Improved k-NN Mapping of Forest Attributes in Northern Canada Using Spaceborne L-Band SAR, Multispectral and LiDAR Data
Abstract
:1. Introduction
2. Materials and Methods
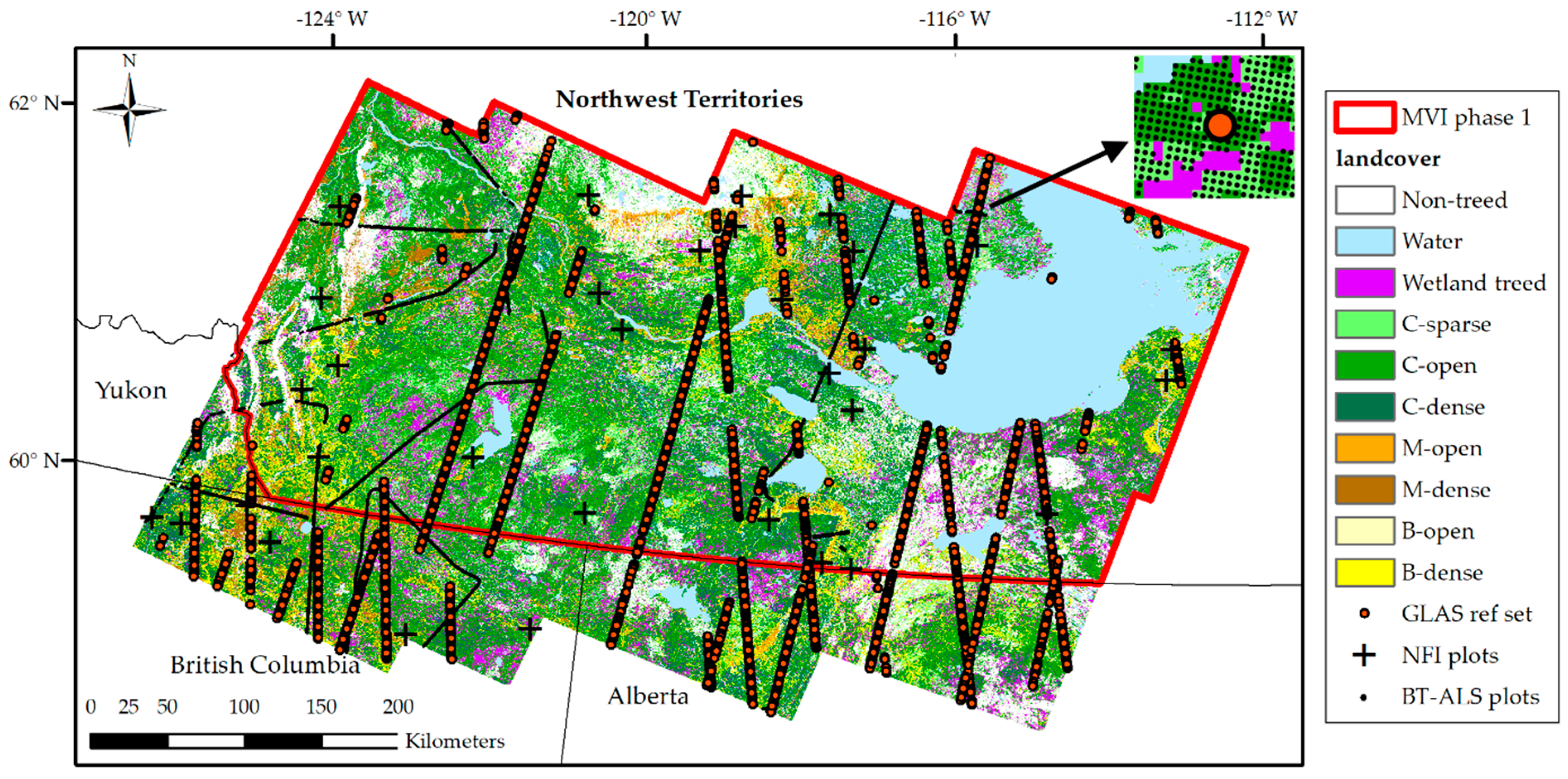
2.1. Study Area
2.2. Datasets
2.2.1. Response Variables
- Stand height (Ht, m): average height of dominant and codominant live trees, i.e., with height ≥ average Lorey’s height, where Lorey’s height is the average height of all trees with diameter at breast height (DBH) ≥5 cm and taller than 1.3 m) weighted by stem cross-section;
- Crown closure (CC, %): percent tree cover;
- Stand volume (Vs, m3·ha−1): sum of volume inside bark of the boles of live trees with height ≥ Lorey’s height;
- Total volume (Vt, m3·ha−1): sum of volume inside bark of the boles of all live trees with DBH ≥ 5 cm;
2.2.2. Feature Variables from Remote Sensing and Other Sources
2.2.3. Ancillary Data
2.2.4. Independent Validation Datasets
- Fifty-two 400 m2 NFI ground plots [51,52] (hereafter NFI plots) for which stand-level forest attributes derived from a combination of ground measurements and allometric equations were available as continuous variables, except for crown closure provided in broad ordinal classes. NFI plots qualify as an independent validation set as they provide a probabilistic sample set but with the caveat that it is a relatively small sample size for the study area;
- Over 1 million Boreal transect ALS 25 m cells (hereafter BT−ALS LiDAR plots) derived from ALS data acquired in the summer of 2010 along 750 m wide transects totalling 1800 km in length with a point sampling density of 2.8 point·m−2 [53,54]. Stand height, Lorey’s height, and crown closure were estimated from ALS models, while stand volume was estimated from stand height, and both total volume and AGB were estimated from average Lorey’s height [5]. However, crown closure estimates were not retained for validation because of a laser power issue preventing the proper transferability of the ALS-based crown closure model to the BT−ALS data [7]. Although the BT−ALS sample set does not provide attribute estimates as accurate as those from the NFI ground plots, and thus qualifies more as a comparison rather than a validation set, we still considered it to be a valuable independent validation dataset. It has a large number of 25 m cells and its extensive spatial extent captures a much wider geographic range of forest conditions across broad forest types than NFI ground plots.
2.2.5. Landsat-Based Forest Attribute Maps
- The ca. 2007 k-NN map of stand height over the same extent from Mahoney et al. [7];
- The large-area 2007 AGB map of Wang et al. [55] covering northwestern Canada and Alaska; this map was part of a 1984–2014 time series of 30 m annual AGB maps derived from the Gradient Boosted Machines machine learning algorithm trained by GLAS-based AGB estimates and using predictors from seasonally fit Landsat time series.
