Retrieval of Crop Variables from Proximal Multispectral UAV Image Data Using PROSAIL in Maize Canopy
Abstract
:1. Introduction
- How well can LAI, LCC and CCC be retrieved from high-resolution UAV multispectral image data for complex canopies such as maize?
- Which inversion scheme, mean reflectance (applying the scheme to a single spectrum averaged per plot) or pixel-based approach (applying the scheme to every pixel and then averaging), leads to more accurate results?
- How does the retrieval accuracy vary within the growing season for different growth stages?
2. Materials and Methods
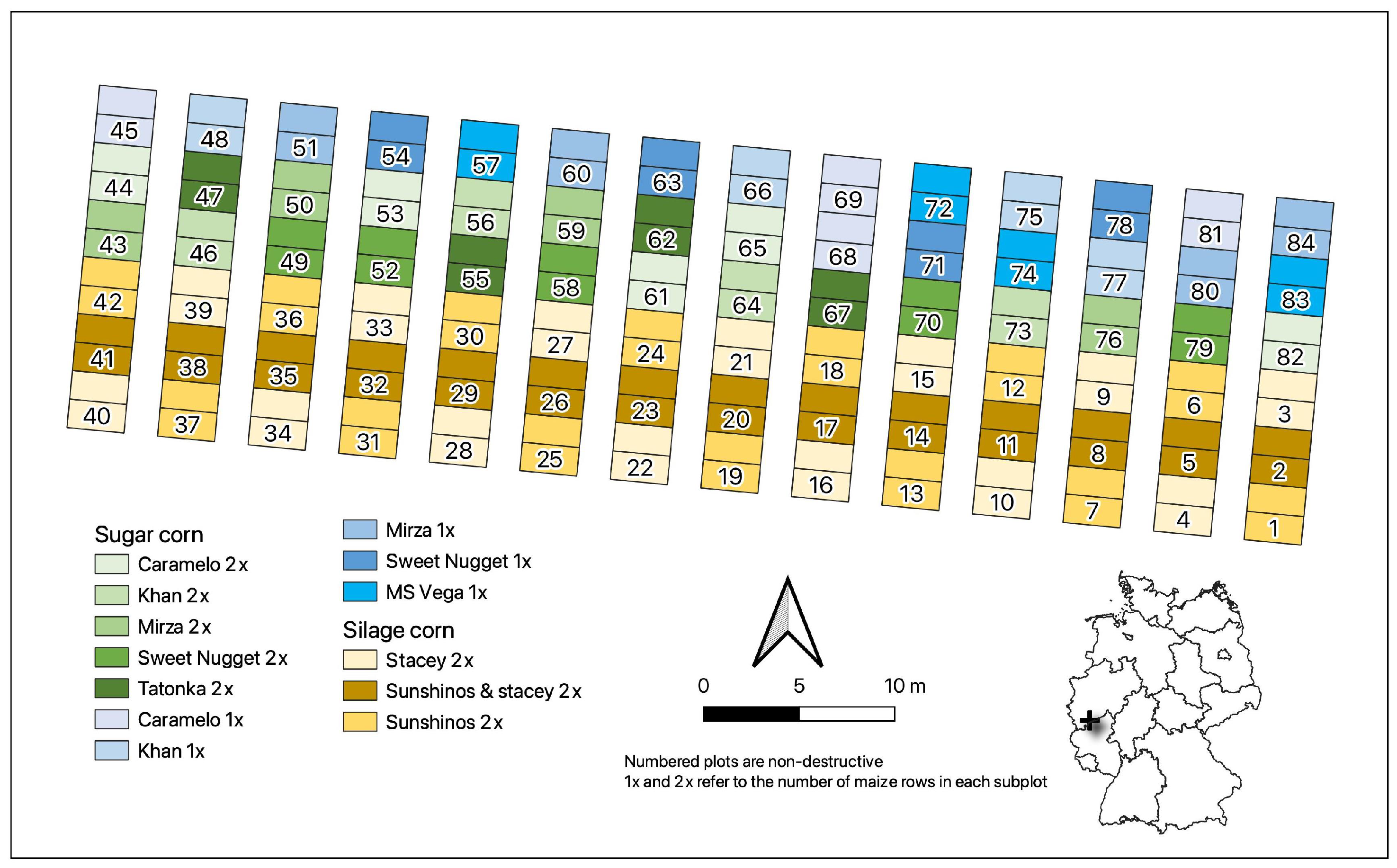
2.1. Study Area
2.2. Aerial Campaigns
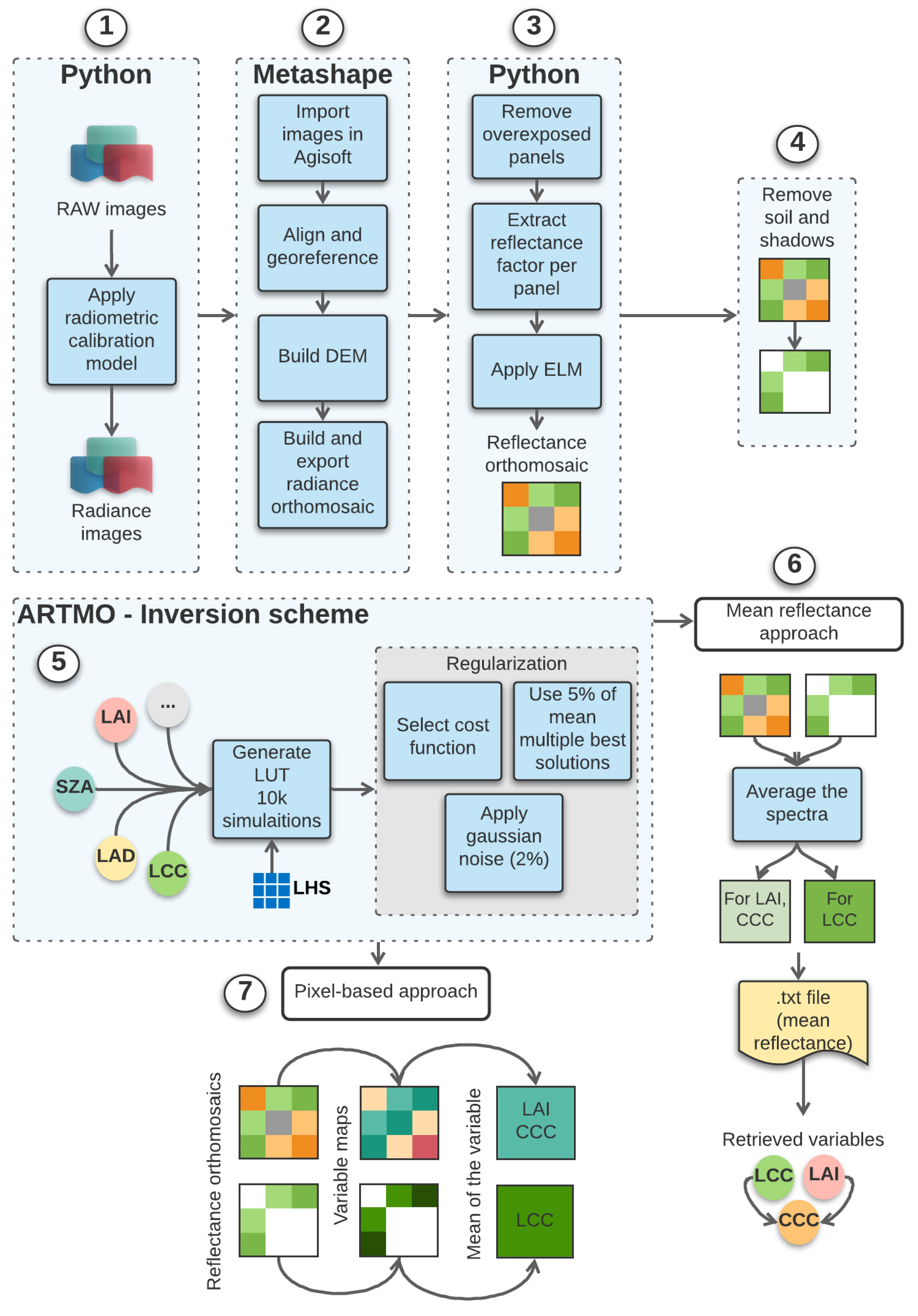
2.3. Image Processing
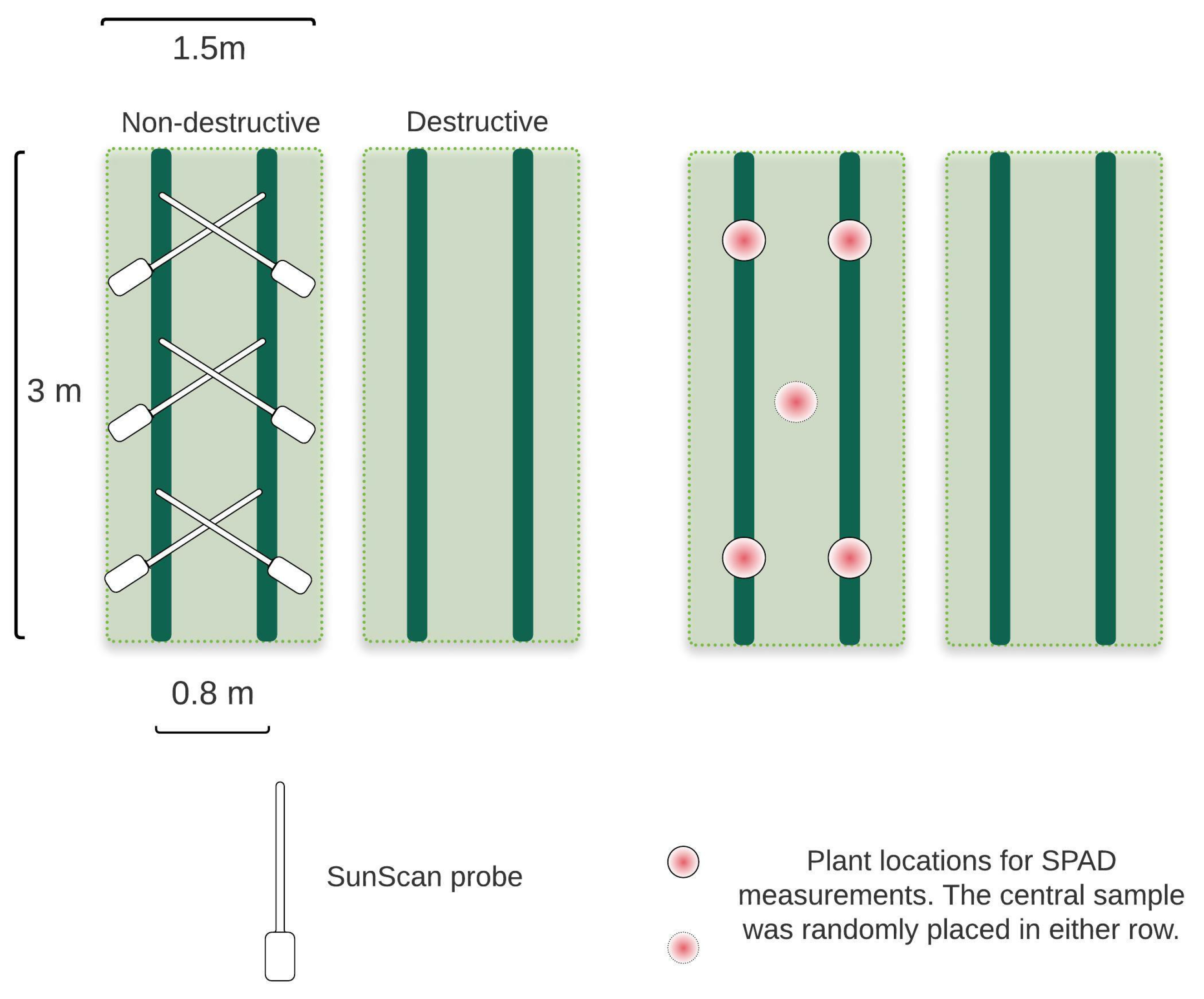
2.4. Field Measurements
2.5. LUT-Based PROSAIL Inversion
2.6. Statistical Analysis
3. Results
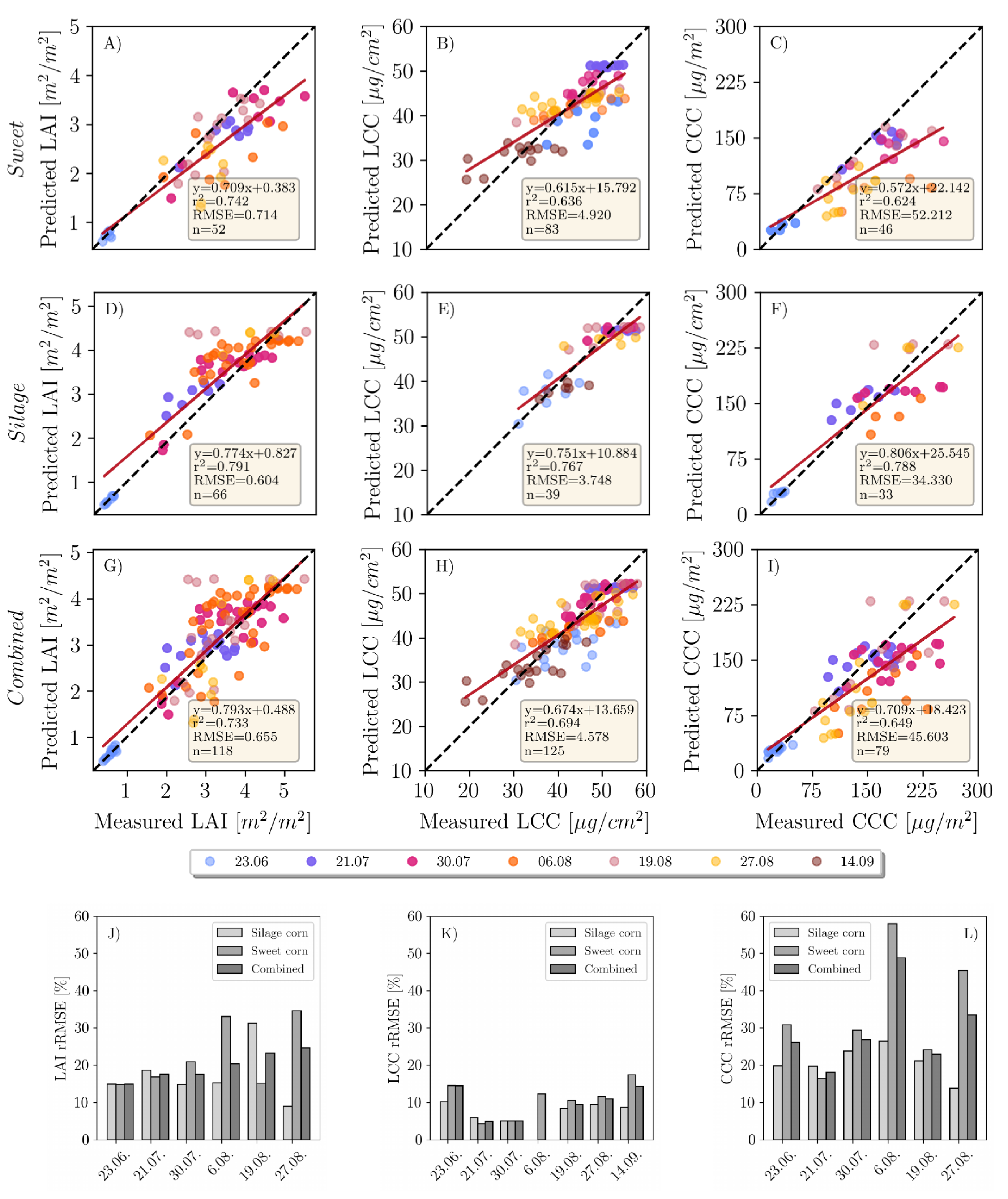
3.1. Variable Retrieval
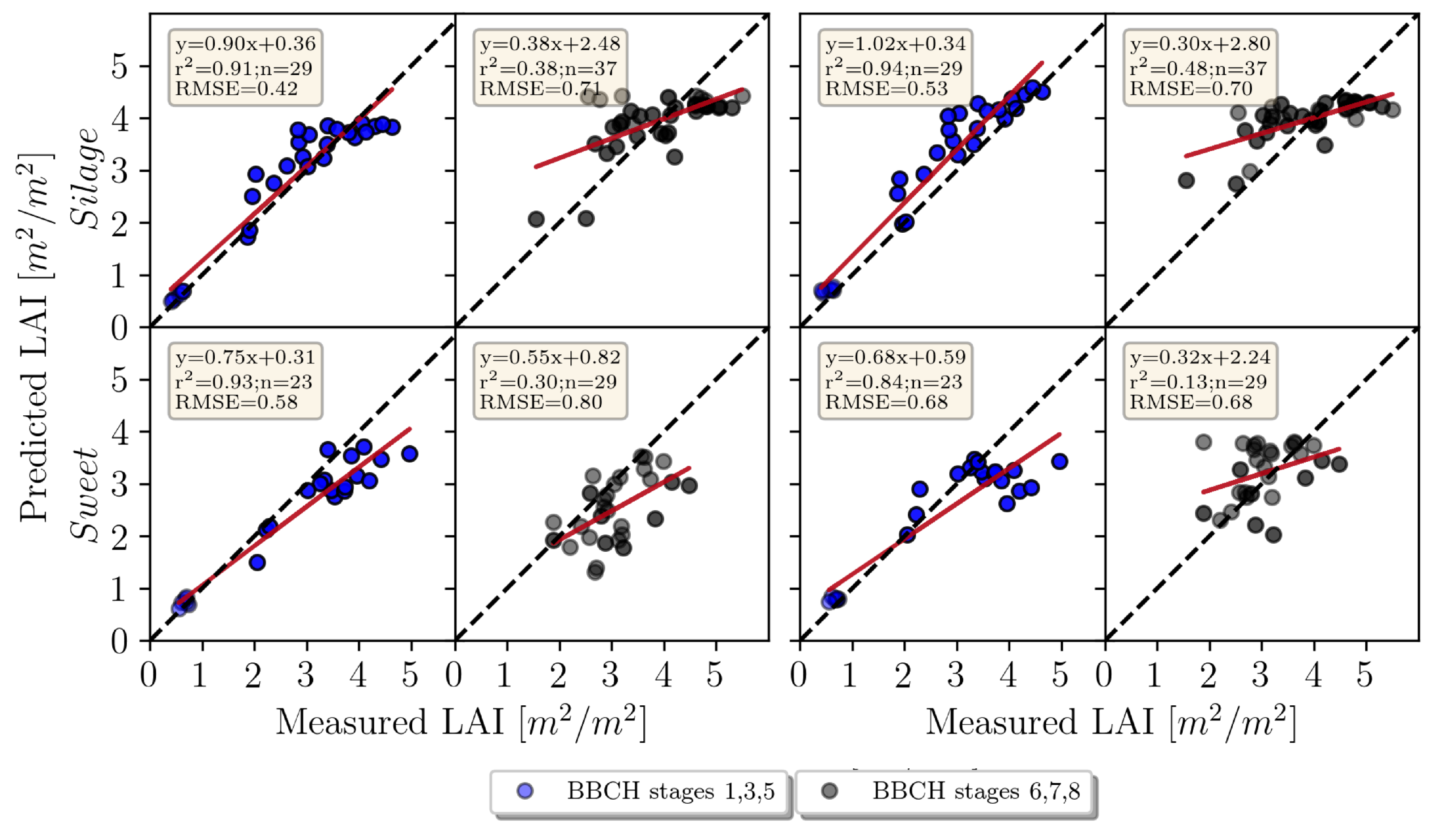
3.1.1. LAI
3.1.2. LCC
3.1.3. CCC
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A

References
- Sankaran, S.; Khot, L.R.; Espinoza, C.Z.; Jarolmasjed, S.; Sathuvalli, V.R.; Vandemark, G.J.; Miklas, P.N.; Carter, A.H.; Pumphrey, M.O.; Knowles, N.R.; et al. Low-altitude, high-resolution aerial imaging systems for row and field crop phenotyping: A review. Eur. J. Agron. 2015, 70, 112–123. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Holman, F.H.; Riche, A.B.; Michalski, A.; Castle, M.; Wooster, M.J.; Hawkesford, M.J. High Throughput Field Phenotyping of Wheat Plant Height and Growth Rate in Field Plot Trials Using UAV Based Remote Sensing. Remote Sens. 2016, 8, 1031. [Google Scholar] [CrossRef]
- Watt, M.; Fiorani, F.; Usadel, B.; Rascher, U.; Muller, O.; Schurr, U. Phenotyping: New Windows into the Plant for Breeders. Annu. Rev. Plant Biol. 2020, 71, 689–712. [Google Scholar] [CrossRef] [PubMed]
- Asrar, G.; Fuchs, M.; Kanemasu, E.T.; Hatfield, J.L. Estimating Absorbed Photosynthetic Radiation and Leaf Area Index from Spectral Reflectance in Wheat1. Agron. J. 1984, 76, 300–306. [Google Scholar] [CrossRef]
