Assessing the Allometric Scaling of Vectorized Branch Lengths of Trees with Terrestrial Laser Scanning and Quantitative Structure Modeling: A Case Study in Guyana
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
2.2. TLS Data
2.3. 3D Reconstruction of Tree Architecture
2.4. Comparison of Scalar and Vector Allometric Models
3. Results
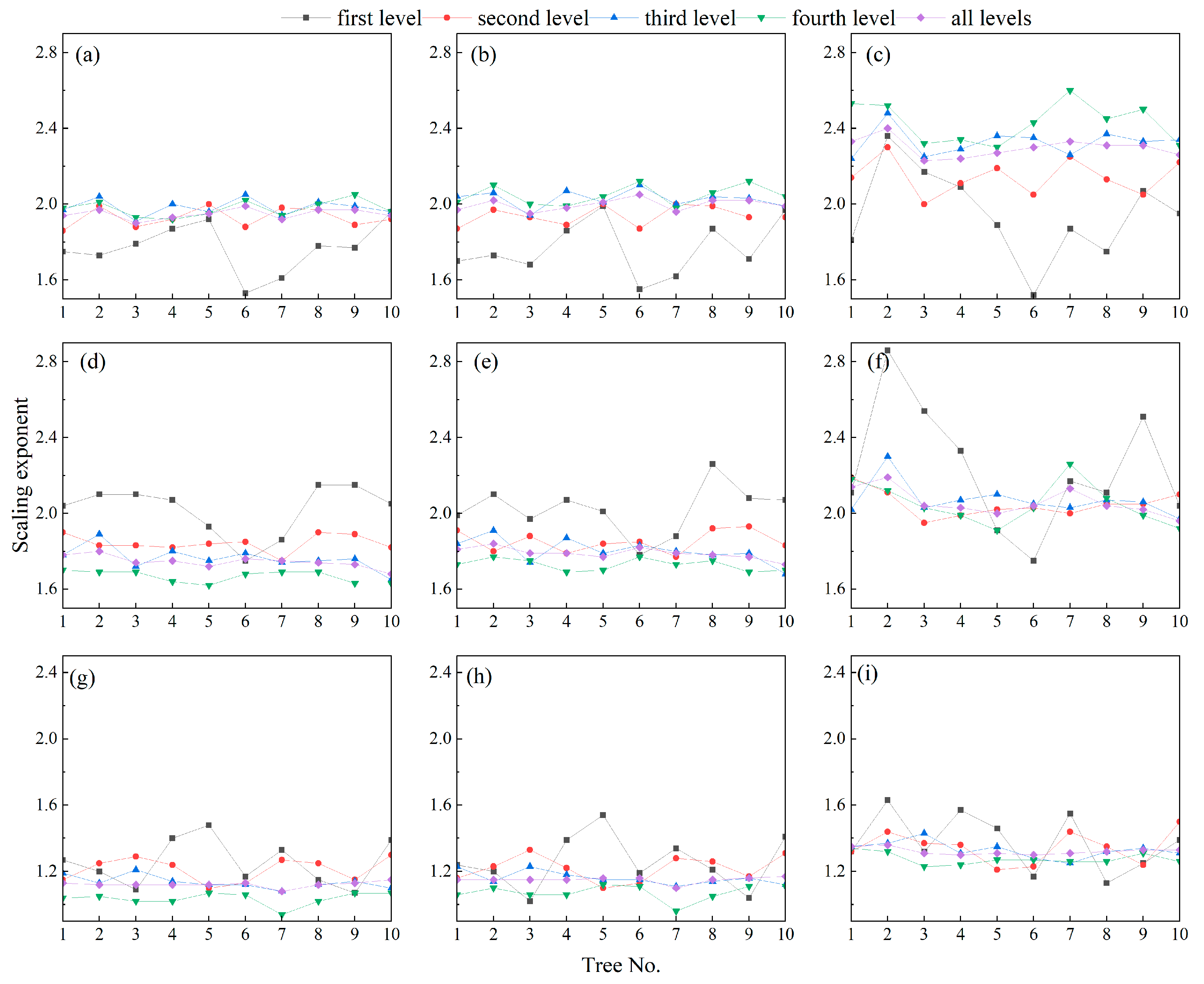
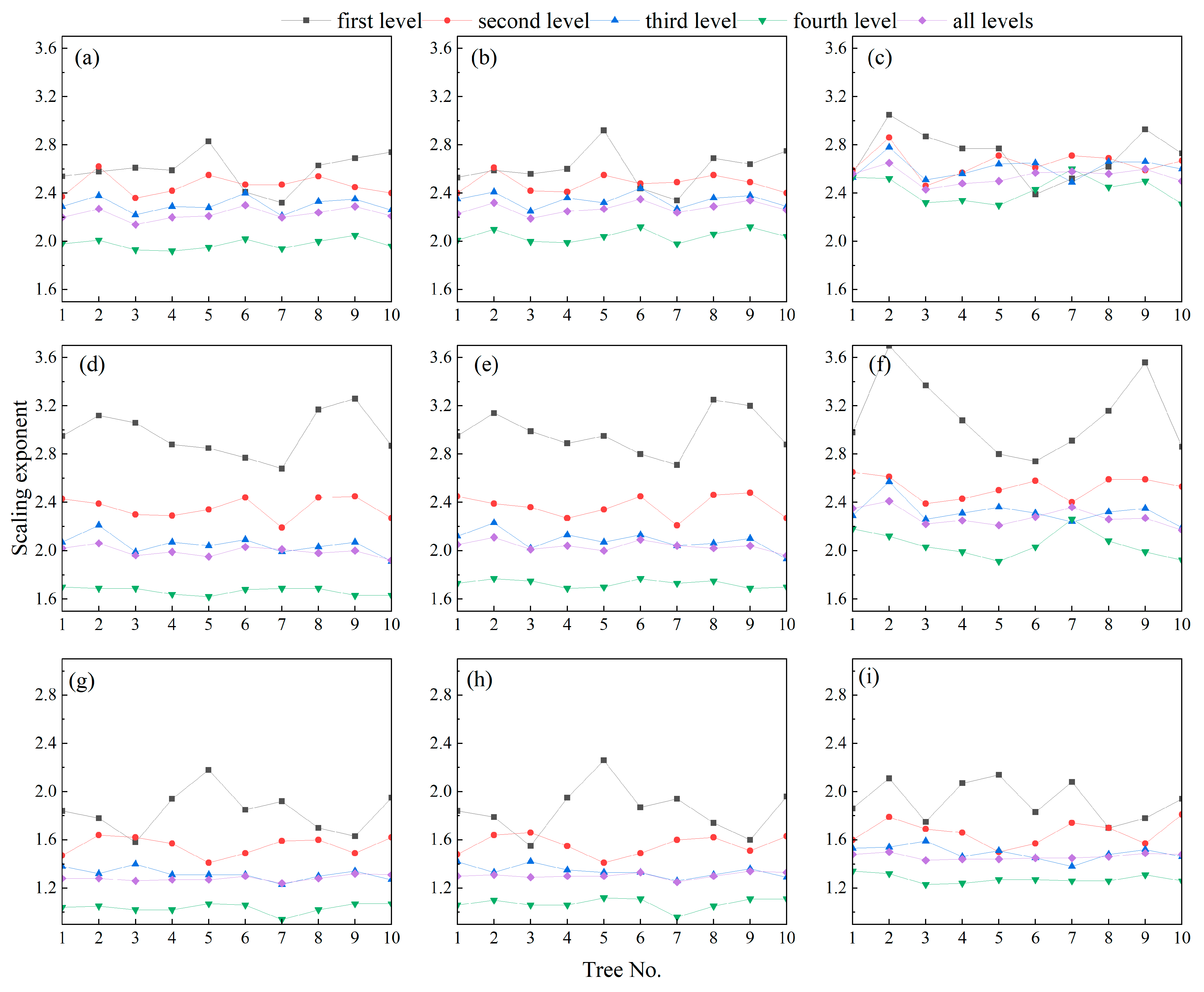
3.1. Tree Branch Information from QSM
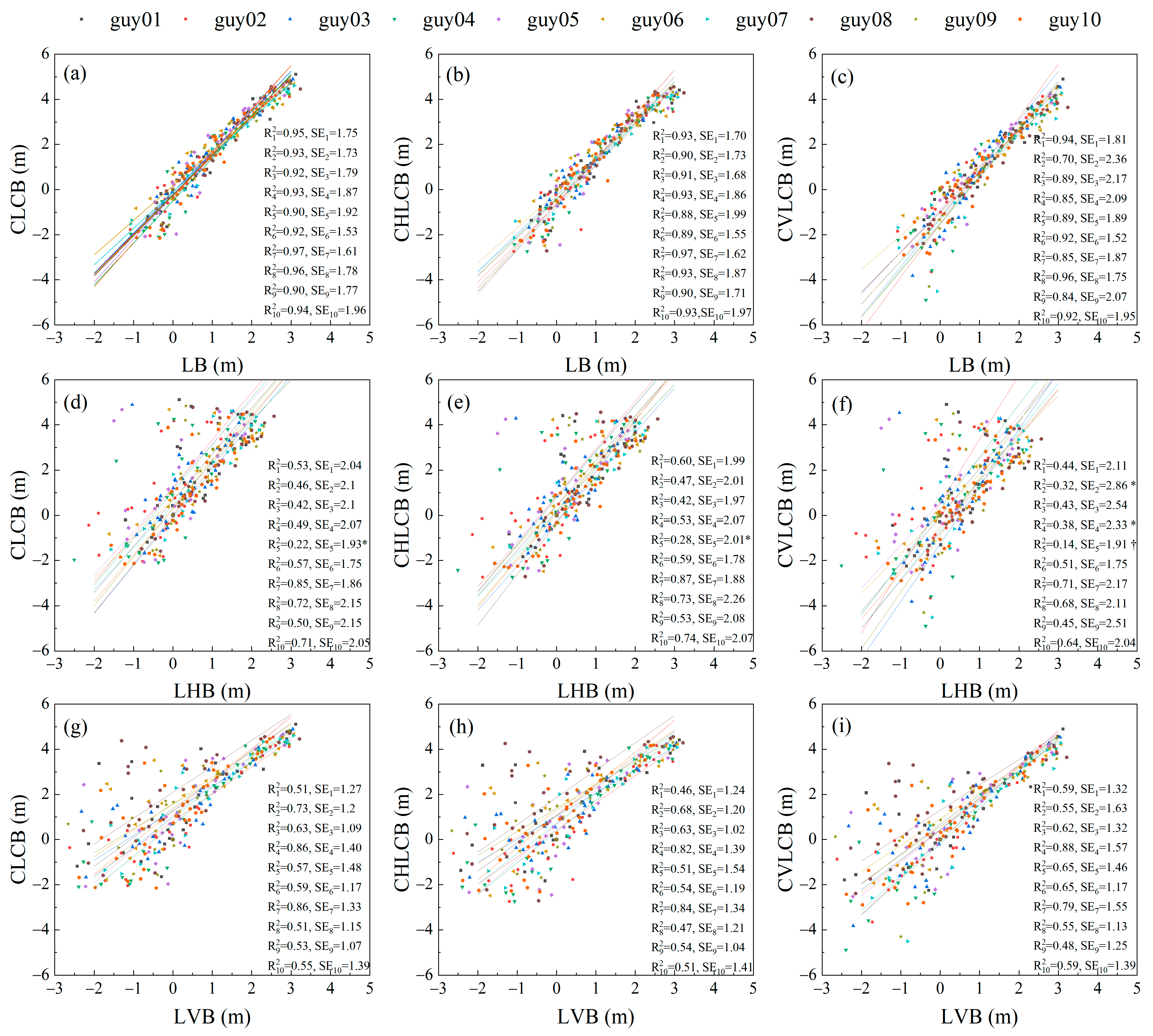
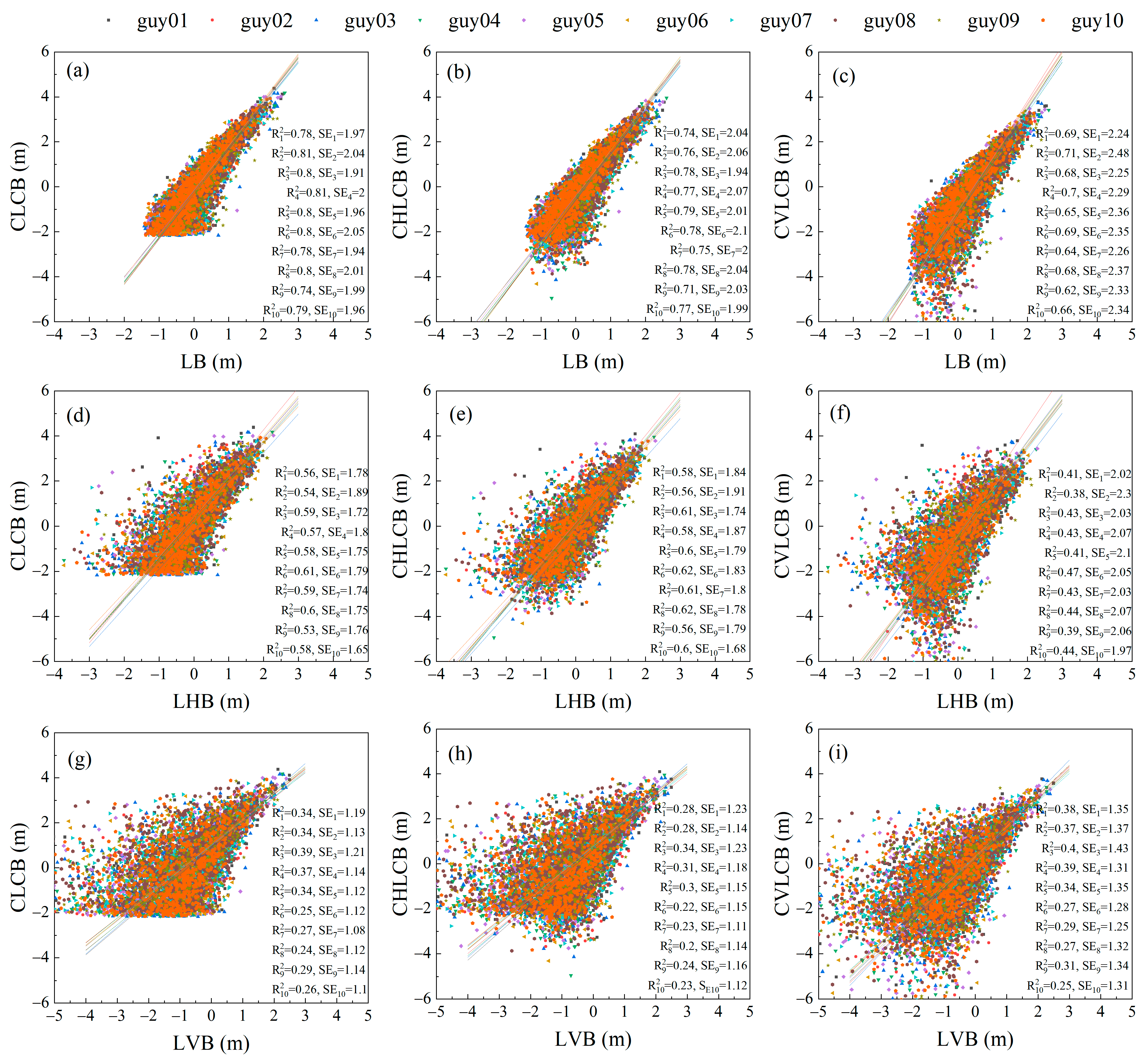
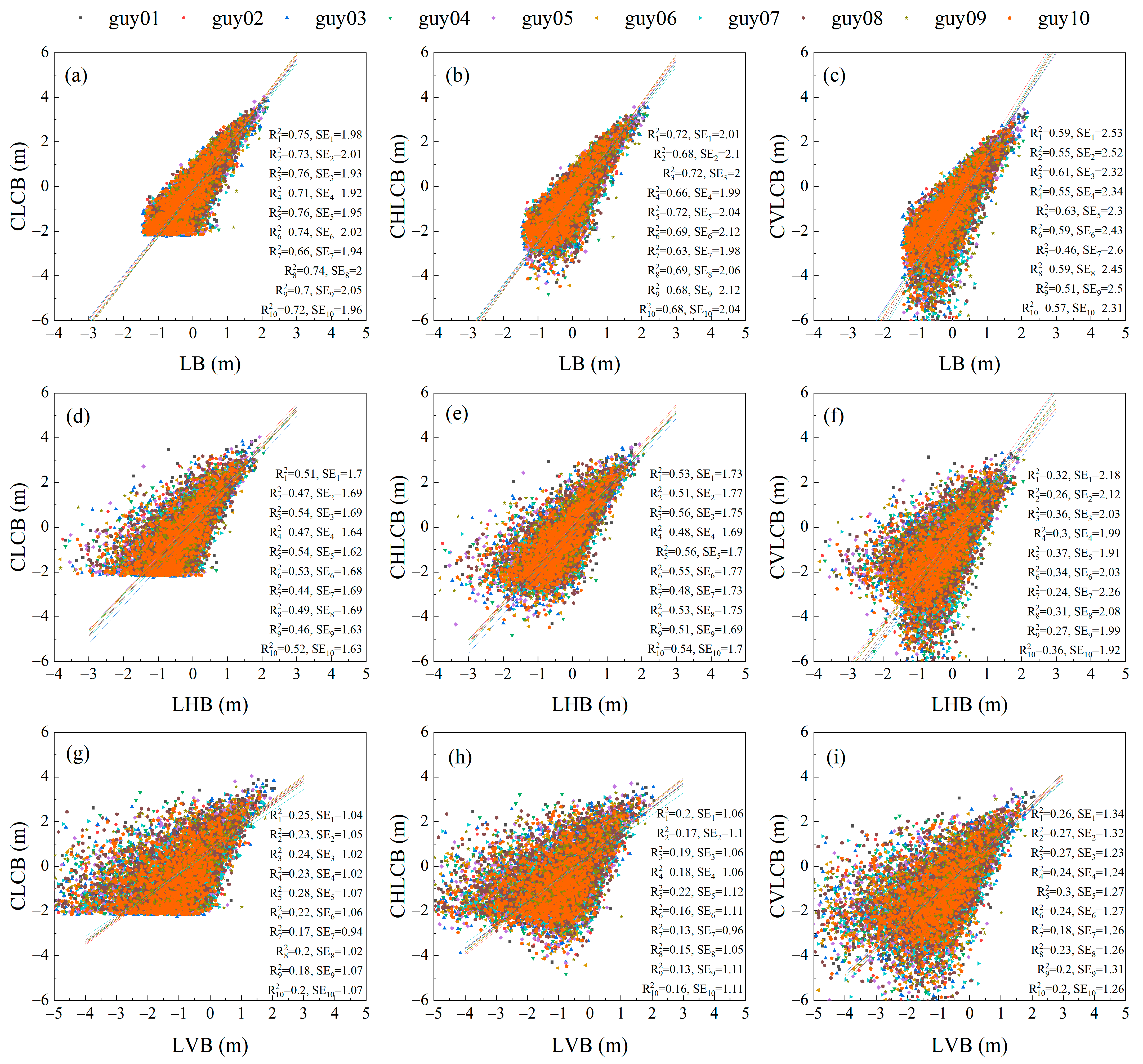
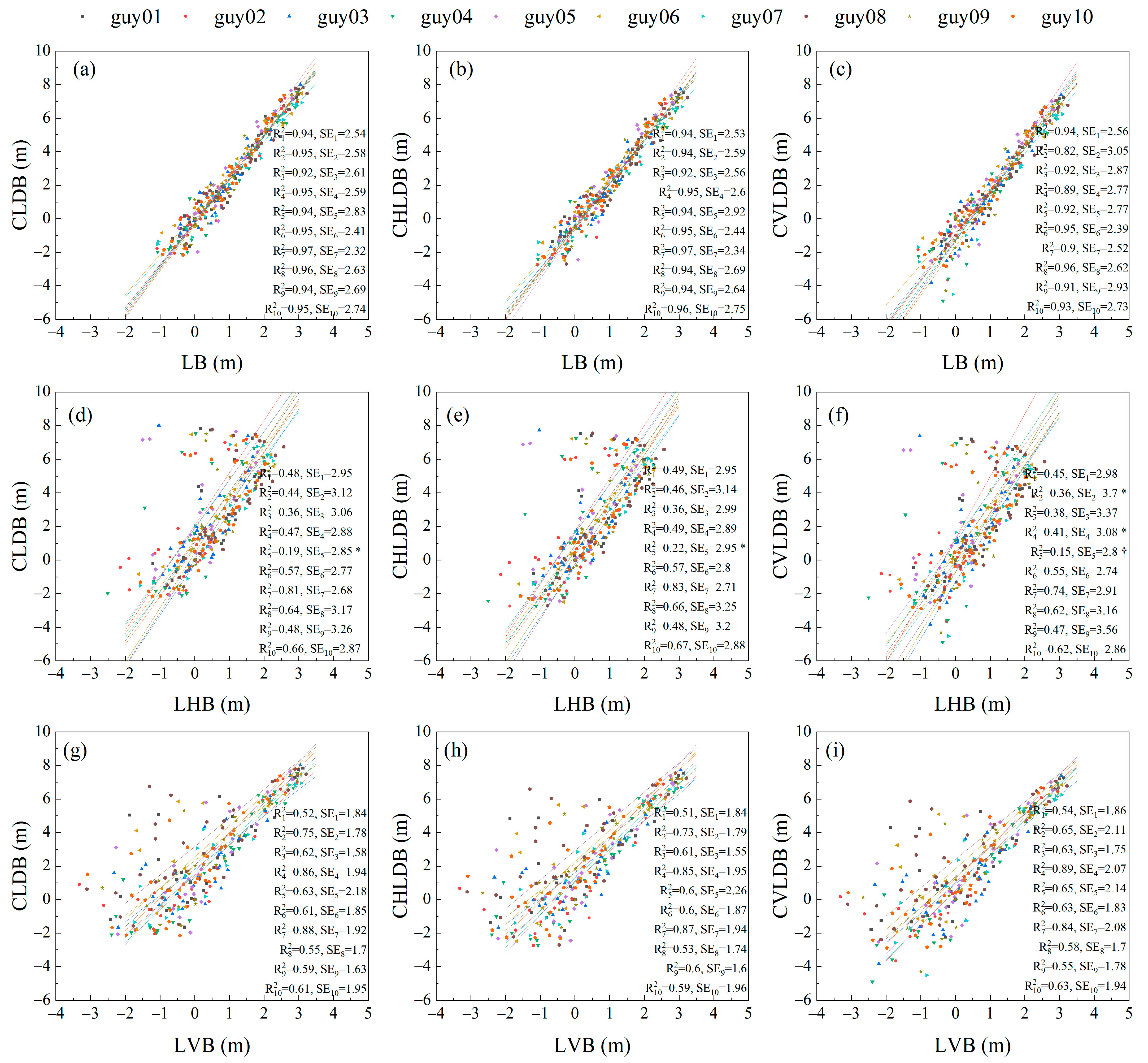
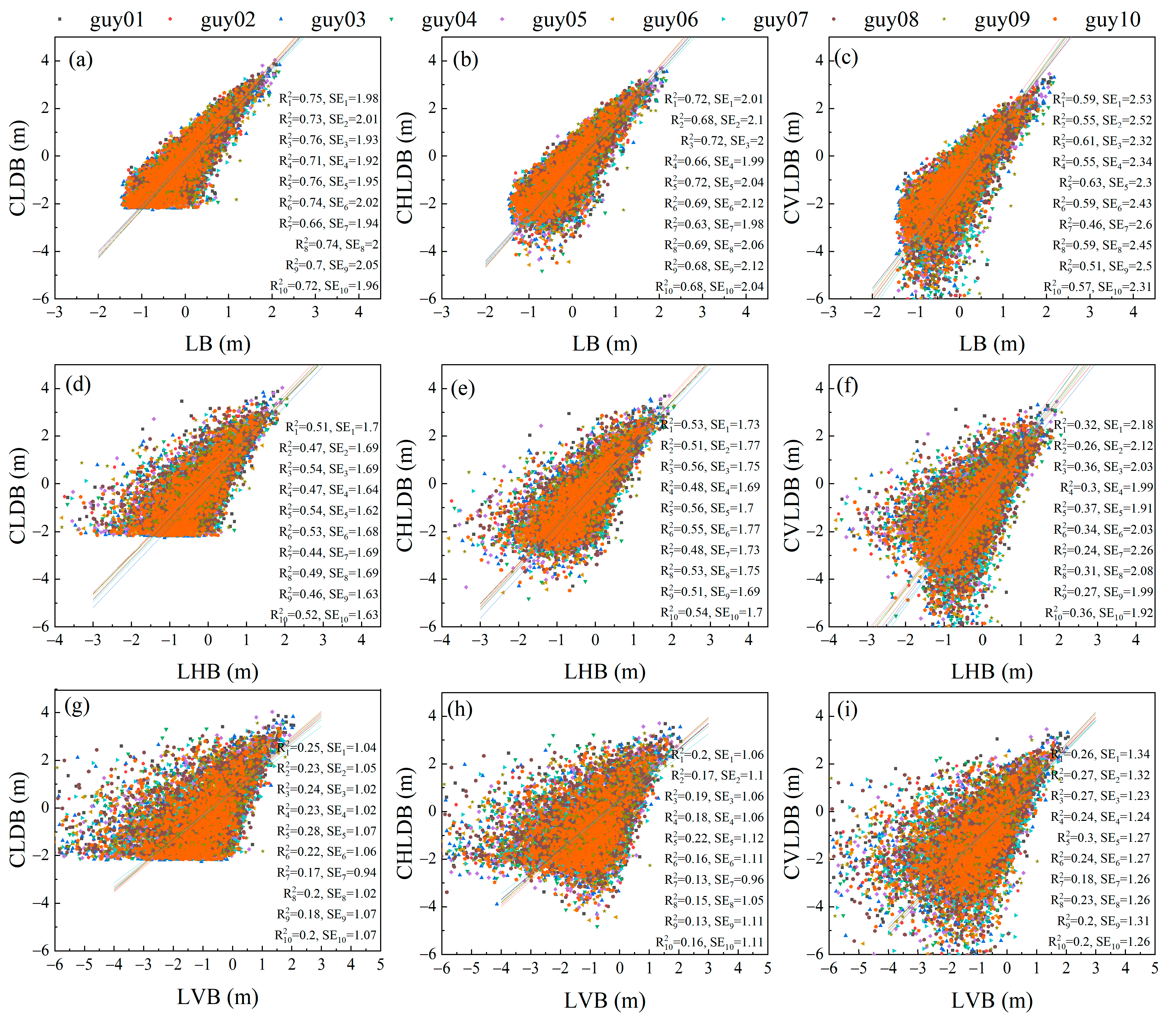
3.2. Comparison of Scalar and Vector Allometry between Branch Lengths and Cumulative Child Branch Lengths
3.3. Comparison of Scalar and Vector Allometry between Branch Lengths and Cumulative Descendant Branch Lengths
4. Discussion
4.1. Possible Influencing Factors of Allometric Model
4.2. Scaling of Branch Lengths and Cumulative Child (Descendant) Branch Lengths
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Duncanson, L.I.; Dubayah, R.O.; Enquist, B.J. Assessing the General Patterns of Forest Structure: Quantifying Tree and Forest Allometric Scaling Relationships in the United States. Glob. Ecol. Biogeogr. 2015, 24, 1465–1475. [Google Scholar] [CrossRef]
- Komiyama, A.; Poungparn, S.; Kato, S. Common Allometric Equations for Estimating the Tree Weight of Mangroves. J. Trop. Ecol. 2005, 21, 471–477. [Google Scholar] [CrossRef]
- Momo Takoudjou, S.; Ploton, P.; Sonké, B.; Hackenberg, J.; Griffon, S.; Coligny, F.; Kamdem, N.G.; Libalah, M.; Mofack, G.I.; Le Moguédec, G.; et al. Using Terrestrial Laser Scanning Data to Estimate Large Tropical Trees Biomass and Calibrate Allometric Models: A Comparison with Traditional Destructive Approach. Methods Ecol. Evol. 2018, 9, 905–916. [Google Scholar] [CrossRef]
- Smith-Martin, C.M.; Xu, X.; Medvigy, D.; Schnitzer, S.A.; Powers, J.S. Allometric Scaling Laws Linking Biomass and Rooting Depth Vary across Ontogeny and Functional Groups in Tropical Dry Forest Lianas and Trees. New Phytol. 2020, 226, 714–726. [Google Scholar] [CrossRef] [PubMed]
- Vieilledent, G.; Vaudry, R.; Andriamanohisoa, S.F.D.; Rakotonarivo, O.S.; Randrianasolo, H.Z.; Razafindrabe, H.N.; Rakotoarivony, C.B.; Ebeling, J.; Rasamoelina, M. A Universal Approach to Estimate Biomass and Carbon Stock in Tropical Forests Using Generic Allometric Models. Ecol. Appl. 2012, 22, 572–583. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Yang, M.; Liu, Z.; Li, P.; Xie, B.; Peng, C. Dynamic Allometric Scaling of Tree Biomass and Size. Nat. Plants 2021, 7, 42–49. [Google Scholar] [CrossRef]