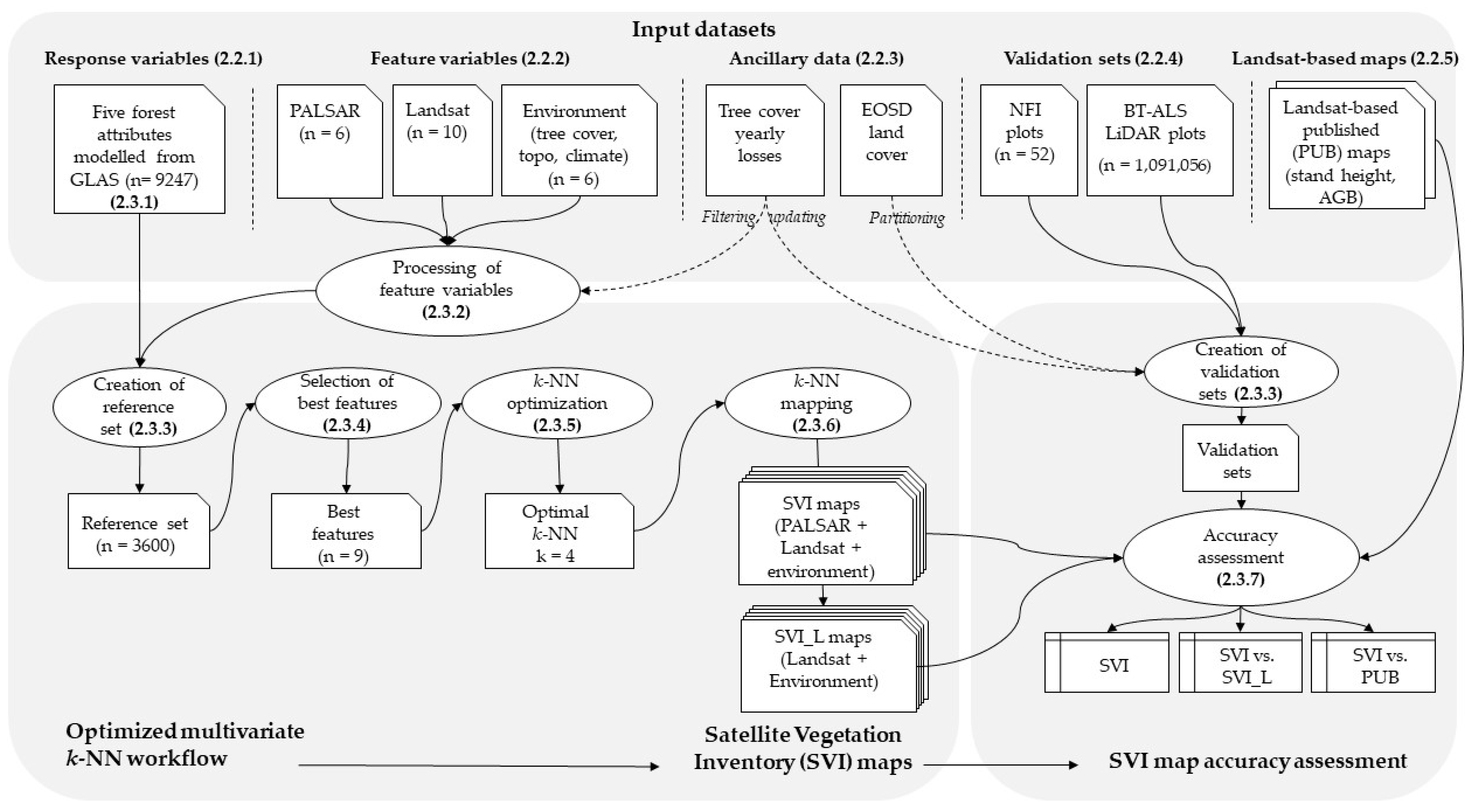
2.3. Methods
2.3.1. GLAS Modelling of Response Variables
2.3.2. Processing of Feature Variables
2.3.3. Creation of Reference and Validation Datasets
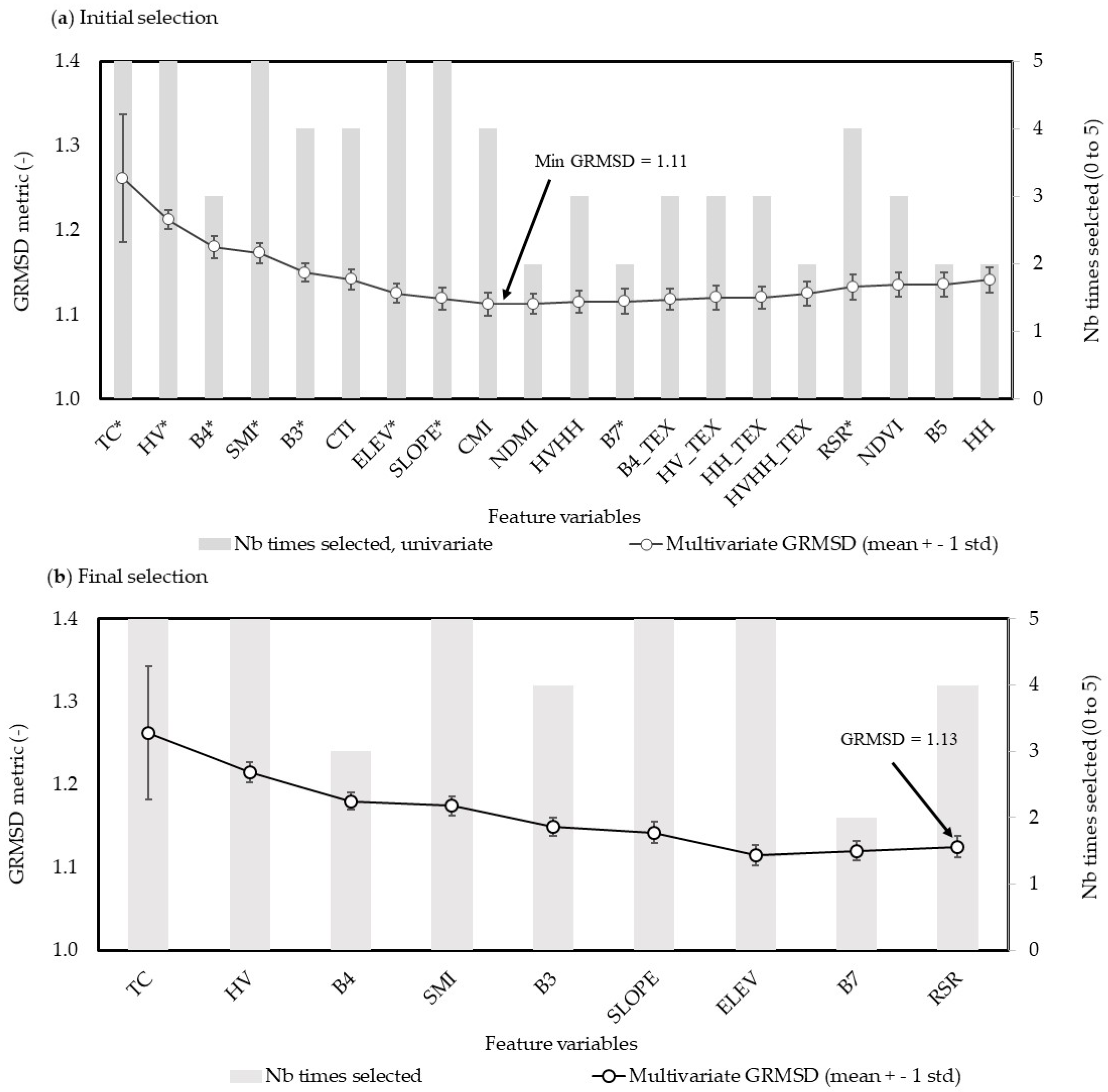
2.3.4. Selection of Best Feature Variables
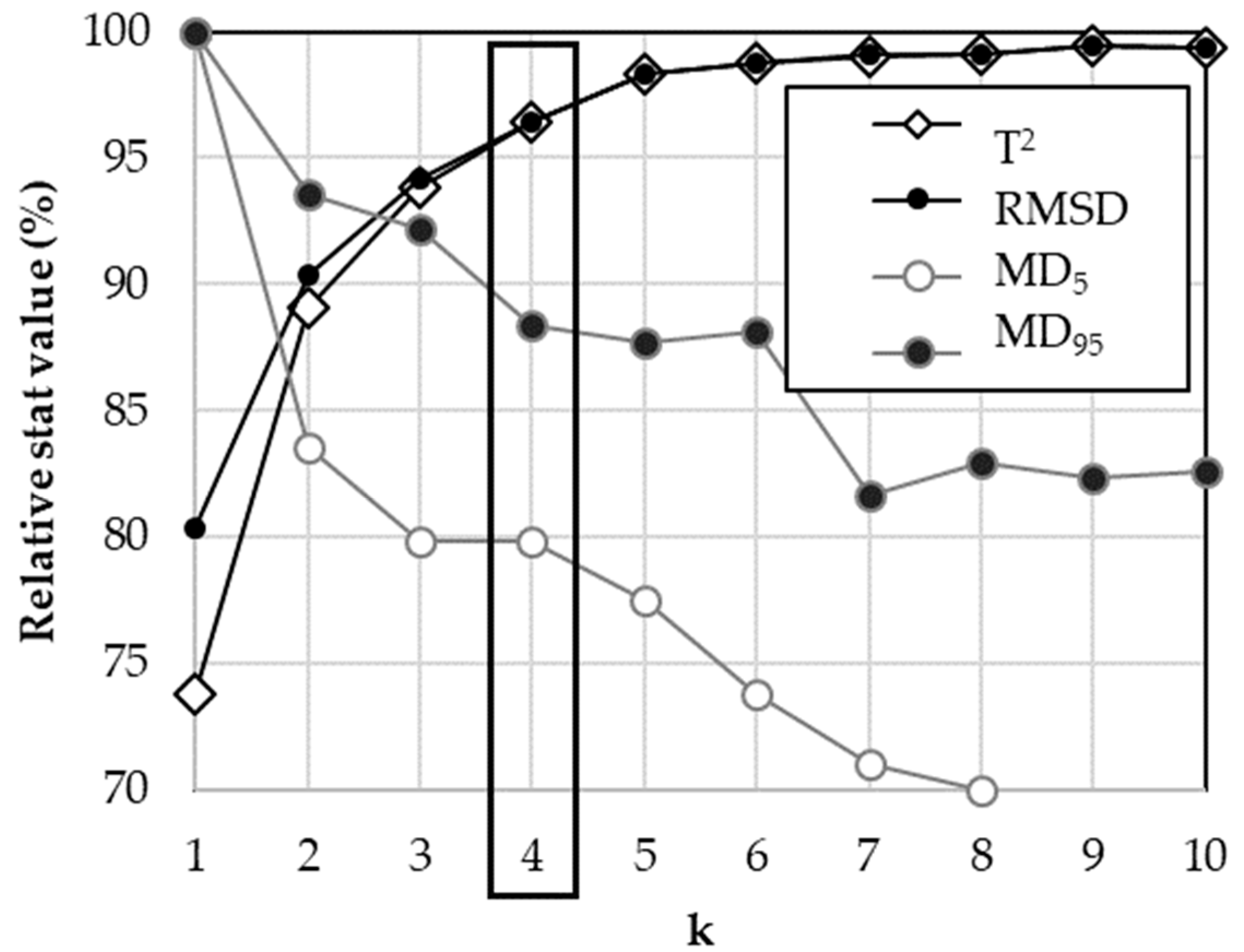
2.3.5. Optimization of k-NN k Parameters
2.3.6. Forest Attribute Maps from k-NN
2.3.7. Accuracy Assessment
- goodness of fit (adj. R2) and coefficients of linear regressions (predictions ~ observations);
- mean error or bias (ME, predicted minus observed, expressed as in Equation (4) for MD) and root mean square error (RMSE, expressed as in Equation (3) for RMSD) as a measure of overall accuracy, both expressed as percent values relative to the observed mean value (ME%, RMSE%);
- mean and standard deviation of prediction error (predicted minus observed) by quartile group across the range of observed NFI attribute values. This is presented along with overall mean prediction error in a plot similar to a Bland Altman diagram [63,64], which provides a visual graphic of the magnitude and distribution of prediction bias and variance across the range of the response variable.
3. Results
3.1. Selection of Best Feature Variables
3.2. Optimization of the k-NN k Parameter
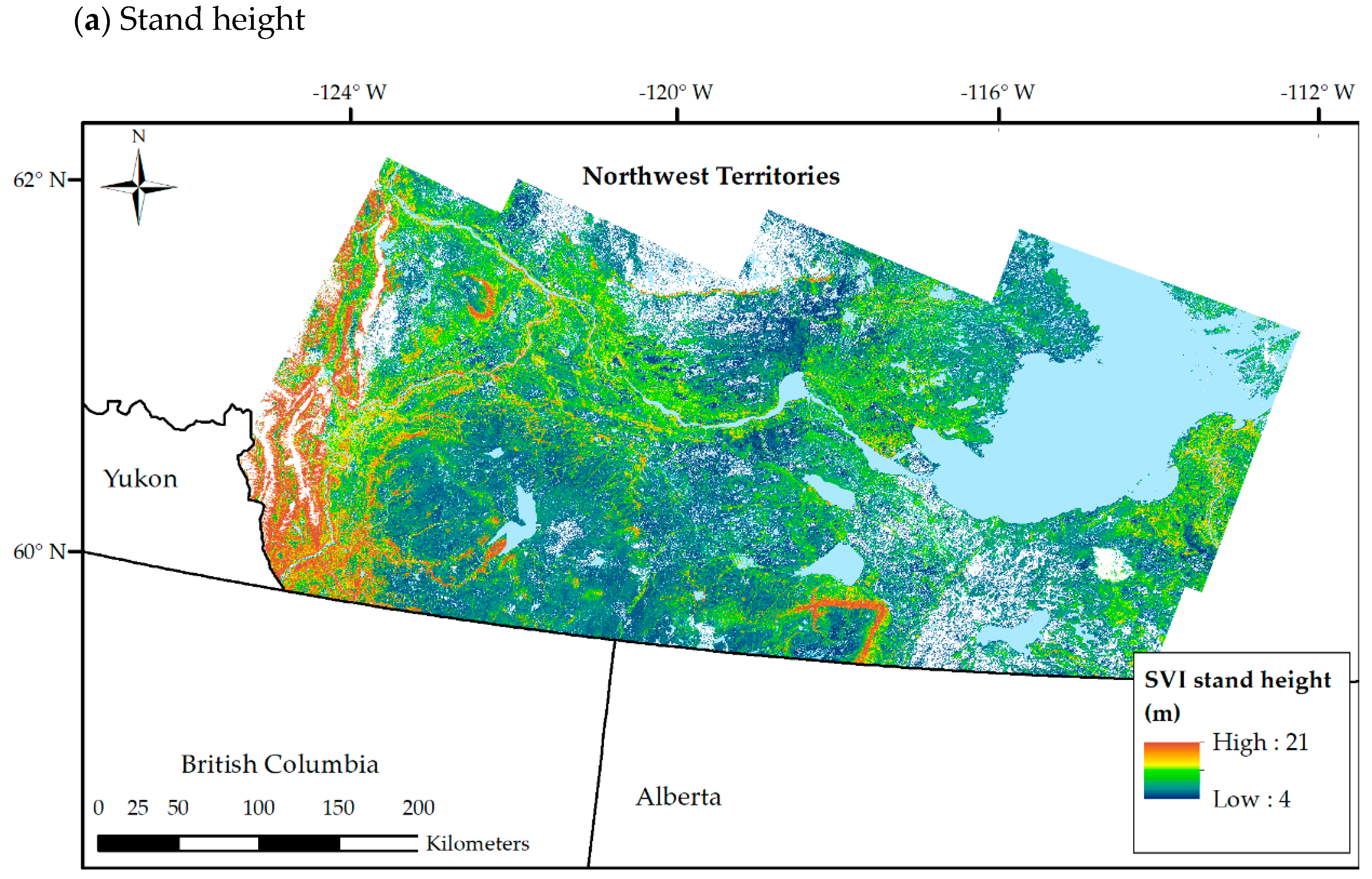
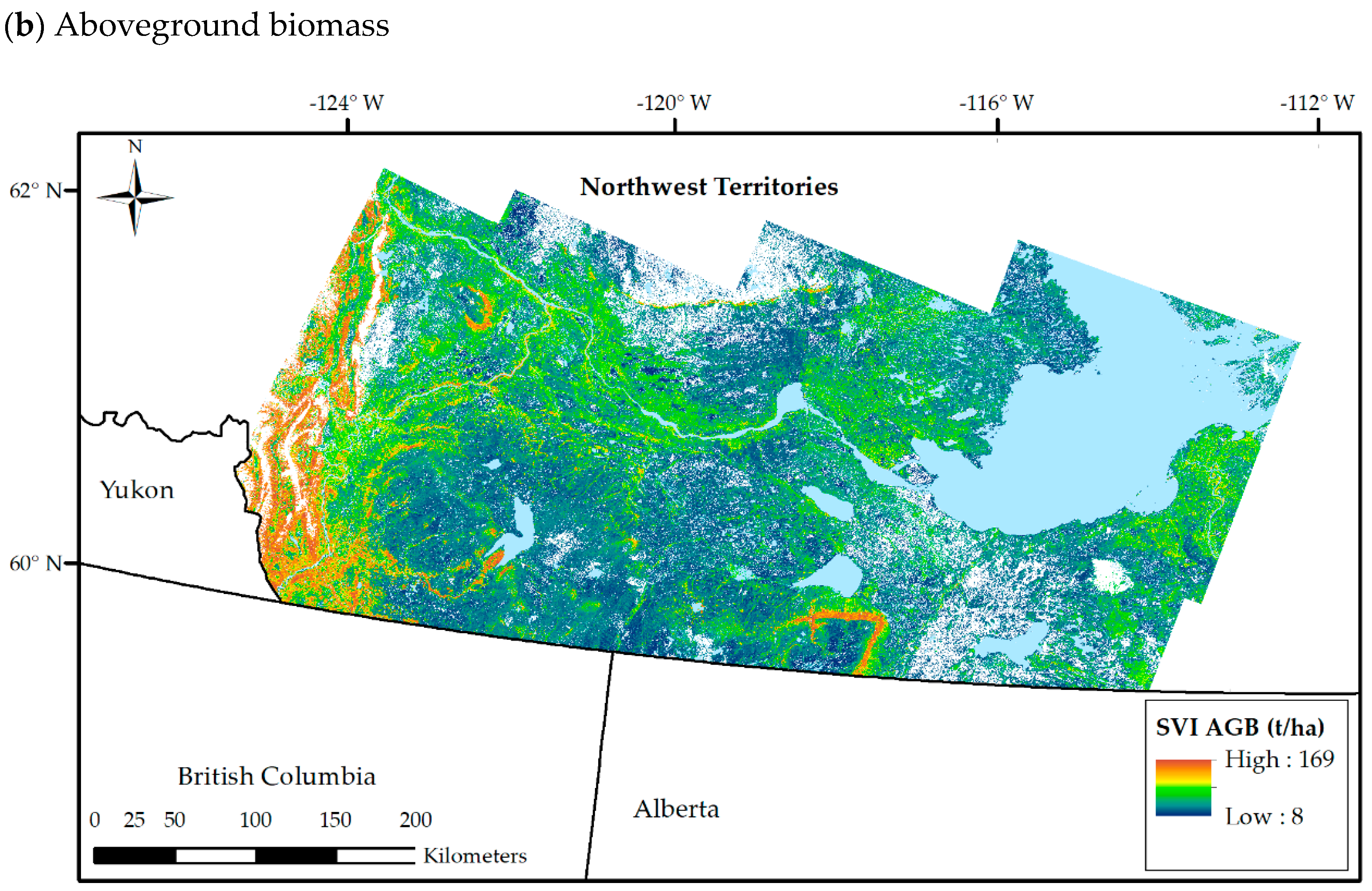
3.3. SVI Maps from k-NN
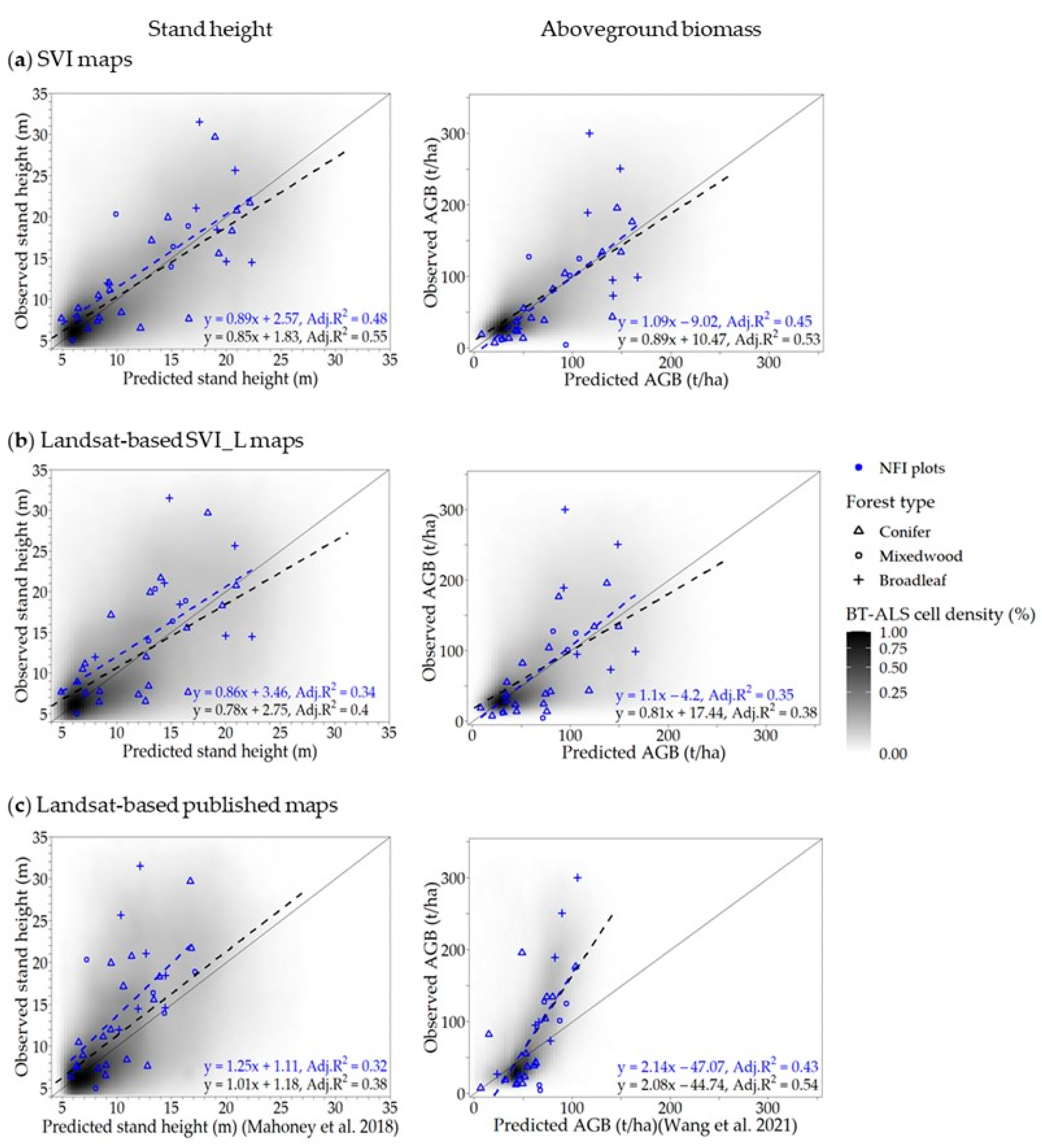
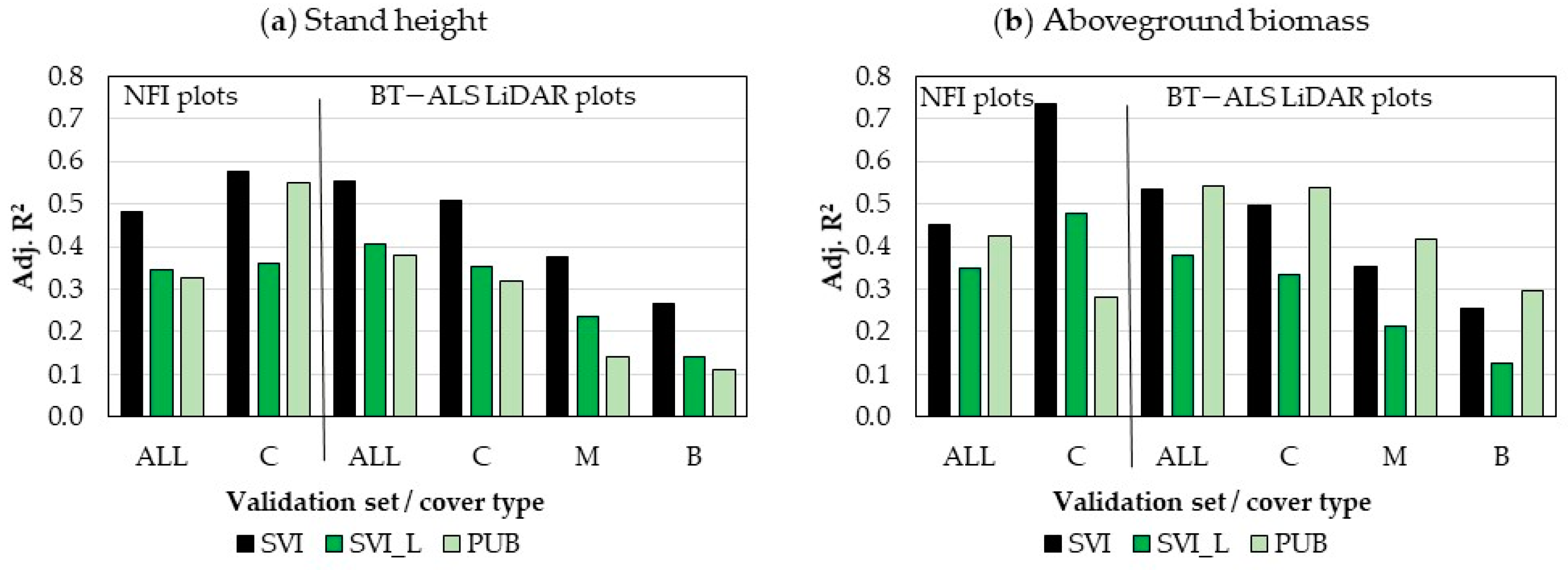
3.4. Accuracy Assessment
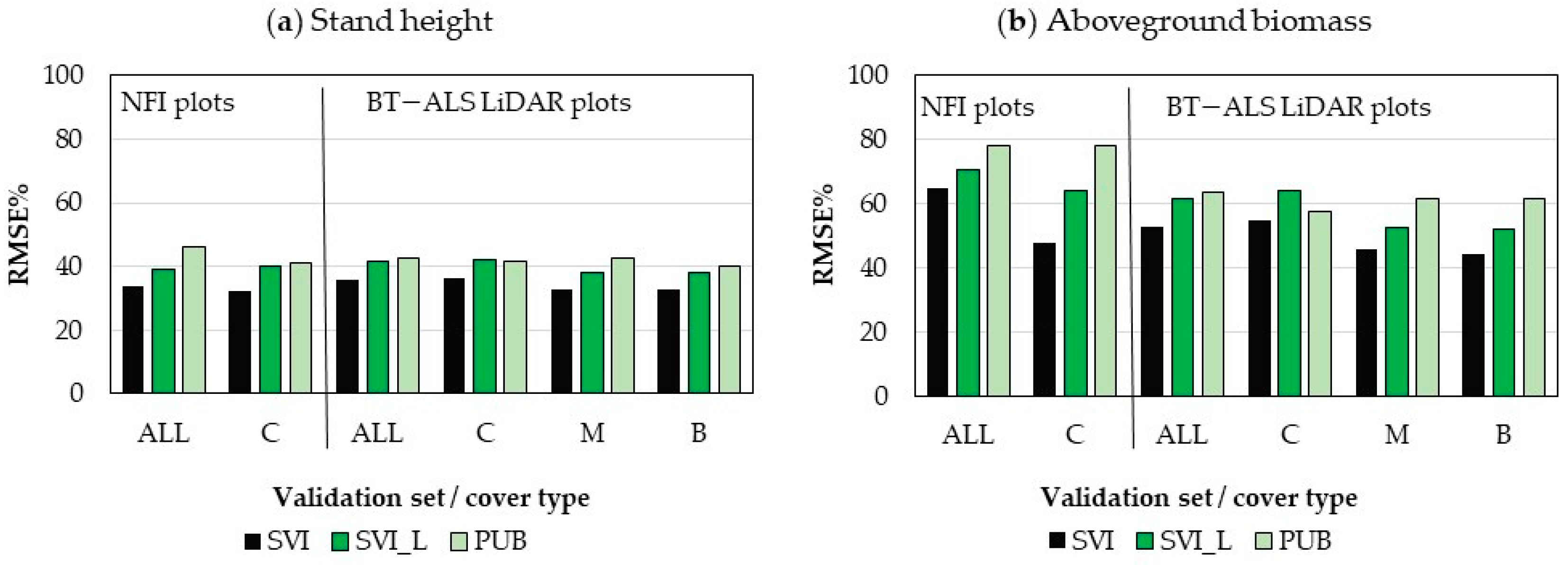
3.4.1. Accuracy of SVI Maps
3.4.2. Accuracy Comparison between SVI Maps and Landsat-Based Maps
4. Discussion
4.1. Primary Results of This Study
4.2. Sources of Errors
4.3. Future Work
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Corona, P. Integration of forest mapping and inventory to support forest management. iForest-Biogeosci. For. 2010, 3, 59–64. [Google Scholar] [CrossRef] [Green Version]
- Brosofske, K.D.; Froese, R.E.; Falkowski, M.J.; Banskota, A. A review of methods for mapping and prediction of inventory attributes for operational forest management. For. Sci. 2014, 60, 733–756. [Google Scholar] [CrossRef]
- Leckie, D.G.; Gillis, M.D. Forest inventory in Canada with emphasis on map production. For. Chron. 1995, 71, 74–88. [Google Scholar] [CrossRef]