- Asner, G.P.; Scurlock, J.M.; A. Hicke, J. Global synthesis of leaf area index observations: Implications for ecological and remote sensing studies. Glob. Ecol. Biogeogr. 2003, 12, 191–205. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.A.; Viña, A.; Ciganda, V.; Rundquist, D.C.; Arkebauer, T.J. Remote estimation of canopy chlorophyll content in crops. Geophys. Res. Lett. 2005, 32. [Google Scholar] [CrossRef] [Green Version]
- Clevers, J.G.P.W.; Kooistra, L. Using Hyperspectral Remote Sensing Data for Retrieving Canopy Chlorophyll and Nitrogen Content. IEEE J.-STARS 2012, 5, 574–583. [Google Scholar] [CrossRef]
- Croft, H.; Chen, J.M.; Luo, X.; Bartlett, P.; Chen, B.; Staebler, R.M. Leaf chlorophyll content as a proxy for leaf photosynthetic capacity. Glob. Chang. Biol. 2017, 23, 3513–3524. [Google Scholar] [CrossRef]
- Verrelst, J.; Malenovský, Z.; Van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.P.; Lewis, P.; North, P.; Moreno, J. Quantifying Vegetation Biophysical Variables from Imaging Spectroscopy Data: A Review on Retrieval Methods. Surv. Geophys. 2018, 40, 589–629. [Google Scholar] [CrossRef] [Green Version]
- Gitelson, A.A. Wide Dynamic Range Vegetation Index for Remote Quantification of Biophysical Characteristics of Vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kross, A.; McNairn, H.; Lapen, D.; Sunohara, M.; Champagne, C. Assessment of RapidEye vegetation indices for estimation of leaf area index and biomass in corn and soybean crops. Int. J. Appl. Earth. Obs. Geoinf. 2015, 34, 235–248. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Muñoz, J.; Alonso, L.; Delegido, J.; Rivera, J.P.; Camps-Valls, G.; Moreno, J. Machine learning regression algorithms for biophysical parameter retrieval: Opportunities for Sentinel-2 and -3. Remote Sens. Environ. 2012, 118, 127–139. [Google Scholar] [CrossRef]
- Siegmann, B.; Jarmer, T. Comparison of different regression models and validation techniques for the assessment of wheat leaf area index from hyperspectral data. Int. J. Remote Sens. 2015, 36, 4519–4534. [Google Scholar] [CrossRef]
