Phenotypic Traits Extraction and Genetic Characteristics Assessment of Eucalyptus Trials Based on UAV-Borne LiDAR and RGB Images
Abstract
:1. Introduction
2. Materials and Methods
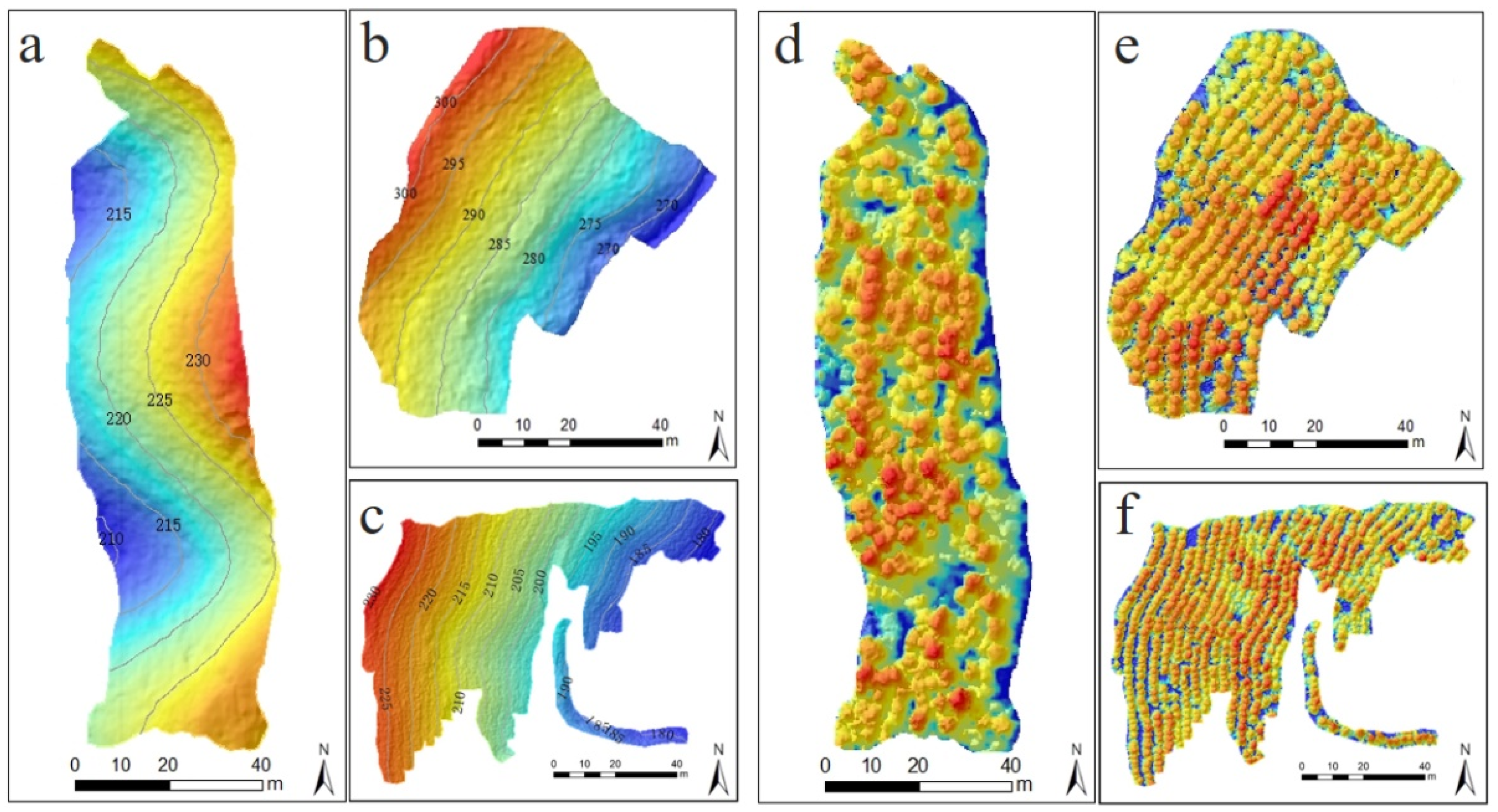
2.1. Study Area
2.2. Study Materials
2.3. Field Data and Manual Measurements
2.4. UAV-Borne LiDAR Data Acquisition and Preprocessing
2.5. UAV-Borne RGB Orthophoto Acquisition and Preprocessing
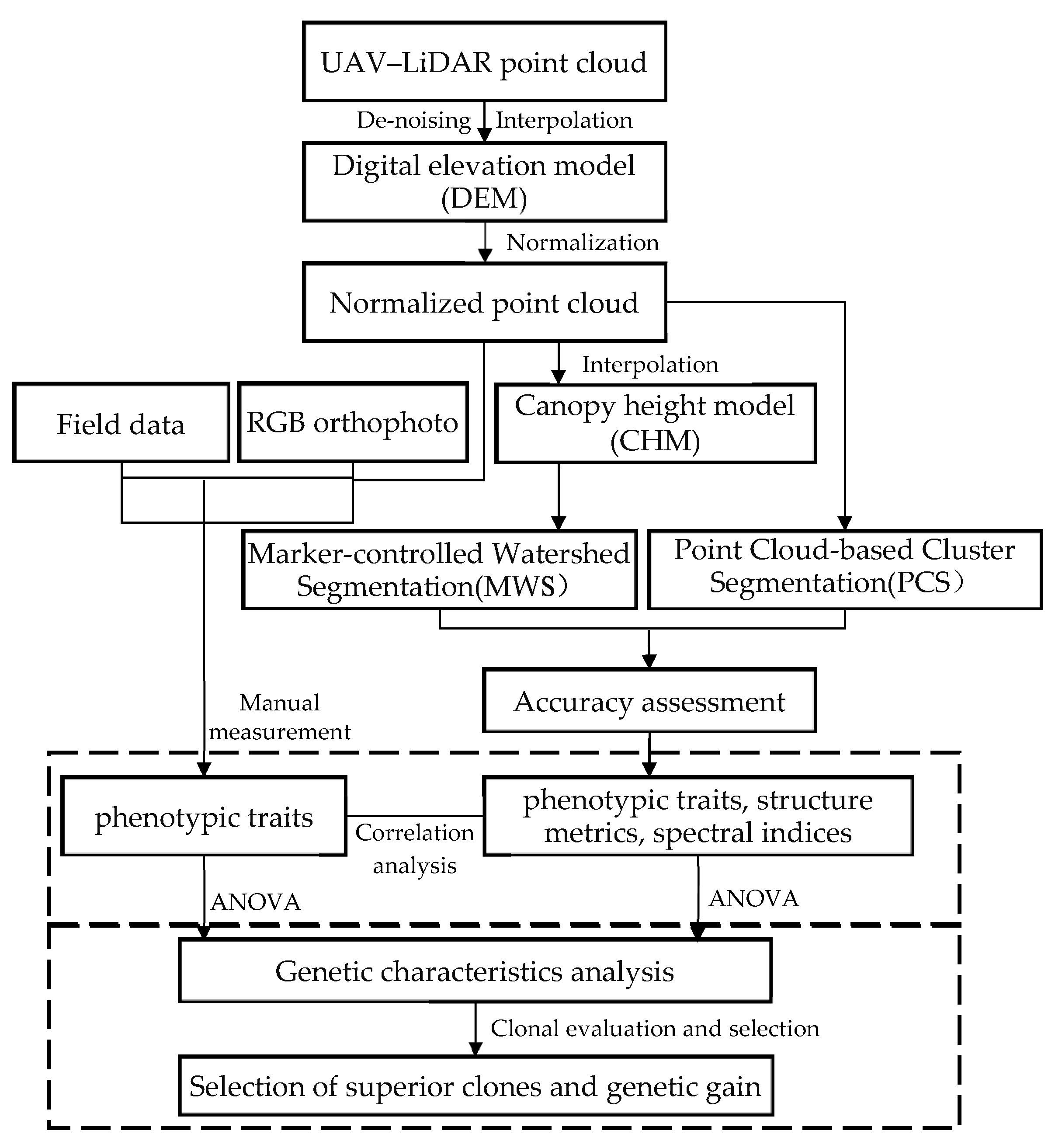
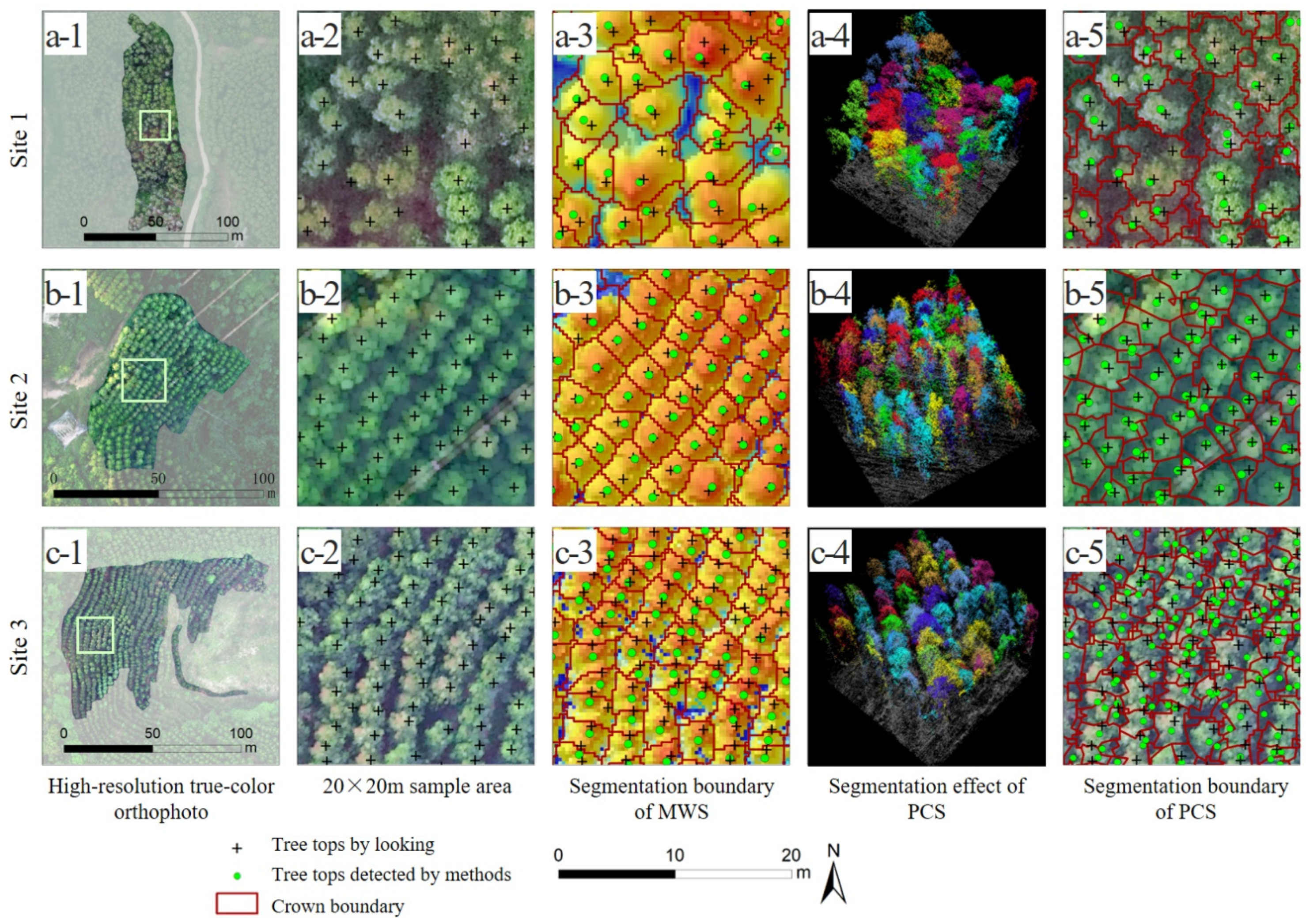
2.6. Individual Tree Segmentation Methods
2.7. Statistical Analysis Methods
3. Results
3.1. Individual Tree Segmentation
3.2. Statistical Analysis
3.2.1. Diameter at Breast Height (DBH)—Tree Height (H) Regression Model
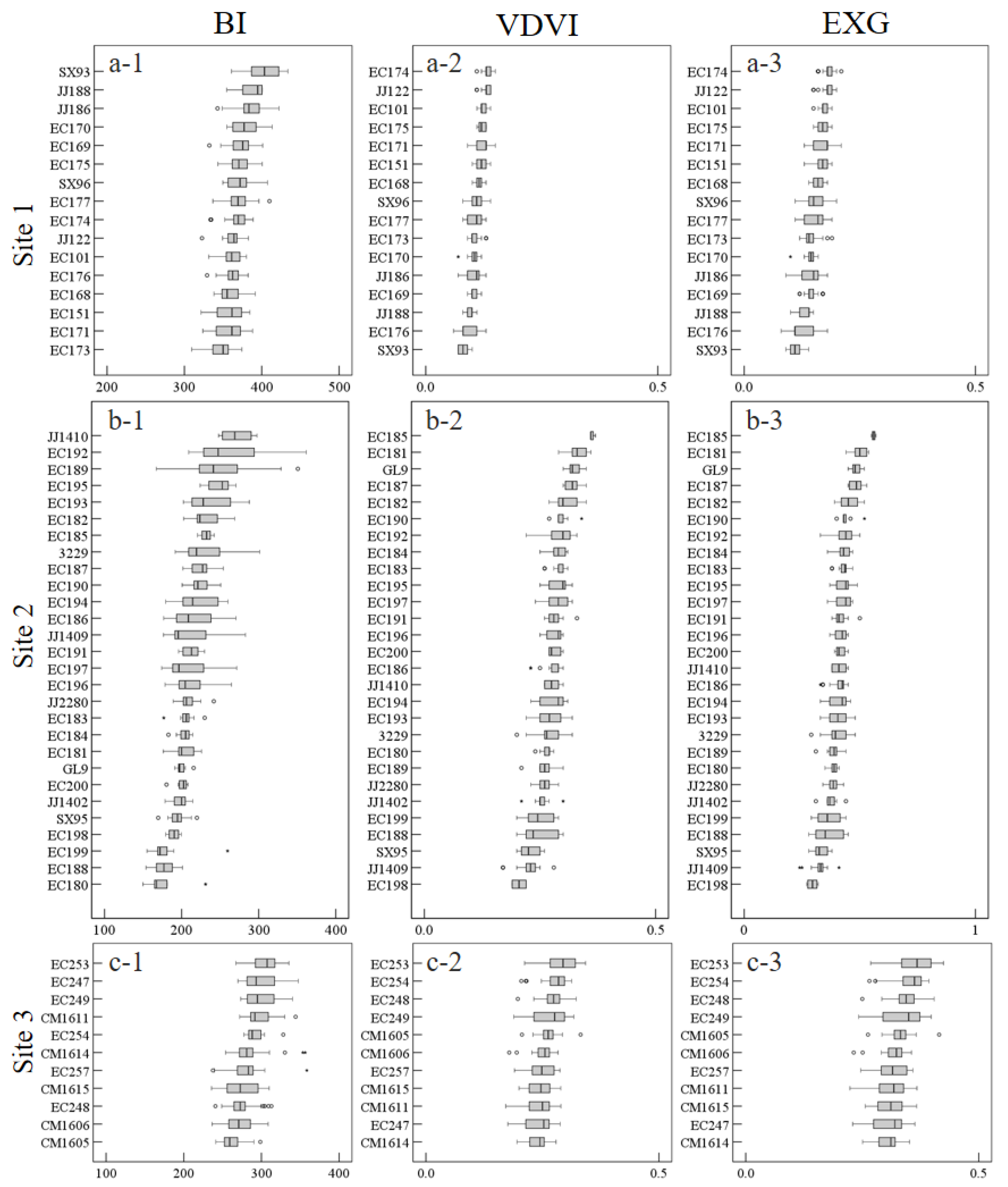
3.2.2. Phenotypic Traits and Analysis
3.2.3. LiDAR Metrics and Vegetation Indices Extraction and Analysis
3.3. Genetic Parameters and Clone Evaluation
4. Discussion
4.1. Individual Tree Segmentation
4.2. Statistical Analysis
4.3. Genetic Characteristic and Clone Evaluation
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A
| Site | Number | Species | Site | Number | Species |
|---|---|---|---|---|---|
| 1 | EC176 | E. wetarensis× (E. urophylla×E. wetarensis) | 2 | EC194 | E. urophylla × E. pellita |
| EC168 | E. wetarensis× E. grandis | EC196 | E. urophylla × E. pellita | ||
| EC173 | E. wetarensis× E. pellita | EC197 | E. urophylla × E. pellita | ||
| EC177 | E. wetarensis× E. pellita | EC200 | E. urophylla × E. pellita | ||
| EC170 | E. wetarensis× E. pellita | EC189 | E. urophylla × E. camaldulensi | ||
| JJ122 | E. urophylla × E. wetarensis | JJ1402 | E. urophylla | ||
| EC174 | E. urophylla | JJ1409 | E. urophylla | ||
| EC175 | E. urophylla | JJ1410 | E. urophylla | ||
| JJ186 | E. urophylla | JJ2280 | E. urophylla | ||
| JJ188 | E. urophylla | EC182 | E. grandis × E. urophylla | ||
| EC151 | E. wetarensis × E. grandis | GL9 | E. grandis × E. urophylla | ||
| EC171 | E. wetarensis × E. grandis | EC184 | E. grandis × E. grandis | ||
| EC101 | E. wetarensis × E. pellita | EC185 | E. grandis × E. grandis | ||
| EC169 | E. wetarensis × E. pellita | EC195 | E. pellita × E. urophylla | ||
| SX93 | E. camaldulensi | EC199 | E. pellita × E. grandis | ||
| SX96 | E. camaldulensi | SX95 | E. camaldulensi | ||
| 2 | EC180 | E. wetarensis× E. urophylla | 3 | EC247 | E. wetarensis × E. urophylla |
| EC188 | E. wetarensis× E. grandis | EC249 | E. urophylla × E. wetarensis | ||
| EC198 | E.wetarensis× E. pellita | CM1605 | E. urophylla × E. ABL | ||
| 3229 | E. urophylla × E. grandis | CM1606 | E. urophylla × E. ABL | ||
| EC181 | E. urophylla × E. grandis | CM1611 | E. urophylla × E. ABL | ||
| EC186 | E. urophylla × E. pellita | CM1614 | E. urophylla × E. ABL | ||
| EC187 | E. urophylla × E. pellita | CM1615 | E. urophylla × E. ABL | ||
| EC190 | E. urophylla × E. pellita | EC254 | E. wetarensis × (E. urophylla*E. pellita) | ||
| EC191 | E. urophylla × E. pellita | EC248 | E. wetarensis × E. grandis | ||
| EC192 | E. urophylla × E. pellita | EC253 | E. wetarensis × E. grandis | ||
| EC193 | E. urophylla × E. pellita | EC257 | E. wetarensis |
| Mstrics | Site-1 | Site-2 | Site-3 | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Mean ± std | Clone | rep | Clone × rep | Mean ± std | Clone | Mean ± std | Clone | rep | Clone × rep | ||
| LiDAR metrics | Hmean | 12.02 ± 1.99 | 3.38 ** | 1.60 | 2.16 | 12.95 ± 1.88 | 15.87 ** | 8.98 ± 0.96 | 3.22 ** | 0.30 | 8.33 ** |
| Hcv | 0.32 ± 0.06 | 2.51 * | 1.34 | 3.81 | 0.31 ± 0.11 | 6.15 ** | 12.78 ± 1.21 | 2.75 * | 3.01 * | 2.40 ** | |
| h25 | 10.14 ± 1.52 | 3.52 ** | 3.59 * | 2.32 | 11.01 ± 3.39 | 6.71 ** | 0.27 ± 0.04 | 2.06 * | 0.52 | 7.68 * | |
| h5 | 13.11 ± 1.73 | 2.69 ** | 1.69 | 4.43 | 14.32 ± 2.04 | 14.03 ** | 7.49 ± 1.04 | 3.02 ** | 0.53 | 8.34 ** | |
| h75 | 15.16 ± 1.85 | 2.587 * | 1.40 | 2.90 | 15.78 ± 1.68 | 29.56 ** | 9.41 ± 1.11 | 4.77 ** | 0.15 | 7.49 ** | |
| h95 | 16.64 ± 1.82 | 3.19 ** | 0.54 | 3.27 | 16.69 ± 1.67 | 37.29 ** | 10.89 ± 1.08 | 7.00 ** | 0.61 | 5.69 ** | |
| d3 | 0.62 ± 0.10 | 8.06 ** | 2.29 | 61.22 | 0.63 ± 0.10 | 8.94 ** | 0.74 ± 0.07 | 1.51 | 2.66 * | 3.40 ** | |
| d5 | 0.52 ± 0.10 | 6.17 ** | 4.69 * | 266.02 * | 0.58 ± 0.12 | 9.36 ** | 0.62 ± 0.09 | 0.90 | 2.47 * | 5.43 ** | |
| d7 | 0.37 ± 0.09 | 4.28 ** | 2.64 | 98.50 | 0.51 ± 0.13 | 11.25 ** | 0.4 ± 0.09 | 1.31 | 3.32 * | 5.68 ** | |
| d9 | 0.13 ± 0.04 | 2.28 * | 2.83 | 0.95 | 0.22 ± 0.09 | 6.33 ** | 0.09 ± 0.04 | 1.48 | 2.57 * | 2.61 ** | |
| OP | 0.21 ± 0.11 | 0.02 * | 3.05 | 15.18 | 0.07 ± 0.15 | 3.22 ** | 0.01 ± 0.04 | 2.76 ** | 0.64 | 3.76 ** | |
