Mapping Key Indicators of Forest Restoration in the Amazon Using a Low-Cost Drone and Artificial Intelligence
Abstract
:1. Introduction
2. Materials and Methods
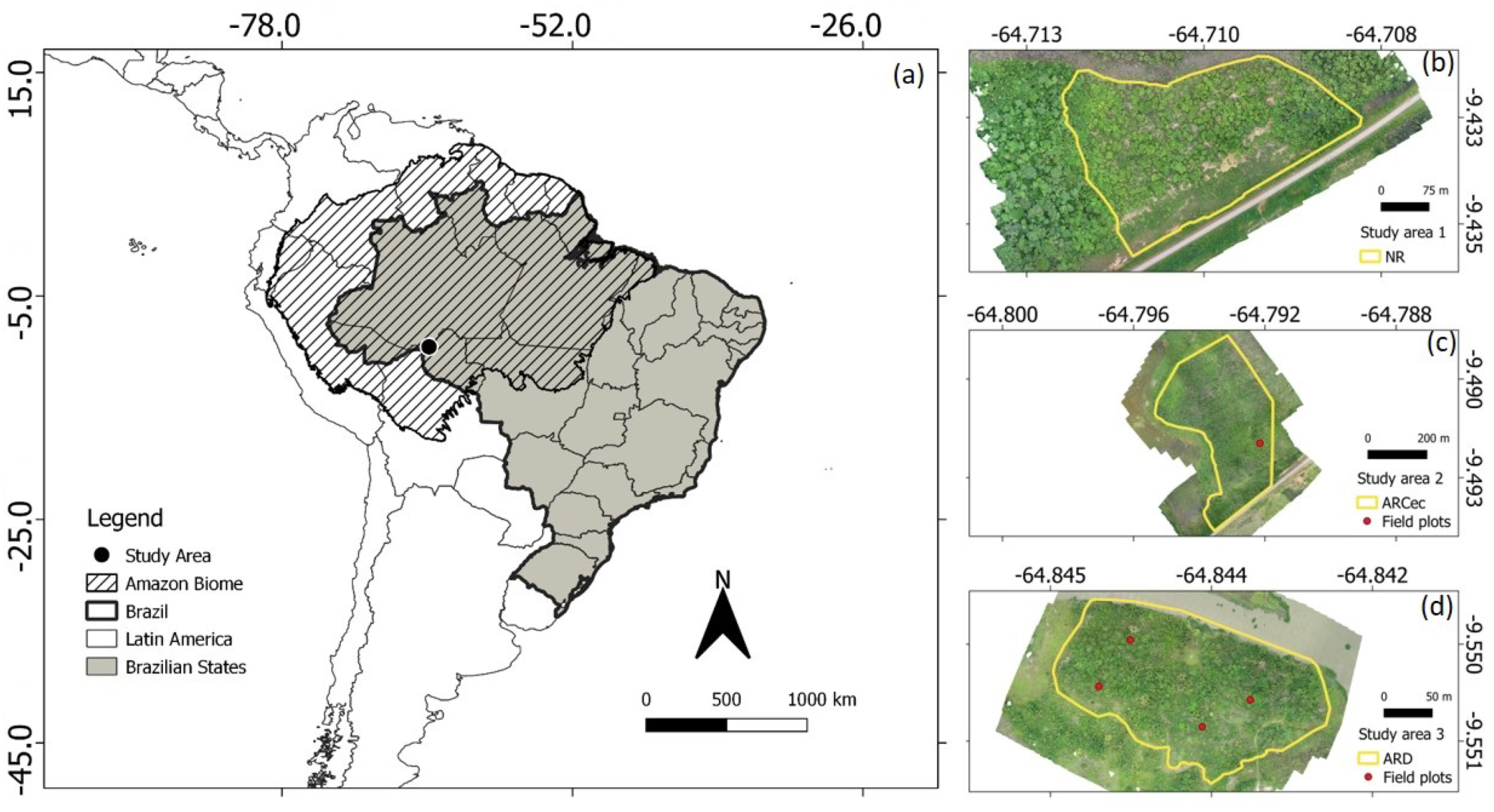
2.1. Study Area
2.2. Materials
2.3. Methods
2.3.1. Flight Patterns
2.3.2. Deep Learning Methods
2.3.3. Regression Analysis for Generating the TCHI after Mapping All Trees
2.3.4. Accuracy Evaluation of the Deep Learning Methods
3. Results
Results Accuracy
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A. Deep Learning Results Illustrated in Whole Study Areas




Appendix B. TCHI Data after Mapping All Trees via Deep Learning
| Species | Plot 1 | Plot 2 | Plot 3 | Plot 4 | Plot 5 |
|---|---|---|---|---|---|
| Adenanthera pavonina | |||||
| Anacardium occidentale | 1 | 2 | |||
| Apocinaceae | |||||
| Astrocaryum aculeatum | 2 | ||||
| Attalea speciosa | 2 | 1 | |||
| Bauhinia sp01 | 1 | ||||
| Belluccia grossularioides | 1 | 2 | 35 | ||
| Bixa orellana | 2 | ||||
| Byrsonima sp02 | 3 | 4 | |||
| Carapa guianensis | |||||
| Cecropia distachya | 5 | 1 | 1 | 4 | |
| Cecropia membranacea | 2 | ||||
| Cecropia purpurascens | 7 | 2 | 3 | 6 | |
| Cedrela fissilis | |||||
| Ceiba samauma | 1 | ||||
| Clitoria fairchildiana | |||||
| Cochlospermum orinocense | 8 | 1 | 4 | 1 | |
| Couratari macrosperma | 2 | ||||
| Croton matourensis | 34 | ||||
| Cupania rubiginosa | 3 | ||||
| Dipteryx odorata | 3 | ||||
| Enterolobium sp | 1 | ||||
| Eriotheca sp | |||||
| Eschweilera coriacea | |||||
| Genipa americana | |||||
| Handroanthus serratifolius | |||||
| Hevea guianensis | 4 | 1 | |||
| Himatanthus sucuuba | 1 | ||||
| Hymenaea courbaril | 2 | ||||
| Inga edulis | 3 | 1 | |||
| Inga heterophylla | 1 | ||||
| Inga sp04 | 3 | ||||
| Isertia hypoleuca | 17 | 9 | |||
| Lindackeria paludosa | 1 | ||||
| Mabea sp | 76 | ||||
| Mangifera indica | 1 | ||||
| Miconia pyrifolia | 11 | 3 | 5 | 3 | 8 |
| Myrcia sp02 | 12 | 102 | 82 | 52 | 10 |
| Myrtaceae 2 | 1 | ||||
| Myrtaceae 3 | 2 | 10 | 1 | 5 | 1 |
| Ocotea sp | 1 | ||||
| Pachira aquatica | |||||
| Pachira sp | |||||
| Parkia multijuga | 1 | ||||
| Physocalymma scaberrimum | 5 | 1 | |||
| Piper aduncum | 1 | 6 | |||
| Protium unifoliolatum | 29 | 19 | |||
| Psidium guajava | 1 | 1 | |||
| Pterodon emarginatus | |||||
| Schizolobium amazonicum | 2 | ||||
| Senna alata | |||||
| Senna multijuga | 1 | ||||
| Simarouba amara | 1 | ||||
| Simarouba versicolor | 3 | 1 | |||
| Solanum spp | |||||
| Stryphnodendron dunckeanum | 1 | ||||
| Swartzia lucida | 3 | 1 | |||
| Tachigali tinctoria | 1 | ||||
| Tapirira guianensis | 1 | 1 | |||
| Trema micrantha | |||||
| Vismia gracilis | 15 | 10 | 39 | ||
| Vismia guianensis | 25 | 1 | 37 | 2 | 16 |
| Vismia sandwithii | 30 | 5 | 9 | 6 | |
| Shannon Index | 2.334775 | 1.678342 | 1.511594 | 1.823349 | 2.394088 |
| Plot | Area | Perimeter | CHMmean | CHMstdev | CHMmin | CHMmax | FTPCA1mean | FTPCA1stde | FTPCA1min | FTPCA1max | FTPCA2mean | FTPCA2stde | FTPCA2min | FTPCA2max |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 65.119 | 39.311 | 5.119435 | 3.280841 | −0.00328 | 12.6938 | 0.404991 | 1.455045 | −0.69597 | 9.669959 | 0.261663 | 2.074722 | −1.71555 | 14.86461 |
| 3 | 28.129 | 26.406 | 3.366363 | 1.901713 | 0 | 7.040863 | −0.31739 | 0.311524 | −0.69597 | 1.197709 | −0.01846 | 0.695184 | −0.66874 | 4.122279 |
| 3 | 3.613 | 7.629 | 3.005856 | 1.947296 | 0 | 5.719925 | −0.19006 | 0.397121 | −0.66932 | 0.848243 | 0.352878 | 0.759364 | −0.41275 | 2.348622 |
| 3 | 9.032 | 17.014 | 7.610554 | 1.196479 | 2.299583 | 9.182724 | 0.427152 | 0.261602 | 0.089069 | 1.550751 | −0.57614 | 0.78659 | −1.09639 | 3.872258 |
