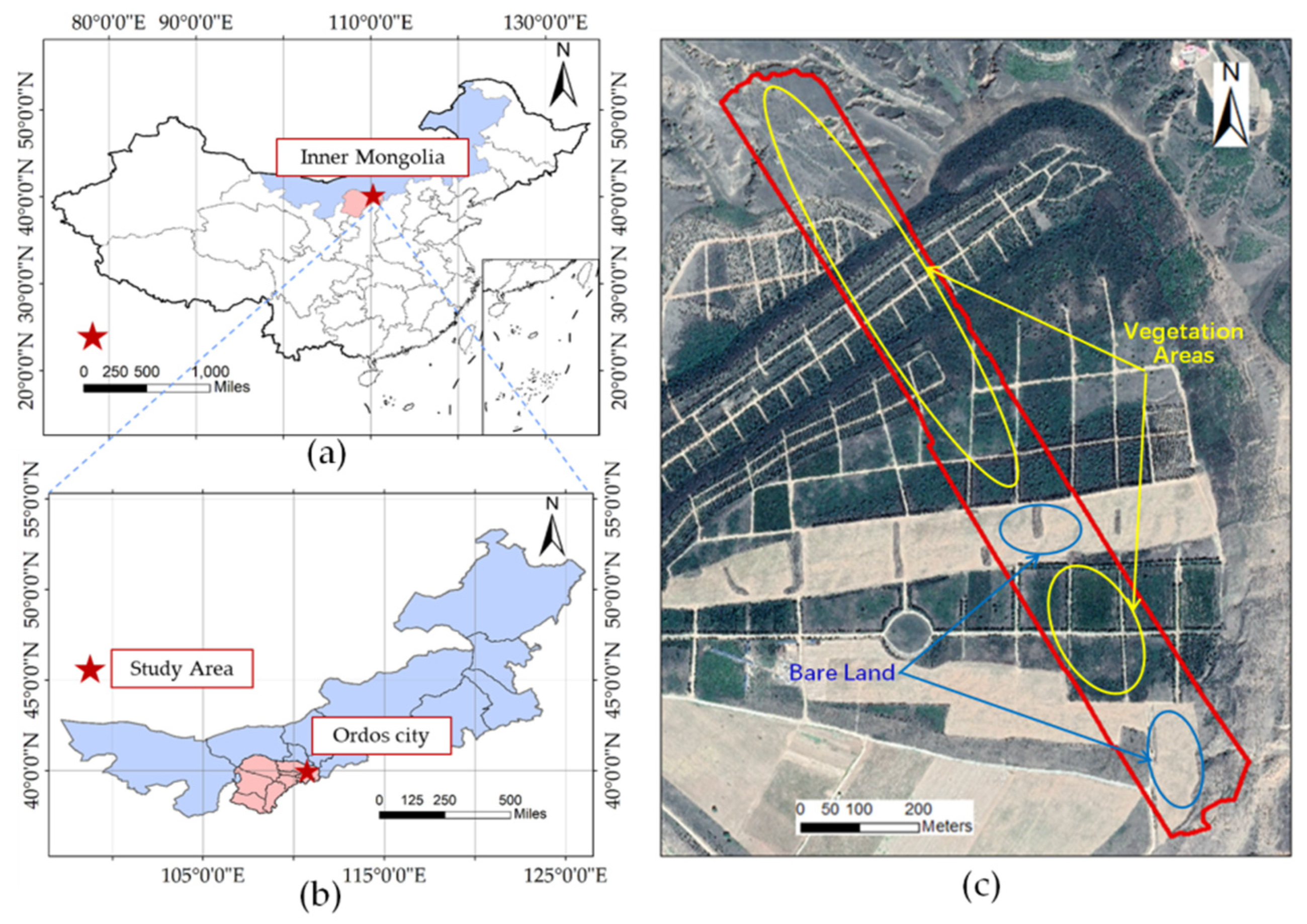
2.2.1. Data
The LiDAR data were acquired by a LiAir 220 UAV LiDAR system on 3 August 2020 using a DJI M600 PRO UAV platform (Dajiang Baiwang Technology Co., Ltd., Shenzhen, Guangdong, China) with a HS40P laser sensor. There were two flight zones planned in the flight area, with a total route of 2762 m, a flight altitude of 90 m, and a flight speed of 5 m/s. During LiDAR data acquisition, the horizontal field of view was 360°, the vertical field of view was greater than 20°, and the average point cloud density was 142 /m2.