| EU | 0.28 ± 0.10 | 0.00 ** | 2.47 | 35.07 | 0.18 ± 0.08 | 4.34 ** | 0.25 ± 0.06 | 1.97 ** | 2.09 | 3.35 ** | |
| OL | 0.46 ± 0.13 | 0.22 | 4.61 * | 1325.86 * | 0.70 ± 0.20 | 3.09 ** | 0.72 ± 0.09 | 1.27 * | 2.18 | 5.38 ** | |
| CL | 0.06 ± 0.06 | 0.01 * | 0.15 | 2.62 | 0.05 ± 0.08 | 3.09 ** | 0.01 ± 0.05 | 2.55 * | 0.93 | 3.00 ** | |
| α | 0.56 ± 0.11 | 5.67 ** | 7.02 ** | 0.62 | 0.38 ± 0.11 | 8.98 ** | 0.37 ± 0.06 | 0.71 * | 2.28 | 7.07 * | |
| β | 1.79 ± 1.05 | 2.44 * | 3.27 | 3.65 | 1.08 ± 0.19 | 6.29 ** | 1.50 ± 0.17 | 1.59 * | 3.44 | 2.73 * | |
| LAI | 2.28 ± 0.63 | 4.04 ** | 3.05 | 2.21 | 2.70 ± 0.15 | - | 2.45 ± 0.19 | 0.84 | 0.32 | 1.71 | |
| CC | 0.64 ± 0.18 | 5.58 ** | 0.89 | 10.96 | 0.687 ± 0.01 | - | 0.70 ± 0.01 | 0.70 | 0.49 | 1.02 | |
| VI | BI | 373.84 ± 16.88 | 4.50 ** | 3.64 * | 0.41 | 345.24 ± 20.22 | 9.75 ** | 373.84 ± 16.88 | 3.93 ** | 5.74 ** | 9.20 ** |
| NG | 0.39 ± 0.01 | 6.23 ** | 0.73 | - | 0.71 ± 0.01 | 7.97 ** | 0.39 ± 0.01 | 3.30 ** | 2.17 | 13.81 ** | |
| RGRI | 0.78 ± 0. 03 | 4.39 ** | 0.04 | 0.94 | 0.85 ± 0. 03 | 16.86 ** | 0.78 ± 0. 03 | 4.29 ** | 5.28 ** | 11.04 ** | |
| GRVI | 0.13 ± 0.02 | 4.34 ** | 0.03 | 0.76 | 0.14 ± 0.02 | 8.34 ** | 0.13 ± 0.02 | 3.60 ** | 2.53 | 13.04 ** | |
| VARI | 0.15 ± 0.03 | 1.61 | 0.76 | 3.80 | 0.15 ± 0.05 | 9.06 ** | 0.15 ± 0.03 | 3.45 ** | 1.73 | 11.82 ** | |
| VDVI | 0.11 ± 0.02 | 6.25 ** | 0.67 | 193.83 | 0.10 ± 0.01 | 16.01 ** | 0.11 ± 0.02 | 4.17 ** | 4.81 ** | 12.13 ** | |
| EXG | 0.16 ± 0.02 | 6.09 ** | 0.71 | 405.46 | 0.16 ± 0.02 | 17.23 ** | 0.16 ± 0.02 | 4.29 ** | 5.35 ** | 11.90 ** | |
| EXG-R | 0.12 ± 0.04 | 5.54 ** | 0.15 | 4.25 | 0.12 ± 0.03 | 11.88 ** | 0.12 ± 0.04 | 3.96 ** | 3.97 ** | 13.59 ** | |
| CIVE | 18.71 ± 0.01 | 6.28 ** | 0.61 | - | 16.48 ± 0.08 | 15.59 ** | 18.71 ± 0.01 | 4.06 ** | 4.71 ** | 11.37 ** | |
| Correlation | Site-1 | Site-2 | Site-3 | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | D1 | D2 | C1 | C2 | H1 | H2 | D1 | D2 | C1 | C2 | H1 | H2 | D1 | D2 | C1 | C2 | ||
| Traits | H1 | 0.95 ** | 0.99 ** | 0.95 ** | 0.52 ** | 0.48 ** | 0.99 ** | 1.00 ** | 0.99 ** | 0.22 ** | 0.26 ** | 0.96 ** | 0.98 ** | 0.96 ** | 0.25 ** | 0.35 ** | |||
| H2 | 0.95 ** | 0.95 ** | 0.99 ** | 0.57 ** | 0.53 ** | 0.99 ** | 0.99 ** | 1.00 ** | 0.22 ** | 0.27 ** | 0.96 ** | 0.96 ** | 1.00 ** | 0.24 ** | 0.34 ** | ||||
| D1 | 0.99 ** | 0.95 ** | 0.95 ** | 0.52 ** | 0.48 ** | 1.00 ** | 0.99 ** | 0.99 ** | 0.22 ** | 0.26 ** | 1.00 ** | 0.96 ** | 0.96 ** | 0.25 ** | 0.35 ** | ||||
| D2 | 0.95 ** | 0.99 ** | 0.95 ** | 0.56 ** | 0.53 ** | 0.99 ** | 1.00 ** | 0.99 ** | 0.68 ** | 0.92 ** | 0.96 ** | 0.98 ** | 0.96 ** | 0.24 ** | 0.34 ** | ||||
| C1 | 0.52 ** | 0.57 ** | 0.52 ** | 0.56 ** | 0.80 ** | 0.22 ** | 0.22 ** | 0.22 ** | 0.68 ** | 0.74 ** | 0.25 ** | 0.24 ** | 0.25 ** | 0.24 ** | 0.63 ** | ||||
| C2 | 0.48 ** | 0.53 ** | 0.48 ** | 0.53 ** | 0.80 ** | 0.26 ** | 0.27 ** | 0.26 ** | 0.92 ** | 0.74 ** | 0.35 ** | 0.34 ** | 0.35 | 0.34 | 0.63 ** | ||||
| LiDAR metrics | Hmean | 0.76 ** | 0.77 ** | 0.52 ** | 0.37 ** | 0.16 ** | 0.17 ** | 0.68 ** | 0.69 ** | 0.39 ** | 0.18 ** | 0.03 | −0.12 * | 0.80 ** | 0.84 ** | 0.51 ** | 0.37 ** | 0.10 | 0.02 |
| Hcv | 0.23 ** | 0.21 ** | 0.24 ** | 0.30 ** | 0.28 ** | 0.19 ** | 0.03 | 0.04 | 0.02 | 0.21 ** | 0.02 | 0.25 ** | 0.14 * | 0.12 * | 0.00 | 0.23 ** | −0.09 | 0.22 ** | |
| h25 | 0.43 ** | 0.46 ** | 0.25 ** | 0.11 | −0.03 | 0.03 | 0.35 ** | 0.04 ** | 0.18 ** | −0.02 | −0.02 | −0.21 ** | 0.63 ** | 0.66 ** | 0.43 ** | 0.23 ** | 0.13 * | −0.06 | |
| h5 | 0.67 ** | 0.69 ** | 0.45 ** | 0.28 ** | 0.08 | 0.14 * | 0.70 ** | 0.71 ** | 0.34 ** | 0.16 ** | −0.05 | −0.16 ** | 0.76 ** | 0.79 ** | 0.47 ** | 0.32 ** | 0.09 | −0.03 | |
| h75 | 0.90 ** | 0.90 ** | 0.65 ** | 0.51 ** | 0.28 ** | 0.27 ** | 0.94 ** | 0.95 ** | 0.56 ** | 0.44 ** | 0.08 | 0.06 | 0.90 ** | 0.93 ** | 0.57 ** | 0.47 ** | 0.07 | 0.10 | |
| h95 | 0.99 ** | 0.95 ** | 0.69 ** | 0.68 ** | 0.46 ** | 0.29 ** | 0.99 ** | 0.99 ** | 0.66 ** | 0.58 ** | 0.18 ** | 0.22 ** | 0.94 ** | 0.99 ** | 0.57 ** | 0.58 ** | 0.08 | 0.20 ** | |
| d3 | 0.20 ** | 0.17 ** | 0.06 | 0.12 | 0.09 | −0.05 | 0.08 | 0.08 | 0.06 | −0.15 ** | 0.02 | −0.23 ** | 0.13 * | 0.14 * | 0.22 ** | −0.08 | 0.20 ** | −0.17 ** | |
| d5 | 0.08 | 0.09 | −0.04 | −0.08 | −0.12 | −0.13 * | 0.10 | 0.10 | 0.04 | −0.20 ** | −0.03 | −0.30 ** | 0.18 ** | 0.18 ** | 0.20 ** | −0.14 * | 0.13 * | −0.27 ** | |
| d7 | 0.03 | 0.09 | −0.03 | −0.22 ** | −0.28 ** | −0.11 | 0.10 | 0.10 | -0.03 | -0.28 ** | −0.11 * | −0.40 ** | 0.16 ** | 0.16 ** | 0.17 ** | −0.20 ** | 0.10 | −0.34 ** | |