- Enquist, B.J. Universal Scaling in Tree and Vascular Plant Allometry: Toward a General Quantitative Theory Linking Plant Form and Function from Cells to Ecosystems. Tree Physiol. 2002, 22, 1045–1064. [Google Scholar] [CrossRef]
- Farnsworth, K.D.; Niklas, K.J. Theories of Optimization, Form and Function in Branching Architecture in Plants. Funct. Ecol. 1995, 9, 355. [Google Scholar] [CrossRef]
- Remphrey, W.R.; Powell, G.R. Crown Architecture of Larix Laricina Saplings: An Analysis of Higher Order Branching. Can. J. Bot. 1987, 65, 268–279. [Google Scholar] [CrossRef]
- Krooks, A.; Kaasalainen, S.; Kankare, V.; Joensuu, M.; Raumonen, P.; Kaasalainen, M. Predicting Tree Structure from Tree Height Using Terrestrial Laser Scanning and Quantitative Structure Models. Silva Fenn. 2014, 48, 1125. [Google Scholar] [CrossRef]
- Bazezew, M.N.; Hussin, Y.A.; Kloosterman, E.H. Integrating Airborne LiDAR and Terrestrial Laser Scanner Forest Parameters for Accurate Above-Ground Biomass/Carbon Estimation in Ayer Hitam Tropical Forest, Malaysia. Int. J. Appl. Earth Obs. Geoinf. 2018, 73, 638–652. [Google Scholar] [CrossRef]
- Gonzalez de Tanago, J.; Lau, A.; Bartholomeus, H.; Herold, M.; Avitabile, V.; Raumonen, P.; Martius, C.; Goodman, R.C.; Disney, M.; Manuri, S.; et al. Estimation of Above-ground Biomass of Large Tropical Trees with Terrestrial LiDAR. Methods Ecol. Evol. 2018, 9, 223–234. [Google Scholar] [CrossRef]
- Malhi, Y.; Jackson, T.; Patrick Bentley, L.; Lau, A.; Shenkin, A.; Herold, M.; Calders, K.; Bartholomeus, H.; Disney, M.I. New Perspectives on the Ecology of Tree Structure and Tree Communities through Terrestrial Laser Scanning. Interface Focus. 2018, 8, 20170052. [Google Scholar] [CrossRef] [PubMed]
- Wilkes, P.; Lau, A.; Disney, M.; Calders, K.; Burt, A.; Gonzalez de Tanago, J.; Bartholomeus, H.; Brede, B.; Herold, M. Data Acquisition Considerations for Terrestrial Laser Scanning of Forest Plots. Remote Sens. Environ. 2017, 196, 140–153. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J.; et al. Nondestructive Estimates of Above-ground Biomass Using Terrestrial Laser Scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Bayer, D.; Seifert, S.; Pretzsch, H. Structural Crown Properties of Norway Spruce (Picea Abies [L.] Karst.) and European Beech (Fagus Sylvatica [L.]) in Mixed versus Pure Stands Revealed by Terrestrial Laser Scanning. Trees 2013, 27, 1035–1047. [Google Scholar] [CrossRef]
- Lin, Y.; Jiang, M. Toward Extending Terrestrial Laser Scanning Applications in Forestry: A Case Study of Broad- and Needle-Leaf Tree Classification. J. Appl. Remote Sens. 2017, 11, 016037. [Google Scholar] [CrossRef]
