.; Li, Z.; Karnieli, A.; Chen, J.; Yan, Y. The superiority of the normalized difference phenology index (NDPI) for estimating grassland aboveground fresh biomass. Remote Sens. Environ. 2021, 264, 112578. [Google Scholar] [CrossRef]
- Qian, B.; Huang, W.; Ye, H. Inversion of winter wheat chlorophyll contents based on improved algorithms for red edge position. Nongye Gongcheng Xuebao/Trans. Chin. Soc. Agric. Eng. 2021, 36, 162–170. [Google Scholar] [CrossRef]
- Zhenchuan, W.; Hu, D.; Tongqing, S.; Wanxia, P.; Fuping, Z.; Zhaoxia, Z.; Hao, Z. Allometric models of major tree species and forest biomass in Guangxi. Acta Ecol. Sin. 2015, 35, 4462–4472. [Google Scholar]
- Jingyun, F.; Anping, C. Dynamic forest biomass carbon pools in China and their significance. Acta Bot. Sin. 2001, 43, 967–973. [Google Scholar]
- Lin, X.; Peng, D.L.; Huang, G.S.; Wang, X.J. Object-oriented classification with multi-scale texture feature based on remote sensing image. Eng. Surv. Mapp. 2016, 25, 22–27. [Google Scholar] [CrossRef]
- Valderrama-Landeros, L.; Flores-Verdugo, F.; Rodríguez-Sobreyra, R.; Kovacs, J.M.; Flores-de-Santiago, F. Extrapolating canopy phenology information using Sentinel-2 data and the Google Earth Engine platform to identify the optimal dates for remotely sensed image acquisition of semiarid mangroves. J. Environ. Manag. 2021, 279, 111617. [Google Scholar] [CrossRef] [PubMed]


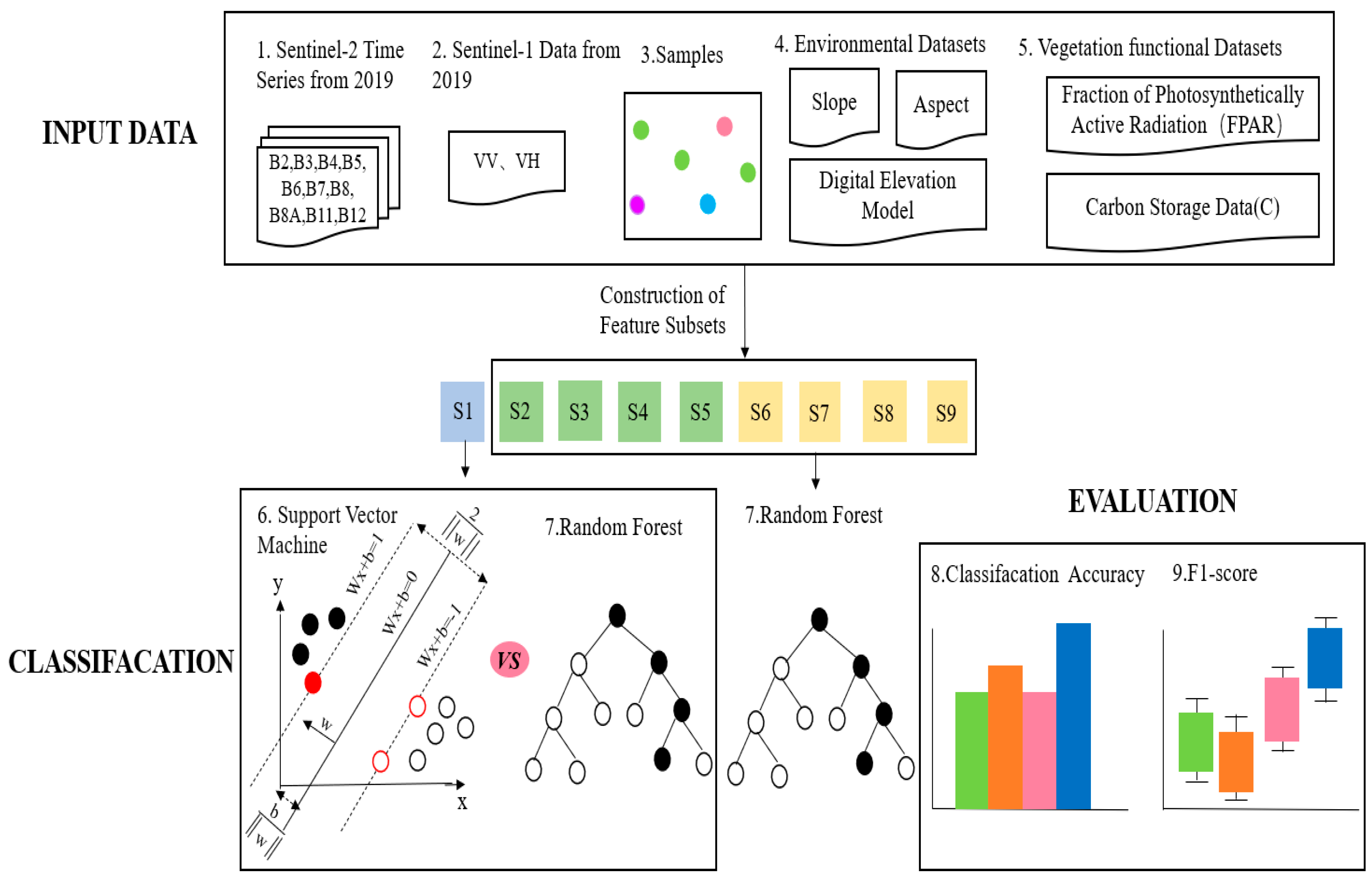


| ID of Schemes | Feature Subsets | Total Features |
|---|---|---|
| S1 | S2 Data a + Vis b | 17 |
| S2 | S2 Data + Vis + DEM c | 18 |
| S3 | S2 Data + Vis + Environmental Datasets d | 20 |
| S4 | S2 Data + Vis + Environmental Datasets + S1 Data e | 22 |
| S5 | S2 Data + Vis + S1 Data | 19 |
| S6 | S2 Data + Vis + FPAR f | 18 |
| S7 | S2 Data + Vis + C f | 18 |
| S8 | S2 Data + Vis + FPAR + C | 19 |
| S9 | S2 Data + Vis + FPAR + C + Environmental Datasets | 22 |
| Index | Calculation | Characteristic and Reference |
|---|---|---|
| NDVI | Highly correlated with canopy leaf area index (LAI) and chlorophyll content, and used to characterize green vegetation [47]. | |
| NDWI | An indicator used to characterize and monitor changes in surface water content. For scattered in the mountain between the small ditch has a relatively good capture effect. | |
| NDBI | To highlight the area of productive building land. NDBI is an effective index to extract urban land use information [48]. | |
| EVI | Helps to avoid the saturation problem of NDVI under high vegetation coverage and further reduces the influence of atmosphere and background [49]. | |
| RVI | An indicator that is sensitive to high vegetation coverage area [50]. | |
| NDPI | Helps to overcome the adverse effects of soil background heterogeneity, and has a high correlation with leaf water content due to the addition of short-wave infrared bands [51]. | |