LiDAR data were pre-processed though track solution, strip splicing, strip redundancy elimination, and point cloud merging [
25]. The morphological point cloud filtering method was used to distinguish the ground information in the LiDAR data to obtain the digital elevation model, the first echo characteristic data of the LiDAR was interpolated, and the ordinary kriging interpolation method was selected to generate the surface model. The crop height model (CHM) was expressed as the difference between the digital surface model and the digital elevation model.
Hyperspectral data were acquired by a S185 hyperspectral sensor (Cubert GmbH, Ulm, Germany) mounted on multi-rotor platform. A total of ten routes were planned in this hyperspectral data acquisition scheme, with an observation height of 100 m and a speed of 7.5 m/s. The sensor had a spectral range of 450–998 nm, a spectral resolution of 8 nm, a spectral sampling interval of 4 nm, and a ground resolution that could reach 5 cm.
The pre-processing of collected hyperspectral data is necessary for improving the quality of the hyperspectral images and the efficiency of image processing [
26]. The whiteboard data obtained during real-time data acquisition were used to calibrate the sensor; then, atmospheric correction, geometric correction, splicing and cutting of the flight belts, image fusion, and other processing steps were completed [
27]. In this study area, 1780 images were collected in a single band, while the mosaic of single-band images was operated through the Agisoft photoscan platform (
www.agisoft.com, accessed on 12 November 2020). During the data pre-processing, radiometric calibration, atmospheric correction, geometric correction, and image band synthesis were all realized with the help of the ENVI platform (Research Systems, Inc., Boulder, CO, USA).
LiDAR data were used to extract the intensity, height, and echo features of vegetation, while the hyperspectral image data were used to extract the vegetation spectrum, vegetation index, and texture features. Statistical analysis and correlation analysis by OriginLab (
www.originlab.com, accessed on 4 March 2021) and IBM SPSS Statistics (
www.ibm.com, accessed on 9 March 2021) were used to explore the relationships between vegetation features. The resampling tool of the ENVI platform (Research Systems, Inc., Boulder, CO, USA) was used to convert the resolution of the LiDAR data and hyperspectral data to 1 × 1 m [
28,
29].
Field survey data were collected simultaneously with the collection of UAV LiDAR and hyperspectral image data. Using random and typical sampling techniques, 10 m × 10 m quadrats were used to sample the vegetation. A total of 52 quadrats were investigated, including recording the vegetation type, canopy coverage, and dominant vegetation in each quadrat, as well as the tree height, diameter at breast height of trees, and the leaf area index of the dominant vegetation. A 1 × 1 m small quadrat was designated at the center of each large quadrat for the measurement of vegetation coverage and gap fraction by the needling method. In addition, 140 sampling points were established mainly for investigating the vegetation type, leaf area, and tree height index of these sampling points; location information for these sampling points was recorded.
2.2.3. Extraction of Feature Factors
An airborne LiDAR system can be used to obtain three-dimensional coordinate information of a target and extract the height feature of vegetation by processing the point cloud, which is also one of the important parameters related to vegetation structure [
31]. A morphological point cloud filtering method was used to distinguish the ground information within the LiDAR data. Based on this elevation model, the point cloud elevation value was subtracted from the value in the corresponding digital elevation model, so that the influence of topographical factors on the estimation of vegetation structure parameters was reduced, allowing the height feature of LiDAR to be extracted.