- Vohland, M.; Mader, S.; Dorigo, W. Applying different inversion techniques to retrieve stand variables of summer barley with PROSPECT + SAIL. Int. J. Appl. Earth Obs. Geoinf. 2010, 12, 71–80. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Berger, K.; Atzberger, C.; Vuolo, F.; Weihs, P.; D’Urso, G. Experimental assessment of the Sentinel-2 band setting for RTM-based LAI retrieval of sugar beet and maize. Can. J. Remote Sens. 2009, 35, 230–247. [Google Scholar]
- Walthall, C.; Dulaney, W.; Anderson, M.; Norman, J.; Fang, H.; Liang, S. A comparison of empirical and neural network approaches for estimating corn and soybean leaf area index from Landsat ETM+ imagery. Remote Sens. Environ. 2004, 92, 465–474. [Google Scholar] [CrossRef]
- Malenovskỳ, Z.; Homolová, L.; Lukeš, P.; Buddenbaum, H.; Verrelst, J.; Alonso, L.; Schaepman, M.E.; Lauret, N.; Gastellu-Etchegorry, J.P. Variability and uncertainty challenges in scaling imaging spectroscopy retrievals and validations from leaves up to vegetation canopies. Surv. Geophys. 2019, 40, 631–656. [Google Scholar] [CrossRef]
- Gastellu-Etchegorry, J.P.; Lauret, N.; Yin, T.; Landier, L.; Kallel, A.; Malenovský, Z.; Bitar, A.A.; Aval, J.; Benhmida, S.; Qi, J.; et al. DART: Recent Advances in Remote Sensing Data Modeling With Atmosphere, Polarization, and Chlorophyll Fluorescence. IEEE J.-STARS 2017, 10, 2640–2649. [Google Scholar] [CrossRef]
- Miraglio, T.; Adeline, K.; Huesca, M.; Ustin, S.; Briottet, X. Monitoring LAI, Chlorophylls, and Carotenoids Content of a Woodland Savanna Using Hyperspectral Imagery and 3D Radiative Transfer Modeling. Remote Sens. 2020, 12, 28. [Google Scholar] [CrossRef] [Green Version]
- Janoutová, R.; Homolová, L.; Novotný, J.; Navrátilová, B.; Pikl, M.; Malenovský, Z. Detailed reconstruction of trees from terrestrial laser scans for remote sensing and radiative transfer modelling applications. Silico Plants 2021, 3, diab026. [Google Scholar] [CrossRef]