| d9 | 0.06 | 0.137 * | 0.04 | −0.27 ** | −0.36 ** | −0.05 | 0.01 | 0.02 | −0.24 ** | −0.46 ** | −0.34 ** | −0.57 ** | 0.07 | 0.08 | 0.03 | −0.25 ** | −0.02 | −0.36 ** | |
| OP | 0.30 ** | 0.23 ** | 0.35 ** | 0.49 ** | 0.51 ** | 0.32 ** | −0.01 | −0.01 | 0.31 ** | 0.48 ** | 0.42 ** | 0.60 ** | 0.08 | 0.08 | 0.14 * | 0.32 ** | 0.11 | 0.36 ** | |
| EU | −0.19 ** | −0.16 ** | −0.15 * | −0.12 * | −0.11 | −0.11 | −0.02 | −0.03 | 0.07 | 0.20 ** | 0.12 * | 0.27 ** | −0.02 ** | −0.14 * | −0.15 ** | 0.09 | −0.08 | 0.19 ** | |
| OL | −0.16 ** | −0.12 * | −0.21 ** | −0.33 ** | −0.34 ** | −0.21 ** | 0.01 | 0.02 | −0.34 ** | −0.54 ** | −0.48 ** | −0.68 ** | 0.06 | 0.05 | 0.00 | −0.30 ** | −0.03 | −0.40 ** | |
| CL | 0.07 | 0.07 | 0.05 | 0.02 | 0.00 | 0.02 | 0.01 | 0.00 | 0.22 ** | 0.28 ** | 0.29 ** | 0.34 ** | 0.01 | 0.00 | 0.06 | 0.14 * | 0.05 | 0.17 ** | |
| α | −0.38 ** | −0.49 ** | −0.21 | −0.38 ** | 0.02 | −0.28 | −0.06 | −0.07 | 0.08 | 0.33 ** | 0.15 ** | 0.44 ** | −0.03 | −0.02 | −0.10 | 0.28 ** | −0.10 | 0.37 ** | |
| β | −0.50 ** | −0.55 ** | −0.44 ** | −0.44 ** | −0.22 | −0.31 * | −0.12 * | 0.00 | 0.16 ** | 0.35 ** | 0.31 ** | 0.51 ** | −0.15 ** | −0.15 ** | −0.07 | 0.21 ** | 0.01 | 0.35 ** | |
| LAI | 0.24 | 0.37 ** | 0.10 | 0.40 ** | −0.06 | 0.36 * | 0.24 | −0.14 * | 0.21 * | 0.21 | −0.05 | 0.32 | 0.26 * | 0.29 * | 0.14 | 0.37 ** | −0.11 | 0.35 ** | |
| CC2 | 0.23 | 0.38 ** | 0.13 | 0.46 ** | −0.02 | 0.43 ** | 0.29 * | 0.32 * | 0.13 | 0.10 | −0.11 | 0.43 ** | 0.24 | 0.40 ** | 0.31 * | 0.49 ** | −0.10 | 0.30 | |
| VI | BI | 0.21 ** | 0.25 ** | 0.21 ** | 0.06 | 0.00 | 0.13 * | −0.08 | −0.07 | 0.05 | 0.08 | 0.12 * | 0.14 * | −0.04 | −0.03 | −0.05 | 0.13 * | −0.03 | 0.19 ** |
| NG | 0.10 | 0.10 | 0.12 | 0.126 * | 0.121 * | 0.08 | 0.08 | 0.08 | 0.01 | 0.07 | −0.04 | 0.06 | 0.07 | 0.07 | −0.16 ** | 0.26 ** | −0.25 ** | 0.29 ** | |
| RGRI | 0.09 | 0.11 | 0.05 | −0.03 | −0.08 | 0.01 | −0.19 ** | −.19 ** | 0.03 | −0.04 | 0.17 ** | 0.04 | −0.05 | −0.04 | 0.20 ** | −0.30 ** | 0.29 ** | −0.35 ** | |
| GRVI | −0.10 | −0.12 | −0.06 | 0.03 | 0.07 | −0.01 | −0.08 | −0.08 | 0.00 | −0.07 | 0.04 | −0.05 | −0.08 | −0.07 | 0.15 ** | −0.26 ** | 0.25 ** | −0.29 ** | |
| VARI | −0.16 ** | −0.18 ** | −0.11 | 0.00 | 0.06 | −0.04 | 0.07 | 0.06 | 0.00 | 0.00 | −0.05 | −0.05 | −0.10 | −0.09 | 0.12 * | −0.24 ** | 0.22 ** | −0.25 ** | |
| VDVI | 0.10 | 0.10 | 0.12 | 0.127 * | 0.121 * | 0.08 | −0.18 ** | −0.18 ** | 0.05 | −0.03 | 0.19 ** | 0.05 | −0.05 | −0.04 | 0.19 ** | −0.30 ** | 0.29 ** | −0.35 ** | |
| EXG | 0.10 | 0.10 | 0.12 | 0.126 * | 0.120 * | 0.08 | −0.18 ** | −0.18 ** | 0.05 | −0.03 | 0.19 ** | 0.06 | −0.05 | −0.04 | 0.20 ** | −0.30 ** | 0.29 ** | −0.35 ** | |
| EXGR | 0.01 | 0.00 | 0.04 | 0.08 | 0.10 | 0.04 | −0.15 ** | −0.15 ** | 0.03 | −0.05 | 0.14 * | 0.01 | −0.06 | −0.05 | 0.18 ** | −0.29 ** | 0.28 ** | −0.33 ** | |
| CIVE | −0.09 | −0.09 | −0.11 | −0.12 * | −0.12 | −0.07 | 0.18 ** | 0.18 ** | −0.06 | 0.03 | −0.19 ** | −0.04 | 0.05 | 0.04 | −0.20 ** | 0.30 ** | −0.29 ** | 0.35 ** | |
References
- Zhou, J.; Tardieu, F.; Pridmore, T.; Doonan, J.; Reynolds, D.; Hall, N.; Griffiths, S.; Cheng, T.; Zhu, Y.; Wang, X.; et al. Plant phenomics: History, present status and challenges. J. Nanjing Agric. Univ. 2018, 41, 580–588. [Google Scholar] [CrossRef]
- Dungey, H.S.; Dash, J.P.; Pont, D.; Clinton, P.W.; Watt, M.S.; Telfer, E.J. Phenotyping Whole Forests Will Help to Track Genetic Performance. Trends Plant Sci. 2018, 23, 854–864. [Google Scholar] [CrossRef]
- Pan, Y. Analysis of Concepts, Categories of Plant Phenome and Phenomics. Acta Agron. Sin. 2015, 41, 175–186. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Annu. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [Green Version]
- Bian, L.; Zhang, H. Application of Phenotyping Techniques in Forest Tree Breeding and Precision Forestry. Sci. Silvae Sin. 2020, 56, 113–126. [Google Scholar] [CrossRef]
- Rincent, R.; Charpentier, J.; Faivre-Rampant, P.; Paux, E.; Le Gouis, J.; Bastien, C.; Segura, V. Phenomic Selection Is a Low-Cost and High-Throughput Method Based on Indirect Predictions: Proof of Concept on Wheat and Poplar. G3 2018, 8, 3961–3972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Araus, J.L.; Kefauver, S.C.; Zaman-Allah, M.; Olsen, M.S.; Cairns, J.E. Translating High-Throughput Phenotyping into Genetic Gain. Trends Plant Sci. 2018, 23, 451–466. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Matese, A.; Toscano, P.; Di Gennaro, S.; Genesio, L.; Vaccari, F.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, Aircraft and Satellite Remote Sensing Platforms for Precision Viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef] [Green Version]