- Thompson, I.D.; Maher, S.C.; Rouillard, D.P.; Fryxell, J.M.; Baker, J.A. Accuracy of forest inventory mapping: Some implications for boreal forest management. For. Ecol. Manag. 2007, 252, 208–221. [Google Scholar] [CrossRef]
- Castilla, G.; Hall, R.J.; Skakun, R.S.; Filiatrault, M.; Beaudoin, A.; Gartrell, M.; Hopkinson, C.; Smith, L.; Groenewegen, K.; van der Sluijs, J. The Multisource Vegetation Inventory (MVI): A satellite-based forest inventory for the Northwest Territories Taiga Plains. Remote Sens. 2022, 14, 1108. [Google Scholar] [CrossRef]
- Beaudoin, A.; Bernier, P.Y.; Guindon, L.; Villemaire, P.; Guo, X.J.; Stinson, G.; Bergeron, T.; Magnussen, S.; Hall, R.J. Mapping attributes of Canada’s forests at moderate resolution through k-NN and MODIS imagery. Can. J. For. Res. 2014, 44, 521–532. [Google Scholar] [CrossRef] [Green Version]
- Mahoney, C.; Hall, R.J.; Hopkinson, C.; Filiatrault, M.; Beaudoin, A.; Chen, Q. A forest attribute mapping framework: A pilot study in a Northern boreal forest, Northwest Territories, Canada. Remote Sens. 2018, 10, 1338. [Google Scholar] [CrossRef] [Green Version]
- Matasci, G.; Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C.; Hobart, G.W.; Zald, H.S. Large-area mapping of Canadian boreal forest cover, height, biomass and other structural attributes using Landsat composites and lidar plots. Remote Sens. Environ. 2018, 209, 90–106. [Google Scholar] [CrossRef]
- Lutz, D.A.; Washington-Allen, R.A.; Shugart, H.H. Remote sensing of boreal forest biophysical and inventory parameters: A review. Can. J. Remote Sens. 2008, 34 (Suppl. S2), S286–S313. [Google Scholar] [CrossRef]
- Lu, D.; Chen, Q.; Wang, G.; Liu, L.; Li, G.; Moran, E. A survey of remote sensing-based aboveground biomass estimation methods in forest ecosystems. Int. J. Digit. Earth 2016, 9, 63–105. [Google Scholar] [CrossRef]
- White, J.C.; Coops, N.C.; Wulder, M.A.; Vastaranta, M.; Hilker, T.; Tompalski, P. Remote sensing technologies for enhancing forest inventories: A review. Can. J. Remote Sens. 2016, 42, 619–641. [Google Scholar] [CrossRef] [Green Version]
- Woods, M.; Lim, K.; Treitz, P. Predicting forest stand variables from LIDAR data in the Great Lakes St. Lawrence Forest of Ontario. For. Chron. 2008, 84, 827–839. [Google Scholar] [CrossRef] [Green Version]