- Féret, J.B.; Gitelson, A.; Noble, S.; Jacquemoud, S. PROSPECT-D: Towards modeling leaf optical properties through a complete lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef] [Green Version]
- Verhoef, W.; Jia, L.; Xiao, Q.; Su, Z. Unified optical-thermal four-stream radiative transfer theory for homogeneous vegetation canopies. IEEE Trans. Geosci. Remote Sens. 2007, 45, 1808–1822. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Verhoef, W.; Baret, F.; Bacour, C.; Zarco-Tejada, P.J.; Asner, G.P.; François, C.; Ustin, S.L. PROSPECT + SAIL models: A review of use for vegetation characterization. Remote Sens. Environ. 2009, 113, S56–S66. [Google Scholar] [CrossRef]
- Berger, K.; Atzberger, C.; Danner, M.; D’Urso, G.; Mauser, W.; Vuolo, F.; Hank, T. Evaluation of the PROSAIL model capabilities for future hyperspectral model environments: A review study. Remote Sens. 2018, 10, 85. [Google Scholar] [CrossRef] [Green Version]
- Baret, F.; Buis, S. Estimating Canopy Characteristics from Remote Sensing Observations: Review of Methods and Associated Problems. In Advances in Land Remote Sensing: System, Modeling, Inversion and Application; Springer: Dordrecht, The Netherlands, 2008; pp. 173–201. [Google Scholar]
- Weiss, M.; Jacob, F.; Duveiller, G. Remote sensing for agricultural applications: A meta-review. Remote Sens. Environ. 2020, 236, 111402. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C. Inversion of a radiative transfer model for estimating vegetation LAI and chlorophyll in a heterogeneous grassland. Remote Sens. Environ. 2008, 112, 2592–2604. [Google Scholar] [CrossRef]
- Danner, M.; Berger, K.; Wocher, M.; Mauser, W.; Hank, T. Retrieval of Biophysical Crop Variables from Multi-Angular Canopy Spectroscopy. Remote Sens. 2017, 9, 726. [Google Scholar] [CrossRef] [Green Version]
- Banskota, A.; Serbin, S.P.; Wynne, R.H.; Thomas, V.A.; Falkowski, M.J.; Kayastha, N.; Gastellu-Etchegorry, J.P.; Townsend, P.A. An LUT-Based Inversion of DART Model to Estimate Forest LAI from Hyperspectral Data. IEEE J.-STARS 2015, 8, 3147–3160. [Google Scholar] [CrossRef]