- Xi, Z.; Hopkinson, C.; Rood, S.B.; Peddle, D.R. See the Forest and the Trees: Effective Machine and Deep Learning Algorithms for Wood Filtering and Tree Species Classification from Terrestrial Laser Scanning. ISPRS J. Photogramm. Remote Sens. 2020, 168, 1–16. [Google Scholar] [CrossRef]
- Raumonen, P.; Kaasalainen, M.; Åkerblom, M.; Kaasalainen, S.; Kaartinen, H.; Vastaranta, M.; Holopainen, M.; Disney, M.; Lewis, P. Fast Automatic Precision Tree Models from Terrestrial Laser Scanner Data. Remote Sens. 2013, 5, 491–520. [Google Scholar] [CrossRef]
- Hackenberg, J.; Spiecker, H.; Calders, K.; Disney, M.; Raumonen, P. SimpleTree —An Efficient Open Source Tool to Build Tree Models from TLS Clouds. Forests 2015, 6, 4245–4294. [Google Scholar] [CrossRef]
- Fan, G.; Nan, L.; Dong, Y.; Su, X.; Chen, F. AdQSM: A New Method for Estimating Above-Ground Biomass from TLS Point Clouds. Remote Sens. 2020, 12, 3089. [Google Scholar] [CrossRef]
- Dalla Corte, A.P.; de Vasconcellos, B.N.; Rex, F.E.; Sanquetta, C.R.; Mohan, M.; Silva, C.A.; Klauberg, C.; de Almeida, D.R.A.; Zambrano, A.M.A.; Trautenmüller, J.W.; et al. Applying High-Resolution UAV-LiDAR and Quantitative Structure Modelling for Estimating Tree Attributes in a Crop-Livestock-Forest System. Land 2022, 11, 507. [Google Scholar] [CrossRef]
- Krishna Moorthy, S.M.; Raumonen, P.; Van den Bulcke, J.; Calders, K.; Verbeeck, H. Terrestrial Laser Scanning for Non-Destructive Estimates of Liana Stem Biomass. For. Ecol. Manag. 2020, 456, 117751. [Google Scholar] [CrossRef]
- Lau, A.; Calders, K.; Bartholomeus, H.; Martius, C.; Raumonen, P.; Herold, M.; Vicari, M.; Sukhdeo, H.; Singh, J.; Goodman, R.C. Tree Biomass Equations from Terrestrial LiDAR: A Case Study in Guyana. Forests 2019, 10, 527. [Google Scholar] [CrossRef]
- Muumbe, T.P.; Tagwireyi, P.; Mafuratidze, P.; Hussin, Y.; van Leeuwen, L. Estimating Above-Ground Biomass of Individual Trees with Terrestrial Laser Scanner and 3D Quantitative Structure Modelling. South. For. A J. For. Sci. 2021, 83, 56–68. [Google Scholar] [CrossRef]
- Lau, A.; Bentley, L.P.; Martius, C.; Shenkin, A.; Bartholomeus, H.; Raumonen, P.; Malhi, Y.; Jackson, T.; Herold, M. Quantifying Branch Architecture of Tropical Trees Using Terrestrial LiDAR and 3D Modelling. Trees 2018, 32, 1219–1231. [Google Scholar] [CrossRef]
- Rodda, S.R.; Nidamanuri, R.R.; Mayamanikandan, T.; Rajashekar, G.; Jha, C.S.; Dadhwal, V.K. Non-Destructive Allometric Modeling for Tree Volume Estimation in Tropical Dry Deciduous Forests of India Using Terrestrial Laser Scanner. J. Indian. Soc. Remote Sens. 2023, 51, 1–15. [Google Scholar] [CrossRef]