| 3 | 22.28 | 23.464 | 2.771878 | 1.024129 | 0 | 7.542839 | −0.50516 | 0.157783 | −0.66554 | 0.454597 | −0.11437 | 0.148365 | −0.34738 | 0.691951 |
| 3 | 6.108 | 11.136 | 6.347235 | 2.709253 | 0.004837 | 9.0868 | 0.886286 | 1.188347 | −0.58509 | 4.260673 | 0.594698 | 2.166958 | −1.06947 | 6.937483 |
| 3 | 22.452 | 28.753 | 5.069082 | 1.075359 | 0 | 10.96689 | −0.12052 | 0.379077 | −0.56762 | 2.985223 | −0.20846 | 0.495652 | −0.80686 | 2.843362 |
| 3 | 48.431 | 36.363 | 3.725492 | 1.71961 | −0.11349 | 6.606369 | −0.31098 | 0.339763 | −0.6836 | 1.804834 | −0.11054 | 0.487495 | −0.50532 | 3.720053 |
| 2 | 6.538 | 11.142 | 7.378312 | 0.835771 | 3.856255 | 8.309235 | 0.335718 | 0.188526 | −0.01269 | 0.813891 | −0.5927 | 0.429563 | −0.87881 | 1.176714 |
| 2 | 74.84 | 47.461 | 7.720049 | 1.813196 | 0.963417 | 11.14235 | 0.525326 | 0.498828 | −0.65947 | 3.16911 | −0.57892 | 0.685009 | −1.44404 | 3.553976 |
| 2 | 4.731 | 9.379 | 7.965543 | 0.782673 | 2.244179 | 9.572548 | 0.419333 | 0.099508 | 0.233903 | 0.596738 | −0.75094 | 0.394346 | −1.00977 | 0.828341 |
| 2 | 57.807 | 35.177 | 6.714935 | 2.757263 | −0.02056 | 11.04849 | 0.481698 | 0.846595 | −0.69597 | 3.786057 | −0.22817 | 1.255041 | −1.55378 | 6.290686 |
| 2 | 32.689 | 34.029 | 4.735298 | 0.778733 | 2.378983 | 6.820885 | −0.28059 | 0.136493 | −0.49576 | 0.151903 | −0.30785 | 0.111566 | −0.54278 | 0.103714 |
| 2 | 5.936 | 12.899 | 4.955179 | 0.492389 | 4.045937 | 6.063095 | −0.27546 | 0.074306 | −0.38744 | −0.11507 | −0.35117 | 0.065346 | −0.50046 | −0.23339 |
| 2 | 2.409 | 7.045 | 7.636636 | 2.943989 | 3.691277 | 12.20493 | 2.063702 | 1.718944 | −0.28772 | 4.599868 | 1.327158 | 1.91322 | −0.47216 | 6.38865 |
| 2 | 64.603 | 42.868 | 8.511374 | 0.764597 | 4.643204 | 12.09242 | 0.574774 | 0.257937 | −0.22508 | 2.294271 | −0.89379 | 0.261569 | −1.32826 | 1.713706 |
| 1 | 63.915 | 45.761 | 4.337894 | 1.651773 | 0.793602 | 12.22772 | −0.30323 | 0.256276 | −0.65293 | 0.56833 | −0.27417 | 0.221224 | −0.98045 | 0.557243 |
| 1 | 5.333 | 9.969 | 0.903739 | 1.251807 | 0 | 3.760948 | −0.47756 | 0.308349 | −0.69597 | 0.392144 | 0.236693 | 0.485465 | −0.15663 | 1.493688 |
| 1 | 4.215 | 9.975 | 12.02187 | 4.007458 | 0 | 15.93677 | 7.834983 | 9.142468 | 1.774665 | 29.69638 | 8.372838 | 15.52596 | −2.68251 | 39.13299 |
| 1 | 35.097 | 31.068 | 8.524904 | 4.193895 | −0.10499 | 15.36579 | 2.141066 | 2.536678 | −0.68205 | 15.8741 | 1.016266 | 4.251897 | −2.58661 | 22.14248 |
| 1 | 4.731 | 10.565 | 11.25474 | 3.037268 | 2.662628 | 13.81744 | 3.407625 | 2.624248 | 1.12727 | 11.38162 | 2.008901 | 6.083042 | −2.28632 | 14.96166 |
| 1 | 3.871 | 8.802 | 7.006234 | 3.731296 | 3.468102 | 12.91788 | 2.527597 | 2.859887 | −0.41815 | 7.528047 | 2.490923 | 4.130253 | −1.795 | 10.32378 |
| 4 | 44.904 | 34.016 | 6.244992 | 1.816363 | 0.996513 | 9.068893 | 0.173356 | 0.41305 | −0.60792 | 1.815155 | −0.29251 | 0.773015 | −1.06127 | 3.919278 |
| 4 | 2.581 | 9.386 | 6.772082 | 3.101002 | 3.562378 | 11.19086 | 2.247702 | 3.012706 | −0.36029 | 7.882115 | 1.863147 | 3.653275 | −1.49495 | 8.587377 |
| 4 | 65.979 | 48.66 | 4.774225 | 2.003087 | 0.424896 | 10.94702 | −0.03879 | 0.750378 | −0.68859 | 6.519537 | −0.04699 | 1.242492 | −0.7953 | 12.68339 |
| 4 | 3.785 | 8.802 | 11.18225 | 2.132175 | 1.550011 | 13.90279 | 2.100087 | 1.444676 | 0.983257 | 8.191 | −0.33297 | 3.448375 | −1.98948 | 14.56447 |
| 4 | 4.043 | 8.796 | 8.847744 | 3.095808 | 0.431656 | 12.70915 | 2.42247 | 2.344196 | −0.60006 | 7.870145 | 1.434274 | 4.169788 | −1.47805 | 15.05686 |
| 4 | 3.871 | 9.379 | 11.56552 | 2.705374 | 1.205254 | 13.88068 | 3.408568 | 2.312897 | 1.695313 | 9.796259 | 0.638469 | 4.27188 | −2.25043 | 12.12099 |