In this study, the following eight height features were selected: height percentile of 95% (HP95), maximum height (MaxH), minimum height (MinH), mean height (MeaH), median height (MedH), standard deviation of height (SDH), root mean square of height (RMSH), and coefficient of variation (CVH).
- 2.
Intensity
The LiDAR intensity quantitatively describes the backscattering of an object. Intensity is a measurement index (collected for each point) that reflects the intensity of the LiDAR pulse echo generated at a certain point. The intensity feature can be used to classify the LiDAR points, and the different features can be distinguished by the intensity signal. Studies have shown that the influence of leaves, branch directions, terrain changes, and laser path length can cause differences in the intensity of LiDAR in forest areas [
32].
The following seven features of LiDAR intensity were selected: intensity percentile of 99% (IP99), maximum intensity (MaxI), minimum intensity (MinI), mean intensity (MeaI), median intensity (MedI), standard deviation of intensity (SDI), and coefficient of variation of intensity (CVI).
- 3.
Echo
The echo feature of LiDAR point cloud data is an important feature that can be used to express the structural parameters of vegetation. A laser pulse emitted may return to the LiDAR sensor in the form of one or more echoes. The first laser pulse returned is the most important echo, and it will be associated with the highest elements on the Earth’s surface (such as treetops). When the first echo represents the ground, the LiDAR system will only detect one echo. Multiple echoes can detect the height of multiple objects within the laser foot point where the laser pulse is emitted. The middle echo usually corresponds to the vegetation structure, and the last echo is related to the exposed terrain model [
33].
The laser sensor used for data acquisition in this study could receive two echoes. There was often no obvious difference between the first echo number (Return1) and the second echo number (Return2). Therefore, we defined the ratio of the first echo to the second echo as the third characteristic factor (Return1/2), and observed the difference between the ratio of the two echoes in dense and sparse vegetation areas in the study area.
- 4.
Spectrum
Different types of vegetation and non-vegetated areas have different absorption and reflection spectral characteristics in different bands [
34]. As shown in
Figure 3, in the visible band, chlorophyll is the most important factor that dominates the spectral response of plants. In the near-infrared band of the spectrum, the spectral features of vegetation are mainly controlled by the internal structure of plant leaves. Between the visible light band and the near-infrared band, i.e., in the range of 700–998 nm, most of the energy is reflected, a small part of the energy is absorbed, and the rest of the energy is completely transmitted, forming a “red edge” phenomenon, which is the most obvious feature of the plant curve [
35].
According to the results of the field survey, the study area was divided into seven vegetation species composition types and the bare land was used for comparison. The region of interest (ROI) data of each object were collected, and the spectral sample data of 138 bands were outputted to form a spectral curve [
36].
Figure 3.
Schematic diagram of plant spectral curve [
37].
Figure 3.
Schematic diagram of plant spectral curve [
37].
- 5.
Vegetation index
Hyperspectral images are easily affected by the environment, atmosphere, and the phenomenon of “different objects with the same spectrum” when researchers attempt to identify different species composition types. The vegetation spectrum shows a complex mixed reaction to vegetation, environmental effects, brightness, soil color, and humidity, which are affected by the spatial–temporal changes in the atmospheric environment. The vegetation index mainly reflects the difference between the reflection of vegetation in the visible light and near-infrared band as well as the soil background [
38]. Under certain conditions, each vegetation index can be used to quantitatively explain the growth status of vegetation, and can qualitatively and quantitatively detect and evaluate the vegetation coverage and its growth [
39]. Therefore, this study selected the following 33 vegetation indices, listed in
Table 1, to study.
Texture is an important type of structural information related to the spatial distribution of ground objects. By measuring the difference between pixels and their surrounding spatial neighborhood, texture can be used as a sufficient auxiliary basis for solving the problem of “different spectra of the same object” or “different objects with the same spectrum”, and can make up for a deficiency of spectral features in hyperspectral remote sensing image classification to a certain extent. Texture has three features: scale, region, and regularity. In this study, eight quadratic statistics, listed in
Table 2, were used as texture feature parameters to extract the textural features of images.