- Wulder, M.A.; White, J.C.; Nelson, R.F.; Næsset, E.; Ørka, H.O.; Coops, N.C.; Hilker, T.; Bater, C.W.; Gobakken, T. Lidar sampling for large-area forest characterization: A review. Remote Sens. Environ. 2012, 121, 196–209. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, R.E.; Ohmann, J.; Gregory, M.; Roberts, H.; Yang, Z.; Bell, D.M.; Kane, V.; Hughes, M.J.; Cohen, W.B.; Powell, S.; et al. An empirical, integrated forest biomass monitoring system. Environ. Res. Lett. 2018, 13, 025004. [Google Scholar] [CrossRef]
- Andersen, H.E.; Strunk, J.; Temesgen, H.; Atwood, D.; Winterberger, K. Using multilevel remote sensing and ground data to estimate forest biomass resources in remote regions: A case study in the boreal forests of interior Alaska. Can. J. Remote Sens. 2011, 37, 596–611. [Google Scholar] [CrossRef]
- Luther, J.E.; Fournier, R.A.; van Lier, O.R.; Bujold, M. Extending ALS-based mapping of forest attributes with medium resolution satellite and environmental data. Remote Sens. 2019, 11, 1092. [Google Scholar] [CrossRef] [Green Version]
- Neigh, C.S.; Nelson, R.F.; Ranson, K.J.; Margolis, H.A.; Montesano, P.M.; Sun, G.; Kharuk, V.; Næsset, E.; Wulder, M.A.; Andersen, H.E. Taking stock of circumboreal forest carbon with ground measurements, airborne and spaceborne LiDAR. Remote Sens. Environ. 2013, 137, 274–287. [Google Scholar] [CrossRef] [Green Version]
- Tomppo, E.; Olsson, H.; Ståhl, G.; Nilsson, M.; Hagner, O.; Katila, M. Combining national forest inventory field plots and remote sensing data for forest databases. Remote Sens. Environ. 2008, 112, 1982–1999. [Google Scholar] [CrossRef]
- Wilson, B.T.; Lister, A.J.; Riemann, R.I. A nearest-neighbor imputation approach to mapping tree species over large areas using forest inventory plots and moderate resolution raster data. For. Ecol. Manage. 2012, 271, 182–198. [Google Scholar] [CrossRef]
- White, J.C.; Wulder, M.A.; Varhola, A.; Vastaranta, M.; Coops, N.C.; Cook, B.D.; Pitt, D.; Woods, M. A best practices guide for generating forest inventory attributes from airborne laser scanning data using an area-based approach. For. Chron. 2013, 89, 722–723. [Google Scholar] [CrossRef] [Green Version]
- Popescu, S.C.; Zhao, K.; Neuenschwander, A.; Lin, C. Satellite lidar vs. small footprint airborne lidar: Comparing the accuracy of aboveground biomass estimates and forest structure metrics at footprint level. Remote Sens. Environ. 2011, 115, 2786–2797. [Google Scholar] [CrossRef]