- Lin, Y.; Hyyppä, J. Towards 3D Basic Theories of Plant Forms. Commun. Biol. 2022, 5, 703. [Google Scholar] [CrossRef]
- Lau, A.; Martius, C.; Bartholomeus, H.; Shenkin, A.; Jackson, T.; Malhi, Y.; Herold, M.; Bentley, L.P. Estimating Architecture-Based Metabolic Scaling Exponents of Tropical Trees Using Terrestrial LiDAR and 3D Modelling. For. Ecol. Manag. 2019, 439, 132–145. [Google Scholar] [CrossRef]
- Warton, D.I.; Wright, I.J.; Falster, D.S.; Westoby, M. Bivariate Line-Fitting Methods for Allometry. Biol. Rev. 2006, 81, 259. [Google Scholar] [CrossRef]



| TreeID | Branching Level | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | |
| GUY01 | 42/163 | 408/705 | 2082/1754 | 4561/2668 | 5498/2779 | 5055/2148 | 3128/1077 | 954/275 | 176/47 | 81/21 |
| GUY02 | 42/119 | 291/429 | 1369/1108 | 3222/1691 | 4108/1639 | 3068/1022 | 1377/397 | 391/100 | 75/18 | - |
| GUY03 | 42/136 | 440/525 | 1746/1284 | 3737/1982 | 5137/2097 | 4103/1472 | 2540/829 | 1176/340 | 384/97 | 96/23 |
| GUY04 | 42/137 | 360/493 | 1559/1201 | 3575/1801 | 3946/1564 | 2670/882 | 1302/394 | 492/134 | 158/41 | - |
| GUY05 | 37/119 | 375/527 | 1925/1370 | 4024/2132 | 5263/2289 | 4631/1825 | 3225/1053 | 1441/405 | 403/104 | 81/21 |
| GUY06 | 33/144 | 354/517 | 1717/1405 | 4094/2279 | 5114/2170 | 3662/1285 | 1666/501 | 498/129 | 92/24 | - |
| GUY07 | 31/169 | 354/655 | 1870/1597 | 3687/2024 | 3344/1413 | 1835/698 | 792/287 | 288/96 | - | - |
| GUY08 | 37/204 | 488/941 | 3017/2597 | 7028/3927 | 7819/3434 | 5070/1866 | 2202/729 | 628/175 | 106/27 | - |
| GUY09 | 36/137 | 353/505 | 1438/1186 | 2930/1653 | 3259/1578 | 2594/1036 | 1255/414 | 338/105 | 80/27 | - |
| GUY10 | 44/141 | 475/515 | 1727/1178 | 3648/1732 | 4063/1534 | 2722/915 | 1406/407 | 462/119 | - | - |
| TreeID | LB-CLCB | LB-CHLCB | LB-CVLCB | LHB-CLCB | LHB-CHLCB | LHB-CVLCB | LVB-CLCB | LVB-CHLCB | LVB-CVLCB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.9/0.95 | 0.87/0.93 | 0.85/0.94 | 0.05/0.53 | 0.16/0.6 | 0/0.44 | 0.84/0.51 | 0.71/0.46 | 0.88/0.59 |
| GUY02 | 0.89/0.93 | 0.82/0.9 | 0.89/0.7 | 0.47/0.46 | 0.49/0.47 | 0.46/0.32 | 0.88/0.73 | 0.81/0.68 | 0.89/0.55 |
| GUY03 | 0.96/0.92 | 0.96/0.91 | 0.94/0.89 | 0.15/0.42 | 0.23/0.42 | 0.12/0.43 | 0.94/0.63 | 0.93/0.63 | 0.93/0.62 |
| GUY04 | 0.92/0.93 | 0.8/0.93 | 0.93/0.85 | 0.34/0.49 | 0.48/0.53 | 0.14/0.38 | 0.87/0.86 | 0.72/0.82 | 0.95/0.88 |
| GUY05 | 0.94/0.9 | 0.89/0.88 | 0.94/0.89 | 0.09/0.22 | 0.17/0.28 | 0.02/0.14 | 0.86/0.57 | 0.77/0.51 | 0.93/0.65 |
| GUY06 | 0.9/0.92 | 0.88/0.89 | 0.88/0.92 | 0.08/0.57 | 0.13/0.59 | 0.04/0.51 | 0.87/0.59 | 0.79/0.54 | 0.93/0.65 |
| GUY07 | 0.96/0.97 | 0.95/0.97 | 0.85/0.85 | 0.48/0.85 | 0.52/0.87 | 0.35/0.71 | 0.92/0.86 | 0.9/0.84 | 0.84/0.79 |