- Kattenborn, T.; Fassnacht, F.E.; Pierce, S.; Lopatin, J.; Grime, J.P.; Schmidtlein, S. Linking plant strategies and plant traits derived by radiative transfer modelling. J. Veg. Sci. 2017, 28, 717–727. [Google Scholar] [CrossRef]
- Koetz, B.; Baret, F.; Poilvé, H.; Hill, J. Use of coupled canopy structure dynamic and radiative transfer models to estimate biophysical canopy characteristics. Remote Sens. Environ. 2005, 95, 115–124. [Google Scholar] [CrossRef]
- Roosjen, P.P.; Brede, B.; Suomalainen, J.M.; Bartholomeus, H.M.; Kooistra, L.; Clevers, J.G. Improved estimation of leaf area index and leaf chlorophyll content of a potato crop using multi-angle spectral data – potential of unmanned aerial vehicle imagery. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 14–26. [Google Scholar] [CrossRef]
- Verger, A.; Vigneau, N.; Chéron, C.; Gilliot, J.M.; Comar, A.; Baret, F. Green area index from an unmanned aerial system over wheat and rapeseed crops. Remote Sens. Environ. 2014, 152, 654–664. [Google Scholar] [CrossRef]
- Jay, S.; Baret, F.; Dutartre, D.; Malatesta, G.; Héno, S.; Comar, A.; Weiss, M.; Maupas, F. Exploiting the centimeter resolution of UAV multispectral imagery to improve remote-sensing estimates of canopy structure and biochemistry in sugar beet crops. Remote Sens. Environ. 2019, 231, 110898. [Google Scholar] [CrossRef]
- Abdelbaki, A.; Schlerf, M.; Retzlaff, R.; Machwitz, M.; Verrelst, J.; Udelhoven, T. Comparison of Crop Trait Retrieval Strategies Using UAV-Based VNIR Hyperspectral Imaging. Remote Sens. 2021, 13, 1748. [Google Scholar] [CrossRef]
- Duan, S.B.; Li, Z.L.; Wu, H.; Tang, B.H.; Ma, L.; Zhao, E.; Li, C. Inversion of the PROSAIL model to estimate leaf area index of maize, potato, and sunflower fields from unmanned aerial vehicle hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 12–20. [Google Scholar] [CrossRef]