- Schutz, B.E.; Zwally, H.J.; Shuman, C.A.; Hancock, D.; DiMarzio, J.P. Overview of the ICESat mission. Geophys. Res. Lett. 2005, 32, L21S01. [Google Scholar] [CrossRef] [Green Version]
- Boudreau, J.; Nelson, R.F.; Margolis, H.A.; Beaudoin, A.; Guindon, L.; Kimes, D.S. Regional aboveground forest biomass using airborne and spaceborne LiDAR in Québec. Remote Sens. Environ. 2008, 112, 3876–3890. [Google Scholar] [CrossRef]
- Margolis, H.A.; Nelson, R.F.; Montesano, P.M.; Beaudoin, A.; Sun, G.; Andersen, H.E.; Wulder, M.A. Combining satellite lidar, airborne lidar, and ground plots to estimate the amount and distribution of aboveground biomass in the boreal forest of North America. Can. J. For. Res. 2015, 45, 838–855. [Google Scholar] [CrossRef] [Green Version]
- McRoberts, R.E.; Tomppo, E.O.; Finley, A.O.; Heikkinen, J. Estimating areal means and variances of forest attributes using the k-Nearest Neighbors technique and satellite imagery. Remote Sens. Environ. 2007, 111, 466–480. [Google Scholar] [CrossRef]
- Chirici, G.; Mura, M.; McInerney, D.; Py, N.; Tomppo, E.O.; Waser, L.T.; Travaglini, D.; McRoberts, R.E. A meta-analysis and review of the literature on the k-Nearest Neighbors technique for forestry applications that use remotely sensed data. Remote Sens. Environ. 2016, 176, 282–294. [Google Scholar] [CrossRef]
- McRoberts, R.E. Estimating forest attribute parameters for small areas using nearest neighbors techniques. For. Ecol. Manag. 2012, 272, 3–12. [Google Scholar] [CrossRef]
- Mäkelä, H.; Hirvelä, H.; Nuutinen, T.; Kärkkäinen, L. Estimating forest data for analyses of forest production and utilization possibilities at local level by means of multi-source National Forest Inventory. For. Ecol. Manag. 2011, 262, 1345–1359. [Google Scholar] [CrossRef]
- Rosenqvist, Å.; Shimada, M.; Ito, N.; Watanabe, M. ALOS PALSAR: A pathfinder mission for global-scale monitoring of the environment. IEEE Trans. Geosci. Remote Sens. 2007, 45, 3307–3316. [Google Scholar] [CrossRef]
- Yu, Y.; Saatchi, S. Sensitivity of L-band SAR backscatter to aboveground biomass of global forests. Remote Sens. 2016, 8, 522. [Google Scholar] [CrossRef] [Green Version]