| 4 | 89.807 | 45.736 | 5.710779 | 3.102415 | 1.64621 | 14.78353 | 0.726542 | 1.88475 | −0.61801 | 8.226952 | 0.533843 | 2.356327 | −1.83763 | 12.82553 |
| 4 | 86.797 | 52.26 | 3.097198 | 1.282993 | 0 | 10.37563 | −0.43387 | 0.436039 | −0.67807 | 3.297346 | −0.0949 | 0.42824 | −1.05175 | 4.798019 |
| 1 | 4.989 | 10.565 | 2.594007 | 0.506873 | 1.295456 | 3.367157 | −0.55758 | 0.033358 | −0.63448 | −0.50407 | −0.14113 | 0.077593 | −0.20492 | 0.173152 |
| 1 | 5.763 | 11.142 | 3.537497 | 0.499803 | 1.566437 | 4.706284 | −0.45407 | 0.051183 | −0.53054 | −0.33943 | −0.18381 | 0.122273 | −0.29835 | 0.203358 |
| 1 | 3.441 | 8.796 | 4.337077 | 0.992384 | 1.794785 | 5.625793 | −0.29708 | 0.053235 | −0.42464 | −0.19068 | −0.24132 | 0.214781 | −0.41681 | 0.244165 |
| 3 | 40.344 | 35.19 | 6.642838 | 0.605936 | 3.950012 | 9.699669 | 0.070372 | 0.147227 | −0.19429 | 0.855601 | −0.58871 | 0.116883 | −1.07072 | −0.07084 |
| 3 | 6.194 | 10.565 | 3.579031 | 1.17162 | 0 | 5.186081 | −0.21053 | 0.448625 | −0.54826 | 1.195263 | 0.110991 | 0.765588 | −0.33293 | 2.763646 |
| 3 | 6.538 | 11.738 | 2.132504 | 0.660017 | 0 | 3.347786 | −0.55308 | 0.125729 | −0.67807 | −0.0338 | −0.02748 | 0.243737 | −0.18173 | 1.039601 |
| 3 | 3.613 | 8.219 | 6.835736 | 0.651863 | 4.250809 | 7.99382 | 0.10962 | 0.117666 | −0.17384 | 0.314418 | −0.51713 | 0.249941 | −0.78584 | 0.28765 |
| 3 | 5.247 | 9.975 | 7.189706 | 0.577249 | 5.691086 | 8.36628 | 0.197599 | 0.120036 | −0.09026 | 0.371255 | −0.66221 | 0.11313 | −0.83086 | −0.40648 |
| 2 | 2.065 | 5.866 | 6.328863 | 0.774717 | 4.998161 | 7.379341 | 0.085219 | 0.084087 | −0.06179 | 0.1692 | −0.44667 | 0.197249 | −0.6946 | −0.1985 |
| 2 | 2.581 | 6.456 | 2.516833 | 0.273776 | 1.735298 | 3.363579 | −0.57799 | 0.019402 | −0.60178 | −0.53215 | −0.13763 | 0.022468 | −0.16155 | −0.08298 |
| 2 | 6.022 | 9.975 | 7.104853 | 1.958472 | 2.622948 | 10.26601 | 0.593139 | 0.496412 | −0.40358 | 1.998008 | −0.32734 | 0.865179 | −1.33871 | 2.749068 |
| 2 | 3.957 | 9.392 | 4.496574 | 1.151446 | 1.601051 | 5.517975 | −0.19725 | 0.261226 | −0.60149 | 0.515177 | −0.14092 | 0.447423 | −0.44077 | 1.183987 |
| 2 | 3.441 | 7.623 | 6.913352 | 1.097175 | 4.765274 | 9.905563 | 0.339027 | 0.466996 | −0.07205 | 1.498015 | −0.32178 | 0.492166 | −0.83903 | 1.172888 |
| 1 | 4.215 | 8.796 | 3.107001 | 0.970136 | 0.442558 | 4.464371 | −0.48844 | 0.082112 | −0.64182 | −0.37683 | −0.15895 | 0.105838 | −0.31404 | 0.047556 |
| 4 | 8.774 | 12.899 | 4.364909 | 1.038066 | 0.549828 | 5.440331 | −0.32587 | 0.148639 | −0.66997 | 0.176937 | −0.18504 | 0.430464 | −0.41643 | 2.120316 |
| 4 | 6.624 | 10.559 | 2.655107 | 0.435818 | 1.801559 | 3.993599 | −0.5565 | 0.057155 | −0.6307 | −0.35925 | −0.15082 | 0.029417 | −0.21058 | −0.06705 |
| 4 | 7.656 | 12.911 | 3.628137 | 1.344258 | 1.239876 | 5.857239 | −0.36409 | 0.165487 | −0.64384 | 0.017518 | −0.11675 | 0.231081 | −0.43421 | 0.321494 |
| 4 | 4.817 | 9.386 | 4.308281 | 1.429484 | 1.84951 | 6.648903 | −0.25823 | 0.187091 | −0.61735 | 0.212086 | −0.07847 | 0.275433 | −0.60457 | 0.520399 |
| 4 | 8.172 | 12.309 | 2.745759 | 1.21562 | 0 | 4.274643 | −0.42806 | 0.174489 | −0.66996 | 0.014023 | −0.02432 | 0.267492 | −0.29852 | 0.903595 |
| 4 | 4.817 | 9.386 | 4.824788 | 1.137 | 2.2743 | 6.535606 | −0.19017 | 0.1711 | −0.55245 | 0.132062 | −0.28883 | 0.21769 | −0.56395 | 0.076414 |
| 4 | 1.29 | 4.693 | 1.99779 | 0.114795 | 1.788002 | 2.433456 | −0.63025 | 0.004002 | −0.6365 | −0.62539 | −0.1262 | 0.002236 | −0.12905 | −0.12288 |
| 4 | 3.871 | 8.79 | 2.232366 | 1.483713 | 0 | 11.91653 | −0.10782 | 1.319545 | −0.63316 | 3.957197 | 0.528906 | 1.699226 | −0.16908 | 6.286868 |