| GUY08 | 0.87/0.96 | 0.88/0.93 | 0.81/0.96 | 0.26/0.72 | 0.29/0.73 | 0.19/0.68 | 0.67/0.51 | 0.66/0.47 | 0.68/0.55 |
| GUY09 | 0.87/0.9 | 0.95/0.9 | 0.73/0.84 | 0.08/0.5 | 0.16/0.53 | 0.02/0.45 | 0.87/0.53 | 0.9/0.54 | 0.79/0.48 |
| GUY10 | 0.92/0.94 | 0.91/0.93 | 0.86/0.92 | 0.35/0.71 | 0.38/0.74 | 0.27/0.64 | 0.87/0.55 | 0.84/0.51 | 0.88/0.59 |
| TreeID | LB-CLCB | LB-CHLCB | LB-CVLCB | LHB-CLCB | LHB-CHLCB | LHB-CVLCB | LVB-CLCB | LVB-CHLCB | LVB-CVLCB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.9/0.86 | 0.84/0.83 | 0.87/0.78 | 0.49/0.63 | 0.55/0.64 | 0.39/0.54 | 0.82/0.49 | 0.73/0.43 | 0.86/0.5 |
| GUY02 | 0.91/0.88 | 0.85/0.87 | 0.85/0.79 | 0.62/0.57 | 0.73/0.61 | 0.45/0.45 | 0.84/0.56 | 0.7/0.51 | 0.88/0.57 |
| GUY03 | 0.88/0.87 | 0.9/0.84 | 0.79/0.84 | 0.35/0.63 | 0.45/0.62 | 0.23/0.58 | 0.83/0.59 | 0.8/0.54 | 0.8/0.59 |
| GUY04 | 0.89/0.88 | 0.88/0.87 | 0.85/0.85 | 0.62/0.75 | 0.68/0.75 | 0.51/0.69 | 0.77/0.43 | 0.71/0.4 | 0.82/0.46 |
| GUY05 | 0.87/0.88 | 0.88/0.87 | 0.78/0.81 | 0.42/0.56 | 0.52/0.59 | 0.27/0.46 | 0.77/0.37 | 0.71/0.33 | 0.77/0.42 |
| GUY06 | 0.9/0.88 | 0.87/0.87 | 0.89/0.81 | 0.62/0.7 | 0.64/0.71 | 0.53/0.62 | 0.73/0.43 | 0.66/0.39 | 0.8/0.44 |
| GUY07 | 0.9/0.87 | 0.88/0.86 | 0.85/0.75 | 0.79/0.68 | 0.79/0.7 | 0.71/0.54 | 0.6/0.38 | 0.56/0.36 | 0.63/0.38 |
| GUY08 | 0.85/0.89 | 0.84/0.88 | 0.78/0.82 | 0.64/0.81 | 0.66/0.81 | 0.53/0.72 | 0.61/0.41 | 0.56/0.38 | 0.66/0.43 |
| GUY09 | 0.88/0.85 | 0.87/0.84 | 0.82/0.77 | 0.58/0.72 | 0.61/0.73 | 0.46/0.62 | 0.77/0.44 | 0.72/0.41 | 0.79/0.45 |
| GUY10 | 0.88/0.88 | 0.89/0.87 | 0.8/0.76 | 0.72/0.78 | 0.77/0.79 | 0.59/0.65 | 0.68/0.45 | 0.65/0.42 | 0.72/0.4 |
| TreeID | LB-CLCB | LB-CHLCB | LB-CVLCB | LHB-CLCB | LHB-CHLCB | LHB-CVLCB | LVB-CLCB | LVB-CHLCB | LVB-CVLCB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.81/0.78 | 0.8/0.74 | 0.77/0.69 | 0.48/0.56 | 0.54/0.58 | 0.35/0.41 | 0.67/0.34 | 0.6/0.28 | 0.75/0.38 |
| GUY02 | 0.85/0.81 | 0.82/0.76 | 0.82/0.71 | 0.51/0.54 | 0.58/0.56 | 0.39/0.38 | 0.66/0.34 | 0.58/0.28 | 0.73/0.37 |
| GUY03 | 0.84/0.8 | 0.82/0.78 | 0.78/0.68 | 0.56/0.59 | 0.61/0.61 | 0.45/0.43 | 0.71/0.39 | 0.65/0.34 | 0.73/0.4 |
| GUY04 | 0.84/0.81 | 0.82/0.77 | 0.83/0.7 | 0.67/0.57 | 0.68/0.58 | 0.6/0.43 | 0.68/0.37 | 0.64/0.31 | 0.74/0.39 |
| GUY05 | 0.83/0.8 | 0.82/0.79 | 0.8/0.65 | 0.61/0.58 | 0.64/0.6 | 0.53/0.41 | 0.63/0.34 | 0.59/0.3 | 0.67/0.34 |
| GUY06 | 0.81/0.8 | 0.8/0.78 | 0.74/0.69 | 0.62/0.61 | 0.65/0.62 | 0.53/0.47 | 0.56/0.25 | 0.52/0.22 | 0.57/0.27 |
| GUY07 | 0.8/0.78 | 0.78/0.75 | 0.75/0.64 | 0.67/0.59 | 0.69/0.61 | 0.57/0.43 | 0.44/0.27 | 0.38/0.23 | 0.49/0.29 |