- Santoro, M.; Eriksson, L.E.B.; Fransson, J.E.S. Reviewing ALOS PALSAR Backscatter Observations for Stem Volume Retrieval in Swedish Forest. Remote Sens. 2015, 7, 4290–4317. [Google Scholar] [CrossRef] [Green Version]
- Peregon, A.; Yamagata, Y. The use of ALOS/PALSAR backscatter to estimate aboveground forest biomass: A case study in Western Siberia. Remote Sens. Environ. 2013, 137, 139–146. [Google Scholar] [CrossRef]
- Suzuki, R.; Kim, Y.; Ishii, R. Sensitivity of the backscatter intensity of ALOS/PALSAR to the aboveground biomass and other biophysical parameters of boreal forest in Alaska. Polar Sci. 2013, 7, 100–112. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez-Veiga, P.; Quegan, S.; Carreiras, J.; Persson, H.J.; Fransson, J.E.S.; Hoscilo, A.; Ziółkowski, D.; Stereńczak, K.; Lohberger, S.; Stängel, M.; et al. Forest biomass retrieval approaches from earth observation in different biomes. Int. J. Appl. Earth Obs. Geoinf. 2019, 77, 53–68. [Google Scholar] [CrossRef]
- Coops, N.C.; Tompalski, P.; Goodbody, T.H.R.; Queinnec, M.; Luther, J.E.; Bolton, D.K.; White, J.C.; Wulder, M.A.; van Lier, O.R.; Hermosilla, T. Modelling lidar-derived estimates of forest attributes over space and time: A review of approaches and future trends. Remote Sens. Environ. 2021, 260, 112477. [Google Scholar] [CrossRef]
- García, M.; Saatchi, S.; Ustin, S.; Balzter, H. Modelling forest canopy height by integrating airborne LiDAR samples with satellite Radar and multispectral imagery. Int. J. Appl. Earth Geoinf. 2018, 66, 159–173. [Google Scholar] [CrossRef]
- Cartus, O.; Kellndorfer, J.; Rombach, M.; Walker, W. Mapping canopy height and growing stock volume using airborne lidar, ALOS PALSAR and Landsat ETM+. Remote Sens. 2012, 4, 3320–3345. [Google Scholar] [CrossRef] [Green Version]
- Cartus, O.; Kellndorfer, J.; Walker, W.; Franco, C.; Bishop, J.; Santos, L.; Fuentes, J.M.M. A National, Detailed Map of Forest Aboveground Carbon Stocks in Mexico. Remote. Sens. 2014, 6, 5559–5588. [Google Scholar] [CrossRef] [Green Version]
- McRoberts, R.E. Diagnostic tools for nearest neighbors techniques when used with satellite imagery. Remote Sens. Environ. 2009, 113, 489–499. [Google Scholar] [CrossRef]
- Ecosystem Classification Group. Ecological Regions of the Northwest Territories–Taiga Plains. Department of Environment and Natural Resources; Government of the Northwest Territories: Yellowknife, NT, Canada, 2007; (rev. 2009).