- Kang, X. Research progress and prospect of forest genetics and tree breeding. J. Nanjing For. Univ. Nat. Sci. Ed. 2020, 44, 1–10. [Google Scholar]
- Zhao, C. Big Data of Plant Phenomics and Its Research Progress. J. Agric. Big Data 2019, 1, 5–18. [Google Scholar] [CrossRef]
- Appels, R. Plant phenome to genome: A mini-review. Funct. Plant Biol. 2012, 39, iii–viii. [Google Scholar] [CrossRef]
- Li, X.; Li, Q.; Lyu, L.; Lan, S. Economic Benefit Analysis of 6-year-old Eucalypt Plantation in Guangxi State-owned Gaofeng Forest Farm. Guangxi For. Sci. 2020, 49, 613–617. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, Y.; Tao, R.; Zhu, Y.; Lan, S. Limitation and Its Comprehensive Measures of High-sustainable Production in Eucalyptus Plantation, Guangxi Province. J. Anhui Agric. Sci. 2020, 48, 125–130. [Google Scholar] [CrossRef]
- Wen, Y.; Zhou, X.; Yu, S.; Zhu, H. The Predicament and Countermeasures of Development of Global Eucalyptus Plantations. Guangxi Sci. 2018, 25, 107–116. [Google Scholar] [CrossRef]
- Zhang, L.; Xiong, T.; Wang, J.; Shi, Q.; Li, L.; Chen, D.; Tang, Z.; Lan, J. Eucalyptus Clonal Breeding at Guangxi Dongmen Forest Farm. Eucalypt Sci. Technol. 2015, 32, 45–49. [Google Scholar] [CrossRef]
- Guo, Q.; Wu, F.; Pang, S.; Zhao, X.; Chen, L.; Liu, J.; Xue, B.; Xu, G.; Li, L.; Jing, H.; et al. Crop 3D: A platform based on LiDAR for 3D high-throughput crop phenotyping. Sci. Sin. 2016, 46, 1210–1221. [Google Scholar] [CrossRef]
- Pieruschka, R.; Poorter, H. Phenotyping plants: Genes, phenes and machines. Funct. Plant Biol. 2012, 39, 813. [Google Scholar] [CrossRef] [PubMed]
- Houle, D.; Govindaraju, D.R.; Omholt, S. Phenomics: The next challenge. Nat. Rev. Genet. 2010, 11, 855–866. [Google Scholar] [CrossRef]
- Cabrera-Bosquet, L.; Crossa, J.; von Zitzewitz, J.; Serret, M.D.; Luis Araus, J. High-throughput Phenotyping and Genomic Selection: The Frontiers of Crop Breeding Converge. J. Integr. Plant Biol. 2012, 54, 312–320. [Google Scholar] [CrossRef] [Green Version]
- Xie, C.; Yang, C. A review on plant high-throughput phenotyping traits using UAV-based sensors. Comput. Electron. Agric. 2020, 178, 105731. [Google Scholar] [CrossRef]
- Pang, Y.; Li, Z.; Chen, E.; Sun, G. Lidar Remote Sensing Technology and Its Application in Forestry. Sci. Silvae Sin. 2005, 41, 129–136. [Google Scholar] [CrossRef]
- Dubayah, R.O.; Sheldon, S.L.; Clark, D.B.; Hofton, M.A.; Blair, J.B.; Hurtt, G.C.; Chazdon, R.L. Estimation of tropical forest height and biomass dynamics using lidar remote sensing at La Selva, Costa Rica. JGR J. Geophys. Res. Biogeosci. 2010, 115. [Google Scholar] [CrossRef]
- Parker, G.G.; Harmon, M.E.; Lefsky, M.A.; Chen, J.; Pelt, R.V.; Weis, S.B.; Thomas, S.C.; Winner, W.E.; Shaw, D.C.; Frankling, J.F. Three-dimensional Structure of an Old-growth Pseudotsuga-Tsuga Canopy and Its Implications for Radiation Balance, Microclimate, and Gas Exchange. Ecosystems 2004, 7, 440–453. [Google Scholar] [CrossRef]
- Wulder, M.A.; Bater, C.W.; Coops, N.C.; Hilker, T.; White, J.C. The role of LiDAR in sustainable forest management. For. Chron. 2008, 84, 807–826. [Google Scholar] [CrossRef] [Green Version]
- Bolton, D.K.; Coops, N.C.; Wulder, M.A. Measuring Forest structure along productivity gradients in the Canadian boreal with small-footprint Lidar. Environ. Monit. Assess. 2013, 185, 6617–6634. [Google Scholar] [CrossRef]
- Monsi, M.; Uchijima, Z.; Oikawa, T. Structure of Foliage Canopies and Photosynthesis. Annu. Rev. Ecol. Syst. 1973, 4, 301–327. [Google Scholar] [CrossRef]
- Rasmussen, J.; Ntakos, G.; Nielsen, J.; Svensgaard, J.; Poulsen, R.N.; Christensen, S. Are vegetation indices derived from consumer-grade cameras mounted on UAVs sufficiently reliable for assessing experimental plots? Eur. J. Agron. 2016, 74, 75–92. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Casadesús, J.; Kaya, Y.; Bort, J.; Nachit, M.M.; Araus, J.L.; Amor, S.; Ferrazzano, G.; Maalouf, F.; Maccaferri, M.; Martos, V.; et al. Using vegetation indices derived from conventional digital cameras as selection criteria for wheat breeding in water-limited environments. Ann. Appl. Biol. 2007, 150, 227–236. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant Remote Sensing Vegetation Indices: A Review of Developments and Applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef] [Green Version]
- Hernández-Clemente, R.; Navarro-Cerrillo, R.M.; Zarco-Tejada, P.J. Carotenoid content estimation in a heterogeneous conifer forest using narrow-band indices and PROSPECT + DART simulations. Remote Sens. Environ. 2012, 127, 298–315. [Google Scholar] [CrossRef]