- Sun, B.; Wang, C.; Yang, C.; Xu, B.; Zhou, G.; Li, X.; Xie, J.; Xu, S.; Liu, B.; Xie, T.; et al. Retrieval of rapeseed leaf area index using the PROSAIL model with canopy coverage derived from UAV images as a correction parameter. Int. J. Appl. Earth Obs. Geoinf. 2021, 102, 102373. [Google Scholar] [CrossRef]
- Chakhvashvili, E.; Siegmann, B.; Bendig, J.; Rascher, U. Comparison of Reflectance Calibration Workflows for a UAV-Mounted Multi-Camera Array System. In Proceedings of the 2021 IEEE International Geoscience and Remote Sensing Symposium IGARSS, Brussels, Belgium , 11–16 July 2021; pp. 8225–8228. [Google Scholar]
- RedEdge Camera Radiometric Calibration Model. Available online: https://support.micasense.com/hc/en-us/articles/115000351194-RedEdge-Camera-Radiometric-Calibration-Model (accessed on 15 October 2021).
- MicaSense RedEdge and Altum Image Processing Tutorials. Available online: https://github.com/micasense/imageprocessing (accessed on 15 October 2021).
- Smith, G.M.; Milton, E.J. The use of the empirical line method to calibrate remotely sensed data to reflectance. Int. J. Remote Sens. 1999, 20, 2653–2662. [Google Scholar] [CrossRef]
- Escadafal, R.; Belghith, A.; Ben Moussa, H. Indices spectraux pour la teledetection de la degradation des milieux naturels en tunisie aride. In Proceedings of the 6th Symposium International Mesures Physiques et Signatures en Teledetection, ISPRS-CNES, Paris, France, 17–21 January 1994; pp. 253–259. [Google Scholar]
- Matias, F.I.; Caraza-Harter, M.V.; Endelman, J.B. FIELDimageR: An R package to analyze orthomosaic images from agricultural field trials. Plant Phenome J. 2020, 3, e20005. [Google Scholar] [CrossRef]
- Geipel, J.; Link, J.; Claupein, W. Combined spectral and spatial modeling of corn yield based on aerial images and crop surface models acquired with an unmanned aircraft system. Remote Sens. 2014, 6, 10335–10355. [Google Scholar] [CrossRef] [Green Version]
- User Manual for the SunScan Canopy Analysis System Type SS1. Available online: https://delta-t.co.uk/wp-content/uploads/2017/02/SSI-UM_v3.3.pdf (accessed on 27 October 2021).