| REPI | An indicator is sensitive to change of vegetation chlorophyll concentration, the widening of absorption characteristics due to the increase of chlorophyll concentration, and the movement of red edge to the long band direction [52]. |
| Model | R | R2 | Adjusted R2 | Errors in Standard Estimates | Statistical Change | ||||
|---|---|---|---|---|---|---|---|---|---|
| R2 Variation | F Variation | Degrees of Freedom 1 | Degrees of Freedom 2 | Change in Significance F | |||||
| A | 0.675 a | 0.456 | 0.452 | 0.12978 | 0.456 | 98.976 | 1 | 118 | 0.000 |
| B | 0.712 b | 0.508 | 0.499 | 0.12401 | 0.051 | 12.235 | 1 | 117 | 0.001 |
| C | 0.726 c | 0.527 | 0.515 | 0.12205 | 0.020 | 4.794 | 1 | 116 | 0.031 |
| Model | Unnormalized Coefficient | Normalization Coefficient | t | p-Value | Collinear Statistics | |||
|---|---|---|---|---|---|---|---|---|
| B | stderr | Beta | Tolerance | VIF | ||||
| A | constant | 0.855 | 0.036 | 23.480 | 0.000 | |||
| B2 | −0.680 | 0.068 | −0.675 | −9.949 | 0.000 | 1.000 | 1.000 | |
| B | constant | 0.750 | 0.0046 | 16.329 | 0.000 | |||
| B2 | −0.610 | 0.068 | −0.606 | −8.930 | 0.000 | 0.914 | 1.094 | |
| SLOPE | 0.226 | 0.065 | 0.237 | 3.498 | 0.001 | 0.914 | 1.094 | |
| C | constant | 0.848 | 0.064 | 13.327 | 0.000 | |||
| B2 | −0.665 | 0.072 | −0.660 | −9.266 | 0.000 | 0.803 | 1.246 | |
| NDWI | −0.164 | 0.075 | −0.155 | −2.189 | 0.031 | 0.813 | 1.231 | |
| SLOPE | 0.251 | 0.065 | 0.264 | 3.886 | 0.000 | 0.885 | 1.129 | |
| ID of Schemes | Feature Subsets | Total Features | OA a | Kappa b |
|---|---|---|---|---|
| S2 | S2 Data c+ VIs d+ DEM e | 18 | 0.7368 | 0.6790 |
| S3 | S2 Data + VIs + Environmental Datasets f | 20 | 0.7699 | 0.7190 |
| S4 | S2 Data + VIs+ Environmental Datasets +S1 Data g | 22 | 0.7577 | 0.7021 |
| S5 | S2 Data + VIs + S1 Data | 19 | 0.7434 | 0.6879 |
| ID of Schemes | Feature Subsets | Total Features | OA a | Kappa b |
|---|---|---|---|---|
| S6 | S2 c Data + VIs d + FPAR | 18 | 0.7651 | 0.7162 |
| S7 | S2 Data + VIs + C e | 18 | 0.7676 | 0.7115 |
| S8 | S2 Data + Vis + FPAR +C | 19 | 0.7774 | 0.7283 |
| S9 | S2 Data + VIs + FPAR + C + Environmental Dataset f | 22 | 0.7953 | 0.7501 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, Z.; Liu, X.; Zhu, L.; Li, J.; Zhang, Y. Remote Sensing Extraction Method of Illicium verum Based on Functional Characteristics of Vegetation Canopy. Remote Sens. 2022, 14, 6248. https://doi.org/10.3390/rs14246248
Zhang Z, Liu X, Zhu L, Li J, Zhang Y. Remote Sensing Extraction Method of Illicium verum Based on Functional Characteristics of Vegetation Canopy. Remote Sensing. 2022; 14(24):6248. https://doi.org/10.3390/rs14246248
Chicago/Turabian StyleZhang, Zhuoyao, Xiangnan Liu, Lihong Zhu, Junji Li, and Yue Zhang. 2022. "Remote Sensing Extraction Method of Illicium verum Based on Functional Characteristics of Vegetation Canopy" Remote Sensing 14, no. 24: 6248. https://doi.org/10.3390/rs14246248