| 4 | 4.129 | 8.808 | 7.194678 | 0.863822 | 5.4515 | 9.093864 | 0.267943 | 0.21689 | −0.05229 | 0.690447 | −0.56302 | 0.229609 | −0.86612 | −0.05738 |
| 4 | 1.979 | 5.872 | 9.90452 | 0.969666 | 7.360344 | 11.57283 | 1.286932 | 0.46469 | 0.872513 | 2.458546 | −0.83582 | 0.520577 | −1.24902 | 0.309813 |
| 5 | 4.051 | 9.596 | 6.993476 | 0.554035 | 2.319511 | 8.072655 | 0.721011 | 0.165133 | 0.362456 | 0.998681 | −0.83646 | 0.204428 | −1.04165 | −0.29032 |
| 5 | 6.302 | 10.802 | 7.177172 | 0.962539 | 3.934128 | 8.710861 | 0.92671 | 0.206999 | 0.336062 | 1.464034 | −0.73948 | 0.448283 | −1.16833 | 0.420686 |
| 5 | 7.022 | 11.399 | 7.740243 | 0.938236 | 4.896156 | 9.538589 | 1.059383 | 0.32214 | 0.621426 | 1.621939 | −0.98677 | 0.351309 | −1.55799 | −0.09234 |
| 5 | 28.54 | 26.983 | 6.871229 | 1.393398 | 0.894493 | 8.841682 | 0.927168 | 0.726795 | −0.37189 | 5.661978 | −0.52068 | 1.287771 | −1.29739 | 7.181975 |
| 5 | 7.743 | 11.996 | 6.367768 | 0.818534 | 3.083389 | 8.828812 | 0.574648 | 0.244304 | 0.04083 | 1.405117 | −0.65111 | 0.348197 | −0.95097 | 0.358819 |
| 5 | 4.411 | 8.999 | 8.144852 | 1.777536 | 2.885094 | 9.614311 | 1.716011 | 0.506895 | 1.163184 | 3.193094 | −0.25323 | 1.67511 | −1.5567 | 3.901798 |
| 5 | 5.942 | 10.199 | 5.172714 | 0.436717 | 3.687805 | 7.992622 | 0.181246 | 0.05757 | 0.048333 | 0.271038 | −0.57408 | 0.091129 | −0.67794 | −0.33007 |
| 5 | 2.881 | 7.201 | 6.094399 | 1.260213 | 3.810753 | 7.862221 | 0.667777 | 0.321439 | −0.13697 | 1.329934 | −0.4243 | 0.59697 | −0.92484 | 0.985291 |
| 5 | 14.765 | 17.412 | 7.514438 | 1.895635 | 3.224686 | 9.724991 | 1.435498 | 0.777824 | −0.03845 | 3.700222 | −0.23322 | 1.390438 | −1.62491 | 3.440152 |
| 5 | 7.563 | 11.99 | 6.417062 | 1.207018 | 4.493248 | 8.258713 | 0.656216 | 0.449859 | −0.00052 | 1.596533 | −0.5972 | 0.422816 | −1.18557 | 0.477079 |
| 5 | 5.492 | 10.205 | 5.385297 | 0.62274 | 4.03315 | 9.073189 | 0.289163 | 0.273601 | −0.01178 | 1.354232 | −0.48864 | 0.35346 | −0.76967 | 0.997614 |
| 5 | 6.662 | 11.399 | 7.782701 | 1.402899 | 3.656174 | 9.601425 | 1.289672 | 0.363723 | 0.078946 | 1.971506 | −0.78493 | 0.833656 | −1.45933 | 1.355572 |
| 5 | 6.212 | 10.802 | 7.36717 | 0.461894 | 4.689758 | 8.755592 | 0.857622 | 0.127292 | 0.704136 | 1.294942 | −0.93603 | 0.145056 | −1.12679 | −0.62726 |
| 5 | 4.772 | 12.002 | 5.167382 | 1.19266 | 3.573372 | 8.392746 | 0.293052 | 0.452028 | −0.08823 | 1.48954 | −0.38059 | 0.31974 | −0.987 | 0.323958 |
| 5 | 5.042 | 10.205 | 7.59755 | 1.479013 | 3.070595 | 9.733353 | 1.38104 | 0.595625 | 0.715453 | 2.795767 | −0.25195 | 1.62803 | −1.58962 | 5.121841 |
| 5 | 2.971 | 7.208 | 4.842277 | 1.418374 | 2.329239 | 8.578911 | 0.307523 | 0.448966 | −0.20342 | 1.22301 | 0.295702 | 0.925839 | −0.61559 | 2.450948 |
| 5 | 5.312 | 9.602 | 8.224847 | 1.599713 | 4.244247 | 9.736839 | 1.533879 | 0.679356 | −0.06244 | 3.439811 | −0.53613 | 1.324322 | −1.57109 | 3.209459 |
| 5 | 6.032 | 10.796 | 4.526514 | 0.863805 | 1.949219 | 6.453194 | 0.086033 | 0.163719 | −0.16705 | 0.433793 | −0.38167 | 0.251908 | −0.64997 | 0.241877 |
| 5 | 5.042 | 10.802 | 8.476414 | 1.178713 | 4.289162 | 11.74305 | 1.45153 | 0.532362 | 0.891494 | 2.879493 | −1.00337 | 0.660389 | −2.13556 | 1.238157 |
| 5 | 3.331 | 9.017 | 10.18775 | 1.185139 | 4.634438 | 12.37226 | 2.297686 | 0.384895 | 1.858209 | 3.104045 | −1.44549 | 0.662927 | −2.04331 | −0.14826 |
| 5 | 36.822 | 28.787 | 6.697485 | 2.38479 | 1.730072 | 11.74391 | 0.999325 | 1.04185 | −0.25401 | 5.554715 | −0.54858 | 1.10566 | −2.04423 | 5.28658 |
| 5 | 7.743 | 12.002 | 5.744165 | 0.359735 | 3.956627 | 6.189232 | 0.307561 | 0.063987 | 0.157081 | 0.448816 | −0.65428 | 0.112285 | −0.77074 | −0.25877 |