- Campbell, G.S.; Norman, J.M. The light environment of plant canopies. In An Introduction to Environmental Biophysics; Springer: Berlin/Heidelberg, Germany, 1998; pp. 247–278. [Google Scholar]
- Verrelst, J.; Romijn, E.; Kooistra, L. Mapping Vegetation Density in a Heterogeneous River Floodplain Ecosystem Using Pointable CHRIS/PROBA Data. Remote Sen. 2012, 4, 2866–2889. [Google Scholar] [CrossRef] [Green Version]
- Verhoef, W. Light scattering by leaf layers with application to canopy reflectance modeling: The SAIL model. Remote Sens. Environ. 1984, 16, 125–141. [Google Scholar] [CrossRef] [Green Version]
- Berger, K.; Atzberger, C.; Danner, M.; Wocher, M.; Mauser, W.; Hank, T. Model-Based Optimization of Spectral Sampling for the Retrieval of Crop Variables with the PROSAIL Model. Remote Sens. 2018, 10, 2063. [Google Scholar] [CrossRef] [Green Version]
- Sacks, J.; Welch, W.J.; Mitchell, T.J.; Wynn, H.P. Design and analysis of computer experiments. Stat. Sci. 1989, 4, 409–423. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Su, W.; Zhang, M.; Bian, D.; Liu, Z.; Huang, J.; Wang, W.; Wu, J.; Guo, H. Phenotyping of Corn Plants Using Unmanned Aerial Vehicle (UAV) Images. Remote Sens. 2019, 11, 2021. [Google Scholar] [CrossRef] [Green Version]
- España, M.; Baret, F.; Chelle, M.; Aries, F.; Andrieu, B. A dynamic model of maize 3D architecture: Application to the parameterisation of the clumpiness of the canopy. Agronomie 1998, 18, 609–626. [Google Scholar] [CrossRef]
- Houborg, R.; McCabe, M.; Cescatti, A.; Gao, F.; Schull, M.; Gitelson, A. Joint leaf chlorophyll content and leaf area index retrieval from Landsat data using a regularized model inversion system (REGFLEC). Remote Sens. Environ. 2015, 159, 203–221. [Google Scholar] [CrossRef] [Green Version]
- Atzberger, C.; Berger, K. Spatially constrained inversion of radiative transfer models for improved LAI mapping from future Sentinel-2 imagery. Remote Sens. Environ. 2012, 120, 208–218. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Bacour, C.; Poilvé, H.; Frangi, J.P. Comparison of Four Radiative Transfer Models to Simulate Plant Canopies Reflectance: Direct and Inverse Mode. Remote Sens. Environ. 2000, 74, 471–481. [Google Scholar] [CrossRef]
- Delegido, J.; Vergara, C.; Verrelst, J.; Gandia, S.; Moreno, J. Remote Estimation of Crop Chlorophyll Content by Means of High-Spectral-Resolution Reflectance Techniques. Agron. J. 2011, 103, 1834–1842. [Google Scholar] [CrossRef]
- Campbell, R.J.; Mobley, K.N.; Marini, R.P.; Pfeiffer, D.G. Growing Conditions Alter the Relationship Between SPAD-501 Values and Apple Leaf Chlorophyll. HortScience 1990, 25, 330–331. [Google Scholar] [CrossRef] [Green Version]
- Richardson, A.D.; Duigan, S.P.; Berlyn, G.P. An evaluation of noninvasive methods to estimate foliar chlorophyll content. New Phytol. 2002, 153, 185–194. [Google Scholar] [CrossRef] [Green Version]
- Berger, K.; Verrelst, J.; Féret, J.B.; Hank, T.; Wocher, M.; Mauser, W.; Camps-Valls, G. Retrieval of aboveground crop nitrogen content with a hybrid machine learning method. Int. J. Appl. Earth Obs. Geoinf. 2020, 92, 102174. [Google Scholar] [CrossRef]
- Bailey, B.N.; Mahaffee, W.F. Rapid measurement of the three-dimensional distribution of leaf orientation and the leaf angle probability density function using terrestrial LiDAR scanning. Remote Sens. Environ. 2017, 194, 63–76. [Google Scholar] [CrossRef] [Green Version]


| Band Names | Centre Wavelength [nm] | Bandwidth [nm] |
|---|---|---|
| Blue444 | 444 | 28 |
| Blue475 | 475 | 32 |
| Green531 | 531 | 14 |
| Green560 | 560 | 27 |
| Red650 | 650 | 16 |
| Red668 | 668 | 14 |
| RE705 | 705 | 10 |
| RE717 | 717 | 12 |
| RE740 | 740 | 18 |
| NIR840 | 842 | 57 |
| Date | Flight Time (CEST) | SZA | BBCH | Weather Conditions |
|---|---|---|---|---|
| 23 June | 13:08–13:25 | 27.81° | 13–34 | sunny |