| GUY08 | 0.79/0.8 | 0.78/0.78 | 0.74/0.68 | 0.6/0.6 | 0.64/0.62 | 0.49/0.44 | 0.45/0.24 | 0.39/0.2 | 0.54/0.27 |
| GUY09 | 0.79/0.74 | 0.79/0.71 | 0.68/0.62 | 0.68/0.53 | 0.72/0.56 | 0.52/0.39 | 0.53/0.29 | 0.48/0.24 | 0.55/0.31 |
| GUY10 | 0.81/0.79 | 0.81/0.77 | 0.76/0.66 | 0.64/0.58 | 0.68/0.6 | 0.55/0.44 | 0.51/0.26 | 0.47/0.23 | 0.56/0.25 |
| TreeID | LB-CLCB | LB-CHLCB | LB-CVLCB | LHB-CLCB | LHB-CHLCB | LHB-CVLCB | LVB-CLCB | LVB-CHLCB | LVB-CVLCB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.79/0.75 | 0.78/0.72 | 0.75/0.59 | 0.57/0.51 | 0.62/0.53 | 0.46/0.32 | 0.56/0.25 | 0.5/0.2 | 0.61/0.26 |
| GUY02 | 0.75/0.73 | 0.74/0.68 | 0.71/0.55 | 0.55/0.47 | 0.6/0.51 | 0.43/0.26 | 0.49/0.23 | 0.43/0.17 | 0.55/0.27 |
| GUY03 | 0.79/0.76 | 0.79/0.72 | 0.73/0.61 | 0.61/0.54 | 0.64/0.56 | 0.52/0.36 | 0.56/0.24 | 0.53/0.19 | 0.58/0.27 |
| GUY04 | 0.79/0.71 | 0.77/0.66 | 0.72/0.55 | 0.68/0.47 | 0.71/0.48 | 0.54/0.3 | 0.37/0.23 | 0.31/0.18 | 0.46/0.24 |
| GUY05 | 0.77/0.76 | 0.77/0.72 | 0.73/0.63 | 0.66/0.54 | 0.69/0.56 | 0.58/0.37 | 0.5/0.28 | 0.46/0.22 | 0.53/0.3 |
| GUY06 | 0.75/0.74 | 0.73/0.69 | 0.72/0.59 | 0.58/0.53 | 0.59/0.55 | 0.49/0.34 | 0.45/0.22 | 0.4/0.16 | 0.51/0.24 |
| GUY07 | 0.67/0.66 | 0.64/0.63 | 0.66/0.46 | 0.56/0.44 | 0.58/0.48 | 0.46/0.24 | 0.32/0.17 | 0.26/0.13 | 0.41/0.18 |
| GUY08 | 0.74/0.74 | 0.72/0.69 | 0.69/0.59 | 0.57/0.49 | 0.59/0.53 | 0.46/0.31 | 0.39/0.2 | 0.34/0.15 | 0.46/0.23 |
| GUY09 | 0.73/0.7 | 0.72/0.68 | 0.67/0.51 | 0.62/0.46 | 0.64/0.51 | 0.51/0.27 | 0.36/0.18 | 0.32/0.13 | 0.41/0.2 |
| GUY10 | 0.77/0.72 | 0.76/0.68 | 0.7/0.57 | 0.59/0.52 | 0.62/0.54 | 0.49/0.36 | 0.45/0.2 | 0.4/0.16 | 0.5/0.2 |
| TreeID | LB-CLDB | LB-CHLDB | LB-CVLDB | LHB-CLDB | LHB-CHLDB | LHB-CVLDB | LVB-CLDB | LVB-CHLDB | LVB-CVLDB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.82/0.94 | 0.83/0.94 | 0.81/0.94 | 0/0.48 | 0/0.49 | 0/0.45 | 0.86/0.52 | 0.86/0.51 | 0.85/0.54 |
| GUY02 | 0.84/0.95 | 0.84/0.94 | 0.83/0.82 | 0.38/0.44 | 0.38/0.46 | 0.38/0.36 | 0.84/0.75 | 0.84/0.73 | 0.83/0.65 |
| GUY03 | 0.8/0.92 | 0.8/0.92 | 0.79/0.92 | 0.01/0.36 | 0.01/0.36 | 0.01/0.38 | 0.83/0.62 | 0.84/0.61 | 0.82/0.63 |
| GUY04 | 0.72/0.95 | 0.73/0.95 | 0.7/0.89 | 0.06/0.47 | 0.07/0.49 | 0.04/0.41 | 0.74/0.86 | 0.75/0.85 | 0.73/0.89 |
| GUY05 | 0.86/0.94 | 0.86/0.94 | 0.85/0.92 | 0.02/0.19 | 0.02/0.22 | 0.01/0.15 | 0.87/0.63 | 0.87/0.6 | 0.87/0.65 |
| GUY06 | 0.8/0.95 | 0.79/0.95 | 0.81/0.95 | 0.02/0.57 | 0.02/0.57 | 0.02/0.55 | 0.85/0.61 | 0.84/0.6 | 0.88/0.63 |
| GUY07 | 0.92/0.97 | 0.93/0.97 | 0.92/0.9 | 0.3/0.81 | 0.3/0.83 | 0.3/0.74 | 0.95/0.88 | 0.96/0.87 | 0.94/0.84 |