- Burkart, A.; Hecht, V.L.; Kraska, T.; Rascher, U. Phenological analysis of unmanned aerial vehicle based time series of barley imagery with high temporal resolution. Precis. Agric. 2018, 19, 134–146. [Google Scholar] [CrossRef]
- Santini, F.; Kefauver, S.C.; Resco De Dios, V.; Araus, J.L.; Voltas, J. Using unmanned aerial vehicle-based multispectral, RGB and thermal imagery for phenotyping of forest genetic trials: A case study in Pinus halepensis. Ann. Appl. Biol. 2019, 174, 262–276. [Google Scholar] [CrossRef] [Green Version]
- Jing, L.; Hu, B.; Noland, T.; Li, J. An individual tree crown delineation method based on multi-scale segmentation of imagery. ISPRS J. Photogramm. 2012, 70, 88–98. [Google Scholar] [CrossRef]
- Hyyppa, J.; Kelle, O.; Lehikoinen, M.; Inkinen, M. A segmentation-based method to retrieve stem volume estimates from 3-D tree height models produced by laser scanners. IEEE Trans. Geosci. Remote Sens. 2001, 39, 969–975. [Google Scholar] [CrossRef]
- Lu, X.; Guo, Q.; Li, W.; Flanagan, J. A bottom-up approach to segment individual deciduous trees using leaf-off lidar point cloud data. ISPRS J. Photogramm. 2014, 94, 1–12. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A New Method for Segmenting Individual Trees from the Lidar Point Cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Shen, X.; Dai, J.; Cao, L. Comparisons and Accuracy Assessments of LiDAR-Based Tree Segmentation Approaches in Planted Forests. Sci. Silvae Sin. 2018, 54, 127–136. [Google Scholar] [CrossRef]
- Li, Y.; He, H.; Huang, X. Analysis on climate change in Nanning city in recent 50 years. J. Guangxi Univ. Nat. Sci. Ed. 2007, 2007, 159–162. [Google Scholar] [CrossRef]
- Bengio, Y. Grandvalet. No Unbiased Estimator of the Variance of K-Fold Cross-Validation. J. Mach. Learn. Res. 2003, 5, 1089–1105. [Google Scholar]
- Zhao, X.; Guo, Q.; Su, Y.; Xue, B. Improved progressive TIN densification filtering algorithm for airborne LiDAR data in forested areas. ISPRS J. Photogramm. 2016, 117, 79–91. [Google Scholar] [CrossRef] [Green Version]
- Næsset, E.; Økland, T. Estimating tree height and tree crown properties using airborne scanning laser in a boreal nature reserve. Remote Sens. Environ. 2002, 79, 105–115. [Google Scholar] [CrossRef]
- Woods, M.; Lim, K.; Treitz, P. Predicting Forest stand variables from LiDAR data in the Great Lakes—St. Lawrence Forest of Ontario. For. Chron. 2008, 84, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Solberg, S.; Næsset, E.; Hanssen, K.H.; Christiansen, E. Mapping defoliation during a severe insect attack on Scots pine using airborne laser scanning. Remote Sens. Environ. 2006, 102, 364–376. [Google Scholar] [CrossRef]
- Coops, N.C.; Hilker, T.; Wulder, M.A.; St-Onge, B.; Newnham, G.; Siggins, A.; Trofymow, J.A.T. Estimating canopy structure of Douglas-fir Forest stands from discrete-return LiDAR. Trees 2007, 21, 295–310. [Google Scholar] [CrossRef] [Green Version]
- Lefsky, M.A.; Cohen, W.B.; Acker, S.A.; Parker, G.G.; Spies, T.A.; Harding, D. Lidar Remote Sensing of the Canopy Structure and Biophysical Properties of Douglas-Fir Western Hemlock Forests. Remote Sens. Environ. 1999, 70, 339–361. [Google Scholar] [CrossRef]
- Fraser, R.; van der Sluijs, J.; Hall, R. Calibrating Satellite-Based Indices of Burn Severity from UAV-Derived Metrics of a Burned Boreal Forest in NWT, Canada. Remote Sens. 2017, 9, 279. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Schaepman, M.E.; Koetz, B.; Kneubühler, M. Angular sensitivity analysis of vegetation indices derived from CHRIS/PROBA data. Remote Sens. Environ. 2008, 112, 2341–2353. [Google Scholar] [CrossRef]
- Meyer, G.E.; Neto, J.C. Verification of color vegetation indices for automated crop imaging applications. Comput. Electron. Agric. 2008, 63, 282–293. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Stark, R.; Rundquist, D. Novel algorithms for remote estimation of vegetation fraction. Remote Sens. Environ. 2002, 80, 76–87. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Wang, M.; Wang, S.; Wu, Y. Extraction of vegetation information from visible unmanned aerial vehicle images. Trans. Chin. Soc. Agric. Eng. 2015, 31, 152–157. [Google Scholar] [CrossRef]
- Neto, J.C. A Combined Statistical-Soft Computing Approach for Classification and Mapping Weed Species in Minimum-Tillage Systems; University of Nebraska: Lincoln, NE, USA, 2006. [Google Scholar]