| 14 July | 13:03–13:21 | 29.52° | 39–55 | partially sunny, few clouds |
| 21 July | 13:21–13:38 | 30.73° | - | sunny |
| 30 July | 13:20–13:37 | 32.71° | 53–65 | sunny, few clouds |
| 6 August | 13:19–13:40 | 34.55° | 65–68 | sunny, few clouds |
| 19 August | 13:35–13:53 | 38.55° | 68–71 | sunny, few clouds |
| 27 August | 13:17–13:33 | 41.33° | 71–73 | sunny, few clouds |
| 14 September | 13:27–13:40 | 48.14° | 83–87 | sunny, few clouds |
| Variable | Maize Type | Stat | 23.06 | 21.07 | 30.07 | 6.08 | 19.08 | 27.08 | 14.09 |
|---|---|---|---|---|---|---|---|---|---|
| LCC [ g/cm2] | Sweet | n | 11 | 11 | 11 | 11 | 6 | 20 | 13 |
| Min | 39.0 | 46.9 | 41.8 | 34.2 | 30.3 | 31.6 | 19.0 | ||
| Max | 53.1 | 54.3 | 53.2 | 54.7 | 53.5 | 53.4 | 42.6 | ||
| Mean | 47.6 | 50.4 | 45.9 | 43.4 | 43.2 | 43.1 | 30.7 | ||
| Stdev | 5.1 | 2.2 | 3.5 | 6.8 | 6.6 | 6.2 | 6.5 | ||
| CV | 0.11 | 0.04 | 0.08 | 0.16 | 0.15 | 0.15 | 0.21 | ||
| Silage | n | 7 | 6 | 7 | - | 7 | 6 | 6 | |
| Min | 30.6 | 48.9 | 46.1 | 35.5 | 41.9 | 40.8 | 35.3 | ||
| Max | 44.2 | 56.9 | 56.5 | 56.8 | 57.9 | 61.4 | 53.6 | ||
| Mean | 38.1 | 52.8 | 52.2 | 46.0 | 50.2 | 50.4 | 42.6 | ||
| Stdev | 6.0 | 3.0 | 3.6 | 7.8 | 5.8 | 5.6 | 5.4 | ||
| CV | 0.16 | 0.06 | 0.07 | 0.17 | 0.12 | 0.11 | 0.13 | ||
| LAI [] | Sweet | n | 6 | 8 | 9 | 8 | 14 | 7 | - |
| Min | 0.5 | 1.5 | 2.1 | 1.9 | 1.4 | 1.6 | 1.4 | ||
| Max | 1.6 | 3.7 | 5.0 | 5.5 | 5.1 | 3.2 | 3.9 | ||
| Mean | 0.9 | 2.9 | 3.5 | 3.5 | 2.9 | 2.6 | 2.9 | ||
| Stdev | 0.3 | 0.8 | 0.8 | 1.0 | 0.8 | 0.5 | 0.6 | ||
| CV | 0.37 | 0.27 | 0.22 | 0.30 | 0.28 | 0.19 | 0.22 | ||
| Silage | n | 7 | 7 | 15 | 27 | 6 | 4 | - | |
| Min | 0.5 | 2.0 | 1.9 | 1.6 | 2.1 | 2.8 | 1.6 | ||
| Max | 0.7 | 3.3 | 4.6 | 5.3 | 5.6 | 5.2 | 5.2 | ||
| Mean | 0.5 | 2.5 | 3.5 | 3.9 | 4.1 | 3.9 | 3.6 | ||
| Stdev | 0.1 | 0.5 | 0.9 | 0.9 | 1.2 | 0.9 | 1.3 | ||
| CV | 0.18 | 0.21 | 0.24 | 0.24 | 0.30 | 0.23 | 0.35 |
| Variable | Description | Range | Distribution |
|---|---|---|---|
| PROSPECT-5 | |||
| N | Leaf structure index | 1.2–1.8 [26] | Uniform |
| [ g/] | Leaf chlorophyll content | 0–70 | Gaussian |
| [ g/] | Leaf carotenoid content | Default value | - |
| [unitless] | Brown pigments | 0–0.5 | Fixed/Uniform |
| [ g/] | Dry matter content | 0.004–0.0075 [26] | Uniform |
| [ g/] | Leaf water content | Default | - |
| 4SAIL | |||
| LAI | Leaf area index | 0–7 | Uniform |
| ALIA [°] | Average leaf inclination angle | 20–70 [26] | Step of 1 |
| Hot | Hot spot parameter | 0.01–0.5 [26] | Uniform |
| [%] | Soil reflectance | Extracted from image | - |
| SZA [°] | Sun zenith angle | Different for each date | - |
| OZA [°] | Observer zenith angle | 0 | - |
| rAA [°] | Relative azimuth angle | 0 | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chakhvashvili, E.; Siegmann, B.; Muller, O.; Verrelst, J.; Bendig, J.; Kraska, T.; Rascher, U. Retrieval of Crop Variables from Proximal Multispectral UAV Image Data Using PROSAIL in Maize Canopy. Remote Sens. 2022, 14, 1247. https://doi.org/10.3390/rs14051247
Chakhvashvili E, Siegmann B, Muller O, Verrelst J, Bendig J, Kraska T, Rascher U. Retrieval of Crop Variables from Proximal Multispectral UAV Image Data Using PROSAIL in Maize Canopy. Remote Sensing. 2022; 14(5):1247. https://doi.org/10.3390/rs14051247
Chicago/Turabian StyleChakhvashvili, Erekle, Bastian Siegmann, Onno Muller, Jochem Verrelst, Juliane Bendig, Thorsten Kraska, and Uwe Rascher. 2022. "Retrieval of Crop Variables from Proximal Multispectral UAV Image Data Using PROSAIL in Maize Canopy" Remote Sensing 14, no. 5: 1247. https://doi.org/10.3390/rs14051247
APA StyleChakhvashvili, E., Siegmann, B., Muller, O., Verrelst, J., Bendig, J., Kraska, T., & Rascher, U. (2022). Retrieval of Crop Variables from Proximal Multispectral UAV Image Data Using PROSAIL in Maize Canopy. Remote Sensing, 14(5), 1247. https://doi.org/10.3390/rs14051247