| GUY08 | 0.81/0.96 | 0.81/0.94 | 0.8/0.96 | 0.07/0.64 | 0.08/0.66 | 0.06/0.62 | 0.8/0.55 | 0.78/0.53 | 0.81/0.58 |
| GUY09 | 0.77/0.94 | 0.79/0.94 | 0.74/0.91 | 0/0.48 | 0.01/0.48 | 0/0.47 | 0.87/0.59 | 0.88/0.6 | 0.84/0.55 |
| GUY10 | 0.71/0.95 | 0.71/0.96 | 0.7/0.93 | 0.2/0.66 | 0.2/0.67 | 0.19/0.62 | 0.77/0.61 | 0.78/0.59 | 0.77/0.63 |
| TreeID | LB-CLDB | LB-CHLDB | LB-CVLDB | LHB-CLDB | LHB-CHLDB | LHB-CVLDB | LVB-CLDB | LVB-CHLDB | LVB-CVLDB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.77/0.86 | 0.76/0.85 | 0.76/0.82 | 0.36/0.63 | 0.38/0.64 | 0.33/0.57 | 0.75/0.5 | 0.73/0.46 | 0.76/0.51 |
| GUY02 | 0.81/0.9 | 0.81/0.89 | 0.78/0.84 | 0.49/0.59 | 0.53/0.62 | 0.42/0.51 | 0.79/0.55 | 0.77/0.52 | 0.79/0.57 |
| GUY03 | 0.6/0.87 | 0.62/0.85 | 0.57/0.85 | 0.12/0.61 | 0.13/0.61 | 0.1/0.58 | 0.64/0.58 | 0.64/0.55 | 0.62/0.59 |
| GUY04 | 0.71/0.89 | 0.71/0.88 | 0.7/0.87 | 0.37/0.74 | 0.38/0.75 | 0.35/0.7 | 0.74/0.45 | 0.73/0.43 | 0.75/0.47 |
| GUY05 | 0.6/0.88 | 0.62/0.88 | 0.59/0.84 | 0.16/0.56 | 0.17/0.58 | 0.14/0.49 | 0.64/0.37 | 0.64/0.34 | 0.64/0.4 |
| GUY06 | 0.76/0.88 | 0.75/0.88 | 0.76/0.84 | 0.45/0.69 | 0.45/0.7 | 0.43/0.64 | 0.7/0.44 | 0.68/0.42 | 0.72/0.45 |
| GUY07 | 0.74/0.88 | 0.74/0.87 | 0.72/0.8 | 0.66/0.7 | 0.67/0.72 | 0.63/0.61 | 0.48/0.37 | 0.47/0.35 | 0.48/0.37 |
| GUY08 | 0.6/0.89 | 0.61/0.88 | 0.58/0.85 | 0.37/0.8 | 0.38/0.8 | 0.34/0.74 | 0.55/0.41 | 0.54/0.4 | 0.57/0.44 |
| GUY09 | 0.76/0.85 | 0.76/0.85 | 0.75/0.81 | 0.45/0.71 | 0.46/0.72 | 0.42/0.65 | 0.7/0.46 | 0.69/0.44 | 0.72/0.47 |
| GUY10 | 0.66/0.88 | 0.67/0.87 | 0.65/0.8 | 0.49/0.77 | 0.5/0.78 | 0.47/0.69 | 0.63/0.44 | 0.62/0.42 | 0.64/0.42 |
| TreeID | LB-CLDB | LB-CHLDB | LB-CVLDB | LHB-CLDB | LHB-CHLDB | LHB-CVLDB | LVB-CLDB | LVB-CHLDB | LVB-CVLDB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.62/0.79 | 0.62/0.76 | 0.6/0.72 | 0.3/0.56 | 0.33/0.57 | 0.25/0.44 | 0.57/0.34 | 0.54/0.3 | 0.61/0.38 |
| GUY02 | 0.74/0.81 | 0.74/0.78 | 0.73/0.74 | 0.41/0.54 | 0.45/0.56 | 0.34/0.42 | 0.61/0.34 | 0.57/0.29 | 0.65/0.37 |
| GUY03 | 0.68/0.81 | 0.68/0.79 | 0.65/0.72 | 0.41/0.59 | 0.43/0.61 | 0.36/0.47 | 0.62/0.39 | 0.6/0.36 | 0.62/0.41 |
| GUY04 | 0.72/0.81 | 0.7/0.79 | 0.73/0.73 | 0.56/0.57 | 0.56/0.58 | 0.54/0.46 | 0.62/0.37 | 0.59/0.32 | 0.66/0.4 |
| GUY05 | 0.7/0.81 | 0.7/0.8 | 0.69/0.69 | 0.48/0.58 | 0.5/0.6 | 0.45/0.44 | 0.57/0.34 | 0.56/0.31 | 0.59/0.36 |
| GUY06 | 0.68/0.81 | 0.68/0.79 | 0.65/0.73 | 0.5/0.61 | 0.51/0.62 | 0.46/0.5 | 0.51/0.26 | 0.49/0.23 | 0.53/0.28 |
| GUY07 | 0.66/0.78 | 0.64/0.76 | 0.66/0.68 | 0.57/0.59 | 0.58/0.61 | 0.53/0.47 | 0.33/0.27 | 0.29/0.23 | 0.38/0.29 |
| GUY08 | 0.65/0.8 | 0.64/0.79 | 0.64/0.72 | 0.49/0.59 | 0.51/0.62 | 0.42/0.47 | 0.39/0.24 | 0.35/0.21 | 0.46/0.28 |