- Kataoka, T.; Kaneko, T.; Okamoto, H.; Hata, S. Crop growth estimation system using machine vision. In Proceedings of the 2003 IEEE/ASME International Conference on Advanced Intelligent Mechatronics, Kobe, Japan, 20–24 July 2003; pp. b1079–b1083. [Google Scholar] [CrossRef]
- Najman, L.; Couprie, M.; Bertrand, G. Watersheds, mosaics, and the emergence paradigm. Discrete Appl. Math. 2005, 147, 301–324. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Baldocchi, D.; Gong, P.; Kelly, M. Isolating Individual Trees in a Savanna Woodland Using Small Footprint Lidar Data. Photogramm. Eng. Remote Sens. 2006, 72, 923–932. [Google Scholar] [CrossRef] [Green Version]
- Goutte, C.; Gaussier, E. A Probabilistic Interpretation of Precision, Recall and F-Score, with Implication for Evaluation. Int. J. Radiat. Biol. Relat. Stud. Phys. Chem. Med. 2005, 3408, 345–359. [Google Scholar] [CrossRef]
- Sokolova, M.; Japkowicz, N.; Szpakowicz, S. Beyond Accuracy, F-Score and ROC: A Family of Discriminant Measures for Performance Evaluation. In Australasian Joint Conference on Artificial Intelligence; Springer: Berlin/Heidelberg, Germany, 2006; pp. 1015–1021. [Google Scholar] [CrossRef] [Green Version]
- Wang, M. The Forest Genetics and Breeding; Forestry Publishing House: Beijing, China, 2001; pp. 203–214. [Google Scholar]
- Lai, M. Genotypic Evaluation and Early Selection of Larix Clones; Chinese Academy of Forestry: Beijing, China, 2014; p. 123. [Google Scholar]
- Wang, J.; Mo, Y.; Shen, L.; Fu, C.; Li, C.; Wei, G. Genetic Variation Analysis and Selection of 23 Eucalyptus Clonesin in Southern Guangxi. Southwest China J. Agric. Sci. 2019, 32, 2174–2179. [Google Scholar] [CrossRef]
- Xie, Y.; Mo, X.; Peng, S.; Deng, H.; Liu, L. Genetic variation analysis and early comprehensive selection of 21 Eucalyptus clones in western Guangdong Province, China. Nanjing For. Univ. Nat. Sci. Ed. 2018, 42, 73–80. [Google Scholar] [CrossRef]
- Xu, J. Application of Repeatability in Tree Breeding. J. Beijing For. Univ. 1988, 1988, 97–102. [Google Scholar] [CrossRef]
- Wu, Y.; Mao, C. Simple Introduction to Heritability, Repeatability and Genetic Gain in Percent in Tree Breeding. Trop. Agric. Sci. Technol. 2012, 35, 47–50. [Google Scholar] [CrossRef]
- Sotelo Montes, C.; Hernández, R.E.; Beaulieu, J.; Weber, J.C. Genetic variation in wood color and its correlations with tree growth and wood density of Calycophyllum spruceanum at an early age in the Peruvian Amazon. New For. 2007, 35, 57–73. [Google Scholar] [CrossRef]
- Li, G.Y.; Xu, J.M.; Lu, Z.H.; Du, Z.H.; Han, C.; Wu, S.J.; Wang, W. Growing and form-quality study on the clones of Eucalyptus in southern Fujian. J. Cent. South Univ. For. Technol. 2012, 32, 21–25. [Google Scholar] [CrossRef]
- Xu, J.; Lu, Z.; Li, G.; Bai, J. Study on Integrated Selection of Provenances-families of Eucalytus tereticornis. For. Res. 2003, 16, 1–7. [Google Scholar] [CrossRef]
- Chen, X.; Shen, X. Forest Tree Breeding; Higher Education Press: Beijing, China, 2005; p. 145. [Google Scholar]
- Vauhkonen, J.; Ene, L.; Gupta, S.; Heinzel, J.; Holmgren, J.; Pitkanen, J.; Solberg, S.; Wang, Y.; Weinacker, H.; Hauglin, K.M.; et al. Comparative testing of single-tree detection algorithms under different types of forest. Forestry 2012, 85, 27–40. [Google Scholar] [CrossRef] [Green Version]
- Yang, Q.; Su, Y.; Jin, S.; Kelly, M.; Hu, T.; Ma, Q.; Li, Y.; Song, S.; Zhang, J.; Xu, G.; et al. The Influence of Vegetation Characteristics on Individual Tree Segmentation Methods with Airborne LiDAR Data. Remote Sens. 2019, 11, 2880. [Google Scholar] [CrossRef] [Green Version]
- Guo, Y.; Liu, Q.; Liu, G.; Huang, C. Individual tree crown extraction of high resolution image based on marker-controlled watershed segmentation method. J. Geo. Inf. Sci. 2016, 18, 1259–1266. [Google Scholar] [CrossRef]
- Bai, S. Research on Single Tree Segmentation and DBH Parameter Extraction Algorithm Based on Point Cloud Data; Beijing University of Civil Engineering and Architecture: Beijing, China, 2020; p. 67. [Google Scholar] [CrossRef]
- Wu, X.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Assessment of Individual Tree Detection and Canopy Cover Estimation using Unmanned Aerial Vehicle based Light Detection and Ranging (UAV-LiDAR) Data in Planted Forests. Remote Sens. 2019, 11, 908. [Google Scholar] [CrossRef] [Green Version]
- Walter, J.D.C.; Edwards, J.; McDonald, G.; Kuchel, H. Estimating Biomass and Canopy Height with LiDAR for Field Crop Breeding. Front. Plant Sci. 2019, 10, 1145. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote Sensing Technologies for Enhancing Forest Inventories: A Review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Camarretta, N.; Harrison, P.A.; Lucieer, A.; Potts, B.M.; Davidson, N.; Hunt, M. From Drones to Phenotype: Using UAV-LiDAR to Detect Species and Provenance Variation in Tree Productivity and Structure. Remote Sens. 2020, 12, 3184. [Google Scholar] [CrossRef]