| 5 | 7.473 | 12.002 | 7.598305 | 1.552404 | 4.781235 | 10.34891 | 1.347795 | 0.689081 | 0.19237 | 2.768367 | −0.44058 | 0.805717 | −1.51418 | 1.784046 |
| 5 | 10.083 | 13.209 | 9.906447 | 1.003685 | 5.13726 | 11.6647 | 2.029642 | 0.357005 | 1.326314 | 3.129253 | −1.30954 | 0.9755 | −2.12405 | 2.578822 |
| 5 | 6.842 | 10.802 | 6.438966 | 0.985115 | 5.438622 | 9.635727 | 0.632272 | 0.516692 | 0.266723 | 2.316442 | −0.56945 | 0.559291 | −1.51076 | 1.258503 |
| Plot | Area | Perimeter | CHMmean | CHMstdev | CHMmin | CHMmax | FTPCA1mean | FTPCA1stde | FTPCA1min | FTPCA1max | FTPCA2mean | FTPCA2stde | FTPCA2min | FTPCA2max | Shannon |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 13.557 | 15.5439 | 5.762497 | 2.084269 | 1.191858 | 9.219016 | 1.333331 | 1.794779 | −0.17786 | 6.402963 | 1.312624 | 3.121833 | −1.17216 | 8.928008 | 2.334775 |
| 2 | 20.58608 | 18.40862 | 6.382908 | 1.2634 | 2.886571 | 8.745109 | 0.314357 | 0.396097 | −0.32853 | 1.457309 | −0.28852 | 0.549242 | −0.8619 | 1.895913 | 1.678342 |
| 3 | 20.54615 | 20.44331 | 4.876593 | 1.42472 | 1.236889 | 7.95645 | −0.00859 | 0.419196 | −0.47375 | 1.959594 | −0.11564 | 0.700278 | −0.75573 | 3.308784 | 1.511594 |
| 4 | 19.66089 | 17.36933 | 5.669507 | 1.626192 | 1.785102 | 9.145865 | 0.516664 | 0.861543 | −0.28378 | 3.348483 | 0.103445 | 1.347034 | −0.93891 | 5.269306 | 1.823349 |
| 5 | 8.36204 | 12.21668 | 6.977465 | 1.157382 | 3.629697 | 9.25874 | 0.958779 | 0.418766 | 0.29713 | 2.217852 | −0.61008 | 0.699209 | −1.31593 | 1.634646 | 2.394088 |
References
- SER. Princípios da Society for Ecological Restoration (SER) International Sobre a Restauração Ecológica. Technical Report, Embrapa Florestas. 2021. Available online: https://cdn.ymaws.com/www.ser.org/resource/resmgr/custompages/publications/SER_Primer/ser-primer-portuguese.pdf (accessed on 11 October 2021).
- Muradian, R.; Corbera, E.; Pascual, U.; Kosoy, N.; May, P.H. Reconciling theory and practice: An alternative conceptual framework for understanding payments for environmental services. Ecol. Econ. 2010, 69, 1202–1208. [Google Scholar] [CrossRef]
- Adams, C.; Rodrigues, S.T.; Calmon, M.; Kumar, C. Impacts of large-scale forest restoration on socioeconomic status and local livelihoods: What we know and do not know. Biotropica 2016, 48, 731–744. [Google Scholar] [CrossRef]
- Brancalion, P.H.S.; Viani, R.A.G.; Rodrigues, R.R.; Gandolfi, S. Avaliação e monitoramento de áreas em processo de restauração. In Restauração Ecológica de Ecossistemas Degradados; Martins, S., Ed.; Editora UFV: Viçosa, Brazil, 2012; pp. 262–293. Available online: http://www.esalqlastrop.com.br/img/aulas/Cumbuca%206(2).pdf (accessed on 31 October 2021).
- PRMA. Protocolo de Monitoramento para Programas e Projetos de Restauração Florestal. Monitoring Protocol for Forest Restoration Programs & Projects. Technical Report, PACTO PELA RESTAURAÇÃO DA MATA ATLÂNTICA. 2013. Available online: http://media.wix.com/ugd/5da841_c228aedb71ae4221bc95b909e0635257.pdf (accessed on 10 July 2021).
- Chaves, R.B.; Durigan, G.; Brancalion, P.H.; Aronson, J. On the need of legal frameworks for assessing restoration projects success: New perspectives from São Paulo state (Brazil). Restor. Ecol. 2015, 23, 754–759. [Google Scholar] [CrossRef]
- McDonald, T.; Gann, G.; Jonson, J.; Dixon, K. International Standards for the Practice of Ecological Restoration—Including Principles and Ley Concepts; Technical Report; Society for Ecological Restoration: Washington, DC, USA, 2016; Available online: http://www.seraustralasia.com/wheel/image/SER_International_Standards.pdf (accessed on 9 August 2021).