- Lambert, M.C.; Ung, C.H.; Raulier, F. Canadian national tree aboveground biomass equations. Can. J. For. Res. 2005, 35, 1996–2018. [Google Scholar] [CrossRef]
- Ung, C.H.; Bernier, P.; Guo, X.J. Canadian national biomass equations: New parameter estimates that include British Columbia data. Can. J. For. Res. 2008, 38, 1123–1132. [Google Scholar] [CrossRef]
- Beaudoin, A.; Bernier, P.; Villemaire, P.; Guindon, L.; Guo, X.J. Tracking forest attributes across Canada between 2001 and 2011 using a k nearest neighbors mapping approach applied to MODIS imagery. Can. J. For. Res. 2017, 48, 85–93. [Google Scholar] [CrossRef] [Green Version]
- Shimada, M.; Itoh, T.; Motooka, T.; Watanabe, M.; Shiraishi, T.; Thapa, R.; Lucas, R. New global forest/non-forest maps from ALOS PALSAR data (2007–2010). Remote Sens. Environ. 2014, 155, 13–31. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R.; et al. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natural Resources Canada. Canadian Digital Elevation Model: Product Specifications-Edition 1.1; Government of Canada: Sherbrooke, QC, Canada, 2016; p. 11.
- Hogg, E.H. Temporal scaling of moisture and the forest-grassland boundary in western Canada. Agric. For. Meteorol. 1997, 84, 115–122. [Google Scholar] [CrossRef] [Green Version]
- Hogg, E.H.; Barr, A.G.; Black, T.A. A simple soil moisture index for representing multi-year drought impacts on aspen productivity in the western Canadian interior. Agric. For. Meteorol. 2013, 178, 173–182. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, S.; Takahashi, T.; Nishizono, T.; Kitahara, F.; Saito, H.; Iehara, T.; Kodani, E.; Awaya, Y. Stand volume estimation using the k-NN technique combined with forest inventory data, satellite image data and additional feature variables. Remote Sens. 2015, 7, 378–394. [Google Scholar] [CrossRef] [Green Version]
- Wulder, M.A.; White, J.C.; Cranny, M.; Hall, R.J.; Luther, J.E.; Beaudoin, A.; Goodenough, D.G.; Dechka, J.A. Monitoring Canada’s forests. Part 1: Completion of the EOSD land cover project. Can. J. Remote Sens. 2008, 34, 549–562. [Google Scholar] [CrossRef]
- Gillis, M.D.; Omule, A.Y.; Brierley, T. Monitoring Canada’s forests: The National Forest Inventory. For. Chron. 2005, 81, 214–221. [Google Scholar] [CrossRef]
- National Forest Inventory. Canada’s National Forest Inventory-National Standard for Ground Plots: Data Dictionary, version 5.1.7; Available online: https://nfi.nfis.org/resources/groundplot/4a-GPDataDictionary5.2.2.pdf (accessed on 25 September 2017).
- Hopkinson, C.; Wulder, M.; Coops, N.; Milne, T.; Fox, A.; Bater, C. Airborne lidar sampling of the Canadian boreal forest: Planning, execution & initial processing. In Proceedings of the 11th International Conference on LiDAR Applications for Assessing Forest Ecosystems, SilviLaser 2011, Hobart, Australia, 16–20 October 2011. [Google Scholar]
- Wulder, M.A.; White, J.C.; Bater, C.W.; Coops, N.C.; Hopkinson, C.; Chen, G. Lidar plots—A new large-area data collection option: Context, concepts, and case study. Can. J. Remote Sens. 2012, 38, 600–618. [Google Scholar] [CrossRef]
- Wang, J.A.; Baccini, A.; Farina, M.; Randerson, J.T.; Friedl, M.A. Disturbance suppresses the aboveground carbon sink in North American boreal forests. Nat. Clim. Change 2021, 11, 435–441. [Google Scholar] [CrossRef]
- Luo, Y.; Trishchenko, A.; Khlopenkov, K. Developing clear-sky, cloud and cloud shadow mask for producing clear-sky composites at 250-meter spatial resolution for the seven MODIS land bands over Canada and North America. Remote Sens. Environ. 2008, 112, 4167–4185. [Google Scholar] [CrossRef]
- Brown, L.; Chen, J.M.; Leblanc, S.G.; Cihlar, J. A shortwave infrared modification to the simple ratio for LAI retrieval in boreal forests: An image and model analysis. Remote Sens. Environ. 2000, 71, 16–25. [Google Scholar] [CrossRef]
- Touzi, R. A review of speckle filtering in the context of estimation theory. IEEE Trans. Geosci. Remote Sens. 2002, 40, 2392–2404. [Google Scholar] [CrossRef]
- Tarboton, D.G. A new method for the determination of flow directions and contributing areas in grid digital elevation models. Water Resour. Res. 1997, 33, 309–319. [Google Scholar] [CrossRef] [Green Version]
- Crookston, N.L.; Finley, A.O.; Coulston, J. Nearest Neighbor Observation Imputation and Evaluation Tools [Online]. 2015 version. Available online: https://cran.r-project.org/web/packages/yaImpute/yaImpute.pdf (accessed on 23 March 2016).
- Crookston, N.L.; Finley, A.O. yaImpute: An R package for k-NN imputation. J. Stat. Softw. 2008, 23, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Mount, D.M.; Arya, S. ANN: A Library for Approximate Nearest Neighbor Searching. 2010. Available online: http://www.cs.umd.edu/~mount/ANN/ (accessed on 15 January 2016).