- Du Toit, F.; Coops, N.C.; Tompalski, P.; Goodbody, T.R.H.; El-Kassaby, Y.A.; Stoehr, M.; Turner, D.; Lucieer, A. Characterizing variations in growth characteristics between Douglas-fir with different genetic gain levels using airborne laser scanning. Trees 2020, 34, 649–664. [Google Scholar] [CrossRef]
- Liu, M.; Yin, S.; Si, D.; Shao, L.; Li, Y.; Zheng, M.; Wang, F.; Li, S.; Liu, G.; Zhao, X. Variation and genetic stability analyses of transgenic TaLEA poplar clones from four different sites in China. Euphytica 2015, 206, 331–342. [Google Scholar] [CrossRef]
- Jiang, L.; Pei, X.; Hu, Y.; Chiang, V.L.; Zhao, X. Effects of environment and genotype on growth traits in poplar clones in Northeast China. Euphytica 2021, 217, 1–14. [Google Scholar] [CrossRef]
- Meena, B.L.; Das, S.P.; Meena, S.K.; Kumari, R.; Devi, A.G.; Devi, H.L. Assessment of GCV, PCV, Heritability and Genetic Advance for Yield and its Components in Field Pea (Pisum sativum L.). Int. J. Curr. Microbiol. App. Sci. 2017, 6, 1025–1033. [Google Scholar] [CrossRef] [Green Version]
- Liziniewicz, M.; Ene, L.T.; Malm, J.; Lindberg, J.; Helmersson, A.; Karlsson, B. Estimation of Genetic Parameters and Selection of Superior Genotypes in a 12-Year-Old Clonal Norway Spruce Field Trial after Phenotypic Assessment Using a UAV. Forest 2020, 11, 992. [Google Scholar] [CrossRef]
- Cao, J.; Luo, J.; Lu, W. Cluster Analysis on Growth Trait of 6-year-old Eucalypt Clone Stands. Eucalypt Sci. Technol. 2012, 29, 37–40. [Google Scholar] [CrossRef]
- Chen, J.; Li, C.; Xiang, D.; Guo, D.; Ren, S.; Deng, Y.; Xu, J. Heterosis test for Eucalyptus hybrid families including Eucalyptus urophylla × Eucalyptus grandis. J. South. Agric. 2017, 48, 1858–1862. [Google Scholar] [CrossRef]
- Xu, Z.; Shen, X.; Cao, L.; Coops, N.C.; Goodbody, T.R.H.; Zhong, T.; Zhao, W.; Sun, Q.; Ba, S.; Zhang, Z.; et al. Tree species classification using UAS-based digital aerial photogrammetry point clouds and multispectral imageries in subtropical natural forests. Int. J. Appl. Earth Obs. 2020, 92, 102173. [Google Scholar] [CrossRef]
- Jin, S.; Su, Y.; Gao, S.; Wu, F.; Hu, T.; Liu, J.; Li, W.; Wang, D.; Chen, S.; Jiang, Y.; et al. Deep Learning: Individual Maize Segmentation from Terrestrial Lidar Data Using Faster R-CNN and Regional Growth Algorithms. Front. Plant Sci. 2018, 9, 866. [Google Scholar] [CrossRef]
- Pont, D.; Dungey, H.S.; Suontama, M.; Stovold, G.T. Spatial Models with Inter-Tree Competition from Airborne Laser Scanning Improve Estimates of Genetic Variance. Front. Plant Sci. 2021, 11, 596315. [Google Scholar] [CrossRef] [PubMed]
- Perich, G.; Hund, A.; Anderegg, J.; Roth, L.; Boer, M.P.; Walter, A.; Liebisch, F.; Aasen, H. Assessment of Multi-Image Unmanned Aerial Vehicle Based High-Throughput Field Phenotyping of Canopy Temperature. Front. Plant Sci. 2020, 11, 150. [Google Scholar] [CrossRef] [PubMed]




| Metrics | Description | Reference |
|---|---|---|
| Distributional metrics | ||
| h25, h50, h75, h95 | The percentiles of the canopy height distributions by first echo (25th, 50th, 75th and 95th). | [42] |
| Hmean | The mean height of all points after normalized. | [42] |
| Hcv | The coefficient of variation of height of all points after normalized (the ratio of the standard deviation to the mean)) | [42] |
| d1, d3, d5, d7, d9 | The proportion of points above the quantiles (10th, 30th, 50th,70th, and 90th) to total number of points | [43] |
| CC2m | The first return points above 2 m accounts for the percentage of all return points | [42] |
| eLAI | Half of the total leaf area per unit ground area | [44] |
| Weibull-fitting metrics | ||
| α/β | The α and β parameter of the Weibull distribution fitted to foliage density profile. | [45] |
| Canopy volume metrics | ||
| Open gap zone (OP)/ Closed gap zone(CL) | The empty voxels located above and below the canopy respectively. | [46] |
| Euphotic zone (EU)/ Oligophotic zone (OL) | The voxels located within an uppermost percentile (65%) of all filled grid cells of that column, and voxels located below the point in the profile | [46] |
| Vegetation Index | Equation | Reference |
|---|---|---|
| Brightness (BI) | G + R + B | [47] |
| NormG (NG) | G/(G + R + B) | [47] |
| RGRI | R/G | [48] |
| GRVI | (G − R)/(G + R) | [49] |
| VARI | (G − R) (G + R-B) | [50] |