| GUY09 | 0.67/0.74 | 0.66/0.73 | 0.66/0.66 | 0.57/0.54 | 0.58/0.56 | 0.53/0.42 | 0.48/0.29 | 0.45/0.25 | 0.52/0.32 |
| GUY10 | 0.65/0.79 | 0.65/0.78 | 0.62/0.7 | 0.51/0.58 | 0.53/0.6 | 0.46/0.47 | 0.42/0.26 | 0.4/0.23 | 0.45/0.26 |
| TreeID | LB-CLDB | LB-CHLDB | LB-CVLDB | LHB-CLDB | LHB-CHLDB | LHB-CVLDB | LVB-CLDB | LVB-CHLDB | LVB-CVLDB |
|---|---|---|---|---|---|---|---|---|---|
| GUY01 | 0.79/0.75 | 0.78/0.72 | 0.75/0.59 | 0.57/0.51 | 0.62/0.53 | 0.46/0.32 | 0.56/0.25 | 0.5/0.2 | 0.61/0.26 |
| GUY02 | 0.75/0.73 | 0.74/0.68 | 0.71/0.55 | 0.55/0.47 | 0.6/0.51 | 0.43/0.26 | 0.49/0.23 | 0.43/0.17 | 0.55/0.27 |
| GUY03 | 0.79/0.76 | 0.79/0.72 | 0.73/0.61 | 0.61/0.54 | 0.64/0.56 | 0.52/0.36 | 0.56/0.24 | 0.53/0.19 | 0.58/0.27 |
| GUY04 | 0.79/0.71 | 0.77/0.66 | 0.72/0.55 | 0.68/0.47 | 0.71/0.48 | 0.54/0.3 | 0.37/0.23 | 0.31/0.18 | 0.46/0.24 |
| GUY05 | 0.77/0.76 | 0.77/0.72 | 0.73/0.63 | 0.66/0.54 | 0.69/0.56 | 0.58/0.37 | 0.5/0.28 | 0.46/0.22 | 0.53/0.3 |
| GUY06 | 0.75/0.74 | 0.73/0.69 | 0.72/0.59 | 0.58/0.53 | 0.59/0.55 | 0.49/0.34 | 0.45/0.22 | 0.4/0.16 | 0.51/0.24 |
| GUY07 | 0.67/0.66 | 0.64/0.63 | 0.66/0.46 | 0.56/0.44 | 0.58/0.48 | 0.46/0.24 | 0.32/0.17 | 0.26/0.13 | 0.41/0.18 |
| GUY08 | 0.74/0.74 | 0.72/0.69 | 0.69/0.59 | 0.57/0.49 | 0.59/0.53 | 0.46/0.31 | 0.39/0.2 | 0.34/0.15 | 0.46/0.23 |
| GUY09 | 0.73/0.7 | 0.72/0.68 | 0.67/0.51 | 0.62/0.46 | 0.64/0.51 | 0.51/0.27 | 0.36/0.18 | 0.32/0.13 | 0.41/0.2 |
| GUY10 | 0.77/0.72 | 0.76/0.68 | 0.7/0.57 | 0.59/0.52 | 0.62/0.54 | 0.49/0.36 | 0.45/0.2 | 0.4/0.16 | 0.5/0.2 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, J.; Lin, Y. Assessing the Allometric Scaling of Vectorized Branch Lengths of Trees with Terrestrial Laser Scanning and Quantitative Structure Modeling: A Case Study in Guyana. Remote Sens. 2023, 15, 5005. https://doi.org/10.3390/rs15205005
Sun J, Lin Y. Assessing the Allometric Scaling of Vectorized Branch Lengths of Trees with Terrestrial Laser Scanning and Quantitative Structure Modeling: A Case Study in Guyana. Remote Sensing. 2023; 15(20):5005. https://doi.org/10.3390/rs15205005
Chicago/Turabian StyleSun, Jingjing, and Yi Lin. 2023. "Assessing the Allometric Scaling of Vectorized Branch Lengths of Trees with Terrestrial Laser Scanning and Quantitative Structure Modeling: A Case Study in Guyana" Remote Sensing 15, no. 20: 5005. https://doi.org/10.3390/rs15205005
APA StyleSun, J., & Lin, Y. (2023). Assessing the Allometric Scaling of Vectorized Branch Lengths of Trees with Terrestrial Laser Scanning and Quantitative Structure Modeling: A Case Study in Guyana. Remote Sensing, 15(20), 5005. https://doi.org/10.3390/rs15205005