- Lovejoy, T.E.; Nobre, C. Amazon Tipping Point. Sci. Adv. 2018, 4, eaat2340. [Google Scholar] [CrossRef] [Green Version]
- Silva-Junior, C.H.L.; Pessôa, A.C.M.; Carvalho, N.S.; Reis, J.B.C.; Anderson, L.O.; Aragão, L.E.O.C. The Brazilian Amazon deforestation rate in 2020 is the greatest of the decade. Nat. Ecol. Evol. 2021, 5, 144–145. [Google Scholar] [CrossRef]
- Rödig, E.; Cuntz, M.; Rammig, A.; Fischer, R.; Taubert, F.; Huth, A. The importance of forest structure for carbon fluxes of the Amazon rainforest. Environ. Res. Lett. 2018, 13, 054013. [Google Scholar] [CrossRef]
- Jakovac, C.C.; Bongers, F.; Kuyper, T.W.; Mesquita, R.C.; Peña-Claros, M. Land use as a filter for species composition in Amazonian secondary forests. J. Veg. Sci. 2016, 27, 1104–1116. [Google Scholar] [CrossRef] [Green Version]
- Poorter, L.; Bongers, F.; Aide, T.M.; Zambrano, A.M.A.; Balvanera, P.; Becknell, J.M.; Boukili, V.; Brancalion, P.H.; Broadbent, E.N.; Chazdon, R.L.; et al. Biomass resilience of Neotropical secondary forests. Nature 2016, 530, 211–214. [Google Scholar] [CrossRef]
- Freitas, M.G.; Rodrigues, S.B.; Campos-Filho, E.M.; do Carmo, G.H.P.; da Veiga, J.M.; Junqueira, R.G.P.; Vieira, D.L.M. Evaluating the success of direct seeding for tropical forest restoration over ten years. For. Ecol. Manag. 2019, 438, 224–232. [Google Scholar] [CrossRef]
- Vieira, D.L.M.; Rodrigues, S.B.; Jakovac, C.C.; da Rocha, G.P.E.; Reis, F.; Borges, A. Active Restoration Initiates High Quality Forest Succession in a Deforested Landscape in Amazonia. Forests 2021, 12, 1022. [Google Scholar] [CrossRef]
- Mesquita, R.C.; Ickes, K.; Ganade, G.; Williamson, G.B. Alternative successional pathways in the Amazon Basin. J. Ecol. 2001, 89, 528–537. [Google Scholar] [CrossRef] [Green Version]
- Bourgoin, C.; Betbeder, J.; Couteron, P.; Blanc, L.; Dessard, H.; Oszwald, J.; Le Roux, R.; Cornu, G.; Reymondin, L.; Mazzei, L.; et al. UAV-based canopy textures assess changes in forest structure from long-term degradation. Ecol. Indic. 2020, 115, 106386. [Google Scholar] [CrossRef] [Green Version]
- Camarretta, N.; Harrison, P.A.; Bailey, T.; Potts, B.; Lucieer, A.; Davidson, N.; Hunt, M. Monitoring forest structure to guide adaptive management of forest restoration: A review of remote sensing approaches. New For. 2020, 51, 573–596. [Google Scholar] [CrossRef]
- Zahawi, R.A.; Dandois, J.P.; Holl, K.D.; Nadwodny, D.; Reid, J.L.; Ellis, E.C. Using lightweight unmanned aerial vehicles to monitor tropical forest recovery. Biol. Conserv. 2015, 186, 287–295. [Google Scholar] [CrossRef] [Green Version]
- Chen, S.; McDermid, G.; Castilla, G.; Linke, J. Measuring vegetation height in linear disturbances in the boreal forest with UAV photogrammetry. Remote Sens. 2017, 9, 1257. [Google Scholar] [CrossRef] [Green Version]
- Wu, X.; Shen, X.; Cao, L.; Wang, G.; Cao, F. Assessment of individual tree detection and canopy cover estimation using unmanned aerial vehicle based light detection and ranging (UAV-LiDAR) data in planted forests. Remote Sens. 2019, 11, 908. [Google Scholar] [CrossRef] [Green Version]
- Belmonte, A.; Sankey, T.; Biederman, J.A.; Bradford, J.; Goetz, S.J.; Kolb, T.; Woolley, T. UAV-derived estimates of forest structure to inform ponderosa pine forest restoration. Remote Sens. Ecol. Conserv. 2020, 6, 181–197. [Google Scholar] [CrossRef]
- Albuquerque, R.W.; Ferreira, M.E.; Olsen, S.I.; Tymus, J.R.C.; Balieiro, C.P.; Mansur, H.; Moura, C.J.R.; Costa, J.V.S.; Branco, M.R.C.; Grohmann, C.H. Forest Restoration Monitoring Protocol with a Low-Cost Remotely Piloted Aircraft: Lessons Learned from a Case Study in the Brazilian Atlantic Forest. Remote Sens. 2021, 13, 2401. [Google Scholar] [CrossRef]
- Almeida, D.R.A.D.; Stark, S.C.; Chazdon, R.; Nelson, B.W.; César, R.G.; Meli, P.; Gorgens, E.; Duarte, M.M.; Valbuena, R.; Moreno, V.S.; et al. The effectiveness of lidar remote sensing for monitoring forest cover attributes and landscape restoration. For. Ecol. Manag. 2019, 438, 34–43. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Zhao, B.; Feng, J.; Wu, X.; Yan, S. A survey on deep learning-based fine-grained object classification and semantic segmentation. Int. J. Autom. Comput. 2017, 14, 119–135. [Google Scholar] [CrossRef]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef] [Green Version]
- Brodrick, P.G.; Davies, A.B.; Asner, G.P. Uncovering ecological patterns with convolutional neural networks. Trends Ecol. Evol. 2019, 34, 734–745. [Google Scholar] [CrossRef]
- Ferreira, M.P.; de Almeida, D.R.A.; de Almeida Papa, D.; Minervino, J.B.S.; Veras, H.F.P.; Formighieri, A.; Santos, C.A.N.; Ferreira, M.A.D.; Figueiredo, E.O.; Ferreira, E.J.L. Individual tree detection and species classification of Amazonian palms using UAV images and deep learning. For. Ecol. Manag. 2020, 475, 118397. [Google Scholar] [CrossRef]
- Moura, M.M.; de Oliveira, L.E.S.; Sanquetta, C.R.; Bastos, A.; Mohan, M.; Corte, A.P.D. Towards Amazon Forest Restoration: Automatic Detection of Species from UAV Imagery. Remote Sens. 2021, 13, 2627. [Google Scholar] [CrossRef]
- Schiefer, F.; Kattenborn, T.; Frick, A.; Frey, J.; Schall, P.; Koch, B.; Schmidtlein, S. Mapping forest tree species in high resolution UAV-based RGB-imagery by means of convolutional neural networks. ISPRS J. Photogramm. Remote Sens. 2020, 170, 205–215. [Google Scholar] [CrossRef]
- Haffer, J. Speciation in Amazonian forest birds. Science 1969, 165, 131–137. [Google Scholar] [CrossRef]
- Prance, G.T. A comparison of the efficacy of higher taxa and species numbers in the assessment of biodiversity in the neotropics. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 1994, 345, 89–99. [Google Scholar]
- Antonelli, A.; Zizka, A.; Carvalho, F.A.; Scharn, R.; Bacon, C.D.; Silvestro, D.; Condamine, F.L. Amazonia is the primary source of Neotropical biodiversity. Proc. Natl. Acad. Sci. USA 2018, 115, 6034–6039. [Google Scholar] [CrossRef] [Green Version]
- Ter Steege, H.; de Oliveira, S.M.; Pitman, N.C.; Sabatier, D.; Antonelli, A.; Andino, J.E.G.; Aymard, G.A.; Salomão, R.P. Towards a dynamic list of Amazonian tree species. Sci. Rep. 2019, 9, 3501. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Santaquiteria, J.; Bueno, G.; Deniz, O.; Vallez, N.; Cristobal, G. Semantic versus instance segmentation in microscopic algae detection. Eng. Appl. Artif. Intell. 2020, 87, 103271. [Google Scholar] [CrossRef]
- Hafiz, A.M.; Bhat, G.M. A survey on instance segmentation: State of the art. Int. J. Multimed. Inf. Retr. 2020, 9, 171–189. [Google Scholar] [CrossRef]
- Braga, J.R.G.; Peripato, V.; Dalagnol, R.; Ferreira, M.P.; Tarabalka, Y.; Aragão, L.E.O.C.; Campos Velho, H.F.; Shiguemori, E.H.; Wagner, F.H. Tree Crown Delineation Algorithm Based on a Convolutional Neural Network. Remote Sens. 2020, 12, 1288. [Google Scholar] [CrossRef] [Green Version]
- He, K.; Gkioxari, G.; Dollár, P.; Girshick, R. Mask r-cnn. In Proceedings of the IEEE International Conference on Computer Vision, Venice, Italy, 22–29 October 2017; pp. 2961–2969. [Google Scholar]
- Spellerberg, I.F.; Fedor, P.J. A tribute to Claude Shannon (1916–2001) and a plea for more rigorous use of species richness, species diversity and the ‘Shannon–Wiener’Index. Glob. Ecol. Biogeogr. 2003, 12, 177–179. [Google Scholar] [CrossRef] [Green Version]
- DJI. Phantom 4PRO. Available online: https://www.dji.com/br/phantom-4-pro (accessed on 12 January 2022).