| VDVI | (2G − (R + B))/(2G + (R + B)) | [51] |
| EXG | (2G − R − B)/(R + G + B) | [52] |
| EXG-R | ((3G − 2.4R − B)/(R + G + B) | [52] |
| CIVE | ((0.411R − 0.811G + 0.385B)/(R + G + B)) + 18.787 | [53] |
| Site | Stem Density (n/ha) | Crown Width (m) | Method | Nt | NO | NC | r | p | F |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 760 | max = 6.67 mean = 4.18 min = 2.06 | MWS | 278 | 98 | 42 | 0.74 | 0.87 | 0.80 |
| PCS | 327 | 49 | 118 | 0.87 | 0.73 | 0.80 | |||
| 2 | 982 | max = 4.54 mean = 3.38 min = 2.14 | MWS | 343 | 20 | 31 | 0.94 | 0.92 | 0.93 |
| PCS | 338 | 25 | 105 | 0.93 | 0.76 | 0.84 | |||
| 3 | 1239 | max = 4.06 mean = 3.05 min = 2.18 | MWS | 632 | 57 | 145 | 0.92 | 0.81 | 0.86 |
| PCS | 581 | 108 | 291 | 0.84 | 0.67 | 0.74 |
| Functions | Models | R2 | RMSE/cm | rRMSE/% | Cross-Validation | ||
|---|---|---|---|---|---|---|---|
| R2 | RMSE/cm | rRMSE/% | |||||
| Linear function | D = 0.7175 H + 1.5791 | 0.80 | 1.50 | 13.74 | 0.77 | 1.54 | 14.12 |
| Quadratic function | D = −0.0187 H2 + 1.2227 H − 1.5491 | 0.78 | 1.54 | 14.10 | 0.76 | 1.55 | 14.22 |
| Power function | D = 1.2152 H0.8547 | 0.80 | 1.50 | 13.74 | 0.76 | 1.53 | 14.03 |
| Exponential function | D = 4.3920 e0.0668H | 0.81 | 1.48 | 13.56 | 0.78 | 1.51 | 13.85 |
| Types of Variation | H1 (m) | H2 (m) | DBH1 (cm) | DBH2 (cm) | C1 (m) | C2 (m) |
|---|---|---|---|---|---|---|
| Low-densitysite | ||||||
| Mean ± std | 17.49 ± 1.9 | 17.08 ± 1.96 | 14.32 ± 1.73 | 13.93 ± 1.73 | 4.22 ± 0.52 | 4.18 ± 0.69 |
| Replication | 0.32 | 0.93 | 0.43 | 1.06 | 2.88 | 0.87 |
| Clone | 2.25 * | 2.63 * | 2.16 * | 2.41 * | 2.11 * | 2.22 * |
| Clone × replication | 3.71 | 10.12 | 3.21 | 8.82 | 61.76 | 0.38 |
| High-density site | ||||||
| Mean ± std | 12.29 ± 1.00 | 12.27 ± 1.05 | 10.02 ± 0.66 | 10.01 ± 0.69 | 3.05 ± 0.39 | 4.09 ± 0.46 |
| Replication | 1.76 | 1.07 | 1.84 | 1.23 | 1.32 | 4.30 ** |
| Clone | 8.59 ** | 9.68 ** | 8.42 ** | 9.22 ** | 2.84 * | 3.59 ** |
| Clone × replication | 3.02 | 2.04 | 3.55 | 1.98 | 3.42 | 2.90 |
| Genetic Parameters | H1 | H2 | DBH1 | DBH2 | C1 | C2 | h95 | α | BI |
|---|---|---|---|---|---|---|---|---|---|
| Low-density site | |||||||||
| PCV | 0.112 | 0.118 (5.1) | 0.125 | 0.128 (2.4) | 0.122 | 0.169 (37.8) | 0.149 | 0.240 | 0.057 |
| GCV | 0.062 | 0.071 (14.5) | 0.067 | 0.074 (9.2) | 0.065 | 0.089 (37.3) | 0.056 | 0.114 | 0.035 |
| RC | 0.568 | 0.629 (10.9) | 0.552 | 0.597 (8.2) | 0.540 | 0.537 (−0.5) | 0.328 | 0.465 | 0.635 |
| Ri | 0.305 | 0.361 (18.7) | 0.291 | 0.331 (13.6) | 0.281 | 0.279 (−0.8) | 0.140 | 0.225 | 0.367 |
| High-density site | |||||||||
| PCV | 0.082 | 0.088 (6.8) | 0.066 | 0.070 (5.9) | 0.101 | 0.150 (49.5) | 0.097 | 0.158 | 0.081 |
| GCV | 0.067 | 0.073 (9.3) | 0.054 | 0.058 (8.0) | 0.058 | 0.100 (72.8) | 0.064 | 0.027 | 0.044 |
| RC | 0.858 | 0.876 (2.0) | 0.855 | 0.869 (1.7) | 0.598 | 0.705 (17.8) | 0.692 | 0.083 | 0.560 |
| Ri | 0.669 | 0.702 (4.9) | 0.662 | 0.689 (4.1) | 0.331 | 0.443 (33.6) | 0.428 | 0.029 | 0.297 |
| Selection Rate | Number of Entries | Genetic Gain (%) | |||||
|---|---|---|---|---|---|---|---|
| H1 | H2 | DBH1 | DBH2 | C1 | C2 | ||
| Low-density site | |||||||
| 10% | 2 | 5.52 | 5.71 | 6.61 | 6.14 | 0.48 | 5.52 |
| 20% | 3 | 5.07 | 5.63 | 5.96 | 6.21 | 1.45 | 3.03 |
| 30% | 5 | 4.25 | 5.18 | 4.75 | 5.48 | 1.95 | 2.72 |
| High-density site | |||||||
| 10% | 1 | 7.08 | 7.05 | 5.70 | 4.74 | 2.57 | 3.50 |
| 20% | 2 | 6.82 | 6.64 | 5.61 | 4.39 | 0.44 | 1.53 |
| 30% | 3 | 6.27 | 6.26 | 5.15 | 4.15 | 1.94 | 2.04 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, L.; Cao, L.; Xie, Y.; Luo, J.; Wang, G. Phenotypic Traits Extraction and Genetic Characteristics Assessment of Eucalyptus Trials Based on UAV-Borne LiDAR and RGB Images. Remote Sens. 2022, 14, 765. https://doi.org/10.3390/rs14030765
Liao L, Cao L, Xie Y, Luo J, Wang G. Phenotypic Traits Extraction and Genetic Characteristics Assessment of Eucalyptus Trials Based on UAV-Borne LiDAR and RGB Images. Remote Sensing. 2022; 14(3):765. https://doi.org/10.3390/rs14030765
Chicago/Turabian StyleLiao, Lihua, Lin Cao, Yaojian Xie, Jianzhong Luo, and Guibin Wang. 2022. "Phenotypic Traits Extraction and Genetic Characteristics Assessment of Eucalyptus Trials Based on UAV-Borne LiDAR and RGB Images" Remote Sensing 14, no. 3: 765. https://doi.org/10.3390/rs14030765
APA StyleLiao, L., Cao, L., Xie, Y., Luo, J., & Wang, G. (2022). Phenotypic Traits Extraction and Genetic Characteristics Assessment of Eucalyptus Trials Based on UAV-Borne LiDAR and RGB Images. Remote Sensing, 14(3), 765. https://doi.org/10.3390/rs14030765