- Bland, J.M.; Altman, D.G. Statistical methods for assessing agreement between two methods of clinical measurement. Lancet 1986, 1, 307–310. [Google Scholar] [CrossRef]
- Watson, P.F.; Petrie, A. Method agreement analysis: A review of correct methodology. Theriogenology 2010, 73, 1167–1179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, M.C.; Ulaby, F.T.; LeToan, T.; Beaudoin, A.; Kasischke, E.S.; Christensen, N. Dependence of radar backscatter on coniferous forest biomass. IEEE Trans. Geosci. Remote Sens. 1992, 30, 412–415. [Google Scholar] [CrossRef]
- Bell, D.M.; Gregory, M.J.; Kane, V.; Kane, J.; Kennedy, R.E.; Roberts, H.M.; Yang, Z. Multiscale divergence between Landsat- and lidar-based biomass mapping is related to regional variation in canopy cover and composition. Carbon Balance Manag. 2018, 13, 15. [Google Scholar] [CrossRef] [Green Version]
- Latifi, H.; Nothdurft, A.; Koch, B. Non-parametric prediction and mapping of standing timber volume and biomass in a temperate forest: Application of multiple optical/LiDAR-derived predictors. Forestry 2010, 83, 395–407. [Google Scholar] [CrossRef] [Green Version]
- Shataee, S.; Kalbi, S.; Fallah, A.; Pelz, D. Forest attribute imputation using machine-learning methods and ASTER data: Comparison of k-NN, SVR and random forest regression algorithms. Int. J. Remote Sens. 2012, 33, 6254–6280. [Google Scholar] [CrossRef]
- Neuenschwander, A.L.; Magruder, L.A. Canopy and terrain height retrievals with ICESat-2: A first look. Remote Sens. 2019, 11, 1721. [Google Scholar] [CrossRef] [Green Version]
- Narine, L.L.; Popescu, S.C.; Malambo, L. Using ICESat-2 to estimate and map forest aboveground biomass: A first example. Remote Sens. 2020, 12, 1824. [Google Scholar] [CrossRef]
- Duncanson, L.; Neuenschwander, A.; Hancock, S.; Thomas, N.; Fatoyinbo, T.; Simard, M.; Silva, C.A.; Armston, J.; Luthcke, S.B.; Hofton, M.; et al. Biomass estimation from simulated GEDI, ICESat-2 and NISAR across environmental gradients in Sonoma County, California. Remote Sens. Environ. 2020, 242, 111779. [Google Scholar] [CrossRef]
- Claverie, M.; Ju, J.; Masek, J.G.; Dungan, J.L.; Vermote, E.F.; Roger, J.C.; Skakun, S.V.; Justice, C. The Harmonized Landsat and Sentinel-2 surface reflectance data set. Remote Sens. Environ. 2018, 219, 145–161. [Google Scholar] [CrossRef]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Matasci, G.; Hermosilla, T.; Wulder, M.A.; White, J.C.; Coops, N.C.; Hobart, G.W.; Bolton, D.K.; Tompalski, P.; Bater, C.W. Three decades of forest structural dynamics over Canada’s forested ecosystems using Landsat time-series and lidar plots. Remote Sens. Environ. 2018, 216, 697–714. [Google Scholar] [CrossRef]



| Feature Category | Description | Label | Units | Year | Pixel Size |
|---|---|---|---|---|---|
| Landsat TM spectral bands, indices and texture (LANDSAT) | Blue band TOA a reflectance | B1 | - | 2006–2008 | 30 m |
| Green band TOA reflectance | B2 | - | |||
| Red band TOA reflectance | B3 * | - | |||
| Near-infrared band TOA reflectance | B4 * | - | |||
| Short-wave infrared band TOA reflectance | B5 | - | |||
| Short-wave infrared band TOA reflectance | B7 * | - | |||
| Normalized Difference Vegetation Index (B4 − B3)/(B4 + B3) | NDVI | - | |||
| Reduced Simple Ratio (B4/B3) ∗ (B5max − B5)/(B5range) | RSR * | - | |||
| Normalized Difference Moisture Index (B4 − B5)/(B4 + B5) | NDMI | - | |||
| Texture: 3 × 3 variance of near-infrared band | B4_TEX | - | |||
| PALSAR dual-polarized backscatter and texture (PALSAR) | HH-polarized L-band backscatter intensity | HH | - | 2007 | 25 m |
| HV-polarized L-band backscatter intensity | HV * | - | |||
| HV/HH backscatter intensity ratio | HVHH | - | |||
| Texture: HH 9 × 9 CV b | HH_ TEX | - | |||
| Texture: HV 9 × 9 CV | HV_TEX | - | |||
| Texture: HV/HH 9 × 9 CV | HVHH_TEX | - | |||
| Environmental c | 2000 percent tree cover map updated to 2007 | TC * | % | 2007 | 30 m |
| Terrain elevation from CDED d | ELEV * | m | variable | 90 m | |
| Terrain slope from CDED | SLOPE * | deg | variable | 90 m | |
| Compound Topographic Index from CDED | CTI | - | variable | 90 m | |
| Average Soil Moisture Index | SMI * | mm | 2001–2010 | 100 m | |
| Average Climatic Moisture Index | CMI | cm | 2001–2010 | 100 m |
| Forest Attribute | Model and Parameters | Adj. R2 | RMSE |
|---|---|---|---|
| Lorey’s height (HL, m) a | HLGLAS = 2.46 + 0.91 × P85 b | 0.89 | 1.1 |
| Stand height (Ht, m) | HtGLAS = 2.30 + 1.10 × P85 | 0.88 | 1.3 |
| Crown closure (CC, %) | CCGLAS = 64.63 × Lz0.25 c | 0.54 | 6.5 |
| Stand volume (Vs, m3·ha−1) | VsGLAS = 0.61 × HtGLAS1.84 | 0.76 | 46.8 |
| Total volume (Vt, m3·ha−1) | VtGLAS = 1.84 × HLGLAS1.69 | 0.81 | 59.3 |
| Aboveground biomass (AGB, t·ha−1) | AGBGLAS = 2.27 × HLGLAS1.45 | 0.76 | 35.7 |
| (a) Reference Set | (b) Validation Sets | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GLAS | NFI a | BT−ALS | ||||||||||||||
| Attribute | Forest Type | n | Min | Max | Mean | SD b | n | Min | Max | Mean | SD | n | Min | Max | Mean | SD |
| Stand height (m) | ALL | 3600 | 3.6 | 34.1 | 9.7 | 5.9 | 31 | 5.0 | 31.5 | 14.8 | 7.0 | 1,080,866 | 2.5 | 35.0 | 11.6 | 6.1 |
| Conifer | 2459 | 3.6 | 33.6 | 8.8 | 4.8 | 19 | 6.5 | 29.7 | 12.9 | 6.6 | 831,619 | 2.5 | 34.8 | 10.0 | 5.0 | |
| Mixedwood | 528 | 3.7 | 34.1 | 13.5 | 7.4 | 7 | 12.0 | 31.5 | 19.7 | 7.0 | 146,738 | 2.6 | 34.9 | 16.5 | 6.7 | |
| Broadleaf | 219 | 3.7 | 34.0 | 15.7 | 8.3 | 5 | 5.0 | 20.4 | 14.9 | 6.1 | 102,509 | 2.6 | 35.0 | 17.4 | 6.1 | |
| AGB (t·ha−1) | ALL | 3600 | 1.2 | 352.1 | 54.2 | 51.6 | 30 | 4.5 | 300.1 | 85.4 | 77.0 | 1,080,734 | 7.9 | 326.4 | 72.1 | 55.1 |
| Conifer | 2459 | 15.1 | 286.5 | 49.2 | 38.6 | 18 | 7.6 | 195.8 | 64.4 | 59.6 | 831,499 | 7.9 | 324.4 | 57.9 | 43.5 | |
| Mixedwood | 528 | 15.9 | 292.6 | 87.2 | 64.8 | 7 | 26.7 | 300.1 | 147.7 | 100.8 | 146,726 | 8.2 | 325.9 | 116.3 | 64.3 | |
| Broadleaf | 219 | 15.9 | 290.6 | 107.0 | 73.1 | 5 | 4.5 | 127.5 | 74.0 | 61.2 | 102,509 | 8.3 | 326.4 | 124.3 | 59.8 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Beaudoin, A.; Hall, R.J.; Castilla, G.; Filiatrault, M.; Villemaire, P.; Skakun, R.; Guindon, L. Improved k-NN Mapping of Forest Attributes in Northern Canada Using Spaceborne L-Band SAR, Multispectral and LiDAR Data. Remote Sens. 2022, 14, 1181. https://doi.org/10.3390/rs14051181
Beaudoin A, Hall RJ, Castilla G, Filiatrault M, Villemaire P, Skakun R, Guindon L. Improved k-NN Mapping of Forest Attributes in Northern Canada Using Spaceborne L-Band SAR, Multispectral and LiDAR Data. Remote Sensing. 2022; 14(5):1181. https://doi.org/10.3390/rs14051181
Chicago/Turabian StyleBeaudoin, André, Ronald J. Hall, Guillermo Castilla, Michelle Filiatrault, Philippe Villemaire, Rob Skakun, and Luc Guindon. 2022. "Improved k-NN Mapping of Forest Attributes in Northern Canada Using Spaceborne L-Band SAR, Multispectral and LiDAR Data" Remote Sensing 14, no. 5: 1181. https://doi.org/10.3390/rs14051181
APA StyleBeaudoin, A., Hall, R. J., Castilla, G., Filiatrault, M., Villemaire, P., Skakun, R., & Guindon, L. (2022). Improved k-NN Mapping of Forest Attributes in Northern Canada Using Spaceborne L-Band SAR, Multispectral and LiDAR Data. Remote Sensing, 14(5), 1181. https://doi.org/10.3390/rs14051181