- SPECTRA GEOSPATIAL. SP60 Product Details. Available online: https://spectrageospatial.com/sp60-gnss-receiver/ (accessed on 12 January 2022).
- DRONESMADEEASY. Map Pilot for DJI. 2020. Available online: https://support.dronesmadeeasy.com/hc/en-us/categories/200739936-Map-Pilot-for-iOS (accessed on 25 February 2021).
- AGISOFT. Discover Intelligent Photogrammetry with Metashape. 2020. Available online: https://www.agisoft.com/ (accessed on 25 February 2021).
- Python Core Team. Python: A dynamic, Open Source Programming Language. Python Softw. Found. Available online: https://www.python.org/ (accessed on 17 June 2021).
- R Core Team. R: A language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2013; Available online: http://www.R-project.org (accessed on 17 June 2021).
- QGIS Development Team. QGIS Geographic Information System. QGIS Association. 2021. Available online: https://www.qgis.org (accessed on 17 June 2021).
- ANAC. Agência Nacional de Aviação Civil. Requisitos Gerais para Aeronaves não Tripuladas de uso Civil. Resolução Número 419, de 2 de maio de 2017. Regulamento Brasileiro da Aviação Civil Especial, RBAC-E Número 94. 2017. Available online: https://www.anac.gov.br/assuntos/legislacao/legislacao-1/rbha-e-rbac/rbac/rbac-e-94/@@display-file/arquivo_norma/RBACE94EMD00.pdf (accessed on 17 June 2021).
- Guariguata, M.R.; Pinard, M.A. Ecological knowledge of regeneration from seed in neotropical forest trees: Implications for natural forest management. For. Ecol. Manag. 1998, 112, 87–99. [Google Scholar] [CrossRef]
- Varma, V.K.; Ferguson, I.; Wild, I. Decision support system for the sustainable forest management. For. Ecol. Manag. 2000, 128, 49–55. [Google Scholar] [CrossRef]
- Minaee, S.; Boykov, Y.Y.; Porikli, F.; Plaza, A.J.; Kehtarnavaz, N.; Terzopoulos, D. Image Segmentation Using Deep Learning: A Survey. IEEE Trans. Pattern Anal. Mach. Intell. 2021. [Google Scholar] [CrossRef]
- Antonelli, A.; Sanmartín, I. Why are there so many plant species in the Neotropics? Taxon 2011, 60, 403–414. [Google Scholar] [CrossRef]
- Ter Steege, H.; Sabatier, D.; Mota de Oliveira, S.; Magnusson, W.E.; Molino, J.F.; Gomes, V.F.; Pos, E.T.; Salomão, R.P. Estimating species richness in hyper-diverse large tree communities. Ecology 2017, 98, 1444–1454. [Google Scholar] [CrossRef]
- Bellinger, C.; Sharma, S.; Japkowicz, N. One-Class versus Binary Classification: Which and When? In Proceedings of the 2012 11th International Conference on Machine Learning and Applications, Boca Raton, FL, USA, 12–15 December 2012; Volume 2, pp. 102–106. [Google Scholar] [CrossRef]
- Deng, X.; Li, W.; Liu, X.; Guo, Q.; Newsam, S. One-class remote sensing classification: One-class vs. binary classifiers. Int. J. Remote. Sens. 2018, 39, 1890–1910. [Google Scholar] [CrossRef]
- Dargan, S.; Kumar, M.; Ayyagari, M.R.; Kumar, G. A survey of deep learning and its applications: A new paradigm to machine learning. Arch. Comput. Methods Eng. 2020, 27, 1071–1092. [Google Scholar] [CrossRef]
- Zhang, J.; Liu, J.; Pan, B.; Shi, Z. Domain Adaptation Based on Correlation Subspace Dynamic Distribution Alignment for Remote Sensing Image Scene Classification. IEEE Trans. Geosci. Remote Sens. 2020, 58, 7920–7930. [Google Scholar] [CrossRef]
- Shannon, C.E. A mathematical theory of communication. Bell Syst. Tech. J. 1948, 27, 379–423. [Google Scholar] [CrossRef] [Green Version]
- PyPI. Fototex. Available online: https://pypi.org/project/fototex/ (accessed on 12 January 2022).