Through combination with the spectral curve images of different vegetation species composition types, it can be seen that the difference in reflectance in the visible light band from different kinds of plants is small. We can consider that the three bands of green, red, and near-infrared can form a pseudo-color image synthesis band that can be used to analyze and extract texture features:
The first two bands of the transformed hyperspectral data are separated by a minimal amount of noise; the pseudo-color bands composed of the green, red, and near-infrared bands were named B1, B2, and B3 when used in the subsequent texture calculations. The central wavelengths of the B1 and B2 bands were 784 nm and 806 nm, respectively. Using a 3 × 3 pixel window, based on the first- and second-order probability statistics, respectively, a gray level co-occurrence matrix of synthetic band images was calculated, and the vegetation texture features were extracted. The naming rules of texture feature factors were defined as: band–texture index–probability statistics order.
- 7.
Single tree segmentation
The watershed segmentation algorithm is a kind of image segmentation algorithm, which is based on the mathematical morphology principle of topology theory. This method is fast and accurate, so it is widely used in image segmentation [
67]. Through a watershed algorithm, a single tree can be segmented, and the position and crown width of a single tree can be obtained. Each pixel in a canopy height model as assigned with an elevation value, resulting in a continuous surface with scattered peaks and valleys. Therefore, the peak point of a continuous surface was defined as the highest point of a single tree. In addition, watershed segmentation can better identify edges, which is more suitable for the high-resolution remote sensing images extracted in this study.
2.2.4. Decision Tree Classifier
A decision tree is a tree data structure based on root, intermediate, and leaf nodes [
68]. According to the experimental samples, the decision tree classifier determined the appropriate discriminant function; next, the branches of the tree were constructed according to the obtained functions, and then sub-branches were constructed according to the needs of each branch to form the final decision tree for herb and bare land extraction [
69]. Based on empirical knowledge, vegetation and non-vegetation are quite different in remote sensing images, and have strong discrimination characteristics. The study area is a mine dump, and the only bare land types are found in non-vegetation regions.
According to the extraction results of the vegetation index, the vegetation index sensitive to herbs and bare land was selected, and the classification rules of vegetated areas and bare land area were defined. According to the numerical distribution of different vegetation species compositions in the vegetation index, the bare land area could be extracted in the first layer. In addition, obvious differences in height features existed among herbs, trees, and shrubs. Therefore, the classification rules of herbs were summarized by analyzing the numerical distribution of height features between herbs and other vegetation species’ compositions; however, overlapping layers may exist between bare land and herbs in the height layer. Therefore, it is necessary to consider removing bare land and extracting the herb layer in the second layer in the herb layer obtained by height features.
2.2.5. Random Forest Classifier
A random forest algorithm is an ensemble learning algorithm based on a classification regression decision tree. By sampling from training samples using the RF Bootstrap Resampling method, a certain number of samples are repeatedly extracted from the training sample set
N, and a new training set of
n samples is generated; each sample generates a classification tree, and the
n classification trees form a random forest. When a certain number of decision trees are produced, the test samples can be used to test the classification effect of each number, so as to vote for the best classification result [
70].
Before using the RF algorithm for classification, the factors of each feature were sorted by random forest; the factors that were more important for the inversion of vegetation species composition types in each feature were screened out [
71]. After the classification features were determined,
n training sets were obtained by using the autonomous sampling method. Each training set needed to be trained to generate a corresponding decision tree model; next, parameters such as the number of base estimators (n_estimators), the maximum depth (max_depth), the minimum sample size (min_samples_split), and the maximum feature number (max_features) of the random forest were tested [
72].
After extracting herbs and bare land, the random forest algorithm was used to invert the tree and shrub species composition types. At first, the image of the study area was classified using the results of canopy extraction and single wood segmentation, i.e., the third layer extracted trees. Finally, the shrubs were classified on the fourth layer.