- Couteron, P.; Barbier, N.; Gautier, D. Textural ordination based on Fourier spectral decomposition: A method to analyze and compare landscape patterns. Landsc. Ecol. 2006, 21, 555–567. [Google Scholar] [CrossRef]
- Pommerening, A. Approaches to quantifying forest structures. For. Int. J. For. Res. 2002, 75, 305–324. [Google Scholar] [CrossRef]
- Alexopoulos, E.C. Introduction to multivariate regression analysis. Hippokratia 2010, 14, 23. [Google Scholar]
- Pal, M.; Bharati, P. Introduction to correlation and linear regression analysis. In Applications of Regression Techniques; Springer: Singapore, 2019; pp. 1–18. [Google Scholar]
- Lewis, S. Regression analysis. Pract. Neurol. 2007, 7, 259–264. [Google Scholar] [CrossRef]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Goutte, C.; Gaussier, E. A probabilistic interpretation of precision, recall and F-score, with implication for evaluation. In European Conference on Information Retrieval; Springer: Berlin/Heidelberg, Germany, 2005; pp. 345–359. [Google Scholar]
- Radoux, J.; Bogaert, P. Good practices for object-based accuracy assessment. Remote Sens. 2017, 9, 646. [Google Scholar] [CrossRef] [Green Version]
- Foody, G.M.; Green, R.M.; Lucas, R.; Curran, P.J.; Honzák, M.; Do Amaral, I. Observations on the relationship between SIR-C radar backscatter and the biomass of regenerating tropical forests. Int. J. Remote Sens. 1997, 18, 687–694. [Google Scholar] [CrossRef]
- Wagner, F.H.; Sanchez, A.; Tarabalka, Y.; Lotte, R.G.; Ferreira, M.P.; Aidar, M.P.; Gloor, E.; Phillips, O.L.; Aragao, L.E. Using the U-net convolutional network to map forest types and disturbance in the Atlantic rainforest with very high resolution images. Remote Sens. Ecol. Conserv. 2019, 5, 360–375. [Google Scholar] [CrossRef] [Green Version]
- Castro, J.; Morales-Rueda, F.; Navarro, F.B.; Löf, M.; Vacchiano, G.; Alcaraz-Segura, D. Precision restoration: A necessary approach to foster forest recovery in the 21st century. Restor. Ecol. 2021, 29, e13421. [Google Scholar] [CrossRef]
- Nuijten, R.J.; Coops, N.C.; Watson, C.; Theberge, D. Monitoring the Structure of Regenerating Vegetation Using Drone-Based Digital Aerial Photogrammetry. Remote Sens. 2021, 13, 1942. [Google Scholar] [CrossRef]
- Guo, G.; Zhang, N. A survey on deep learning based face recognition. Comput. Vis. Image Underst. 2019, 189, 102805. [Google Scholar] [CrossRef]
- Brandt, M.; Tucker, C.J.; Kariryaa, A.; Rasmussen, K.; Abel, C.; Small, J.; Chave, J.; Rasmussen, L.V.; Hiernaux, P.; Diouf, A.A.; et al. An unexpectedly large count of trees in the West African Sahara and Sahel. Nature 2020, 587, 78–82. [Google Scholar] [CrossRef]
- Weinstein, B.G.; Marconi, S.; Aubry-Kientz, M.; Vincent, G.; Senyondo, H.; White, E.P. DeepForest: A Python package for RGB deep learning tree crown delineation. Methods Ecol. Evol. 2020, 11, 1743–1751. [Google Scholar] [CrossRef]
- Zhu, L.; Ma, L. Class centroid alignment based domain adaptation for classification of remote sensing images. Pattern Recognit. Lett. 2016, 83, 124–132. [Google Scholar] [CrossRef]












| Delineation Target | Study Area | Epochs | Training Samples | Validation Samples | Test Samples | Samples in Synthetic Images | Total of Synthetic Train Images | Total of Synthetic Validation Images |
|---|---|---|---|---|---|---|---|---|
| Vismia sp. | Naturally regenerating site (NR) | 150 | 144 | 48 | 48 | 1 | 6000 | 2000 |
| Cecropia sp. | Actively restored site with Cecropia (ARCec) | 150 | 240 | 80 | 80 | 5 | 1800 | 600 |
| Actively restored diverse site (ARD) | - | - | - | 50 | - | - | - | |
| All Trees | Actively restored site with Cecropia (ARCec) | 150 | 369 | 123 | 50 | 50 | 9000 | 3000 |
| Actively restored diverse site (ARD) | 110 | 150 | 50 | 50 | 50 | 27,000 | 9000 |
| Cecropia: ARCec | Vismia: NR | Trees: ARCec | Trees: ARD | Cecropia: ARD (Test Only) | ||
|---|---|---|---|---|---|---|
| Delineation Accuracy | Identified trees | 91.25% | 72.92% | 72.00% | 56.00% | 80.00% |
| Tree crowns correctly delineated | 0.918 | 0.086 | 0.667 | 0.607 | 1.000 | |
| IoU | 0.772 | 0.202 | 0.563 | 0.558 | 0.790 | |
| Precision | 0.937 | 0.221 | 0.730 | 0.764 | 0.989 | |
| Recall | 0.820 | 0.888 | 0.764 | 0.738 | 0.798 | |
| F1 | 0.875 | 0.354 | 0.746 | 0.751 | 0.883 | |
| Area Accuracy | Overall Accuracy | 0.993 | 0.926 | 0.902 | 0.642 | 0.981 |
| Precision | 0.976 | 0.760 | 0.893 | 0.932 | 0.943 | |
| Recall | 0.752 | 0.796 | 0.616 | 0.565 | 0.669 | |
| F1 | 0.849 | 0.777 | 0.729 | 0.704 | 0.783 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Albuquerque, R.W.; Vieira, D.L.M.; Ferreira, M.E.; Soares, L.P.; Olsen, S.I.; Araujo, L.S.; Vicente, L.E.; Tymus, J.R.C.; Balieiro, C.P.; Matsumoto, M.H.; et al. Mapping Key Indicators of Forest Restoration in the Amazon Using a Low-Cost Drone and Artificial Intelligence. Remote Sens. 2022, 14, 830. https://doi.org/10.3390/rs14040830
Albuquerque RW, Vieira DLM, Ferreira ME, Soares LP, Olsen SI, Araujo LS, Vicente LE, Tymus JRC, Balieiro CP, Matsumoto MH, et al. Mapping Key Indicators of Forest Restoration in the Amazon Using a Low-Cost Drone and Artificial Intelligence. Remote Sensing. 2022; 14(4):830. https://doi.org/10.3390/rs14040830
Chicago/Turabian StyleAlbuquerque, Rafael Walter, Daniel Luis Mascia Vieira, Manuel Eduardo Ferreira, Lucas Pedrosa Soares, Søren Ingvor Olsen, Luciana Spinelli Araujo, Luiz Eduardo Vicente, Julio Ricardo Caetano Tymus, Cintia Palheta Balieiro, Marcelo Hiromiti Matsumoto, and et al. 2022. "Mapping Key Indicators of Forest Restoration in the Amazon Using a Low-Cost Drone and Artificial Intelligence" Remote Sensing 14, no. 4: 830. https://doi.org/10.3390/rs14040830
APA StyleAlbuquerque, R. W., Vieira, D. L. M., Ferreira, M. E., Soares, L. P., Olsen, S. I., Araujo, L. S., Vicente, L. E., Tymus, J. R. C., Balieiro, C. P., Matsumoto, M. H., & Grohmann, C. H. (2022). Mapping Key Indicators of Forest Restoration in the Amazon Using a Low-Cost Drone and Artificial Intelligence. Remote Sensing, 14(4), 830. https://doi.org/10.3390/rs14040830







