Forecasting Table Beet Root Yield Using Spectral and Textural Features from Hyperspectral UAS Imagery
Abstract
1. Introduction
2. Materials and Methods
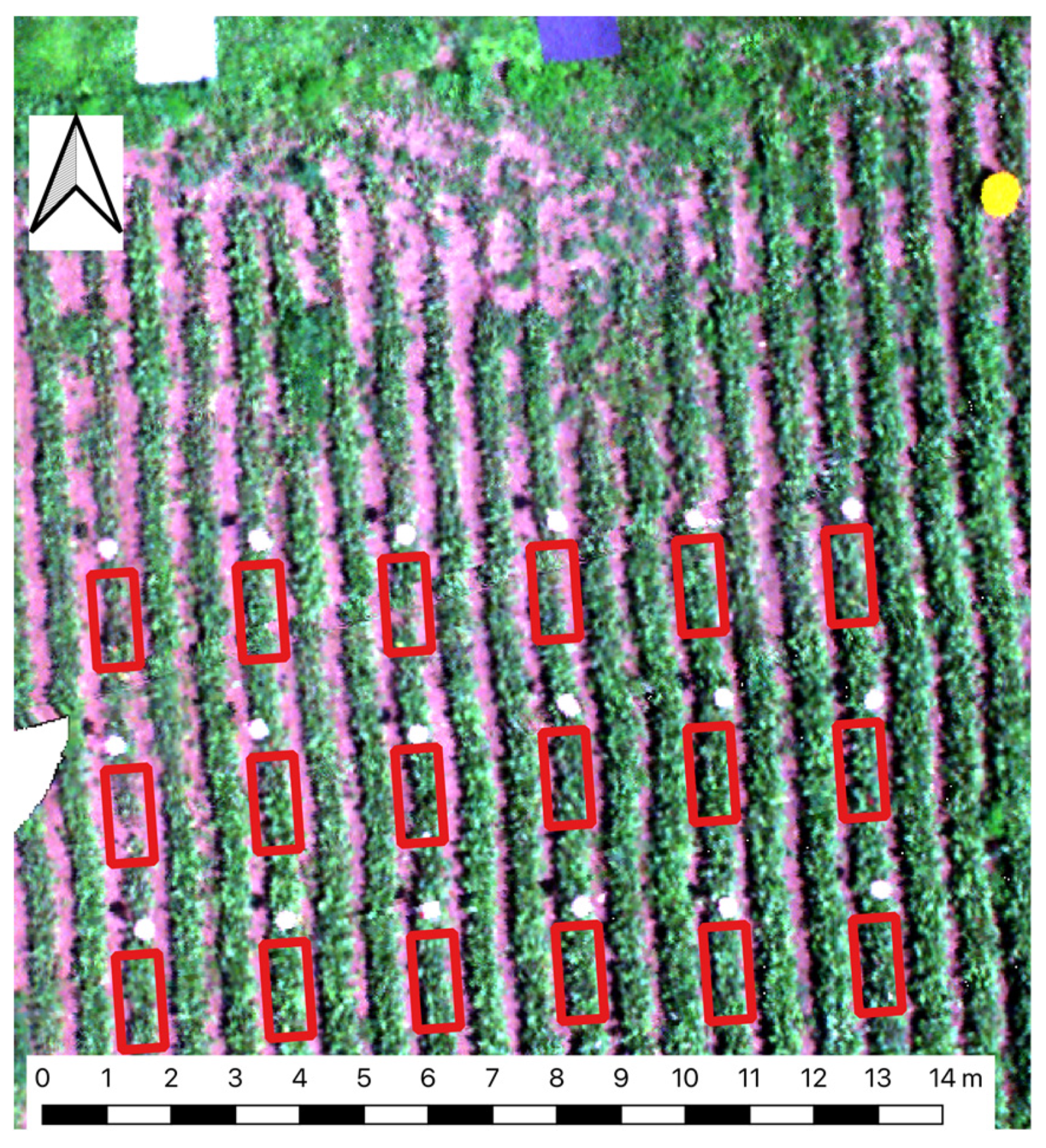
2.1. Data Collection
2.2. Data Preprocessing
2.3. Denoising
2.4. Feature Extraction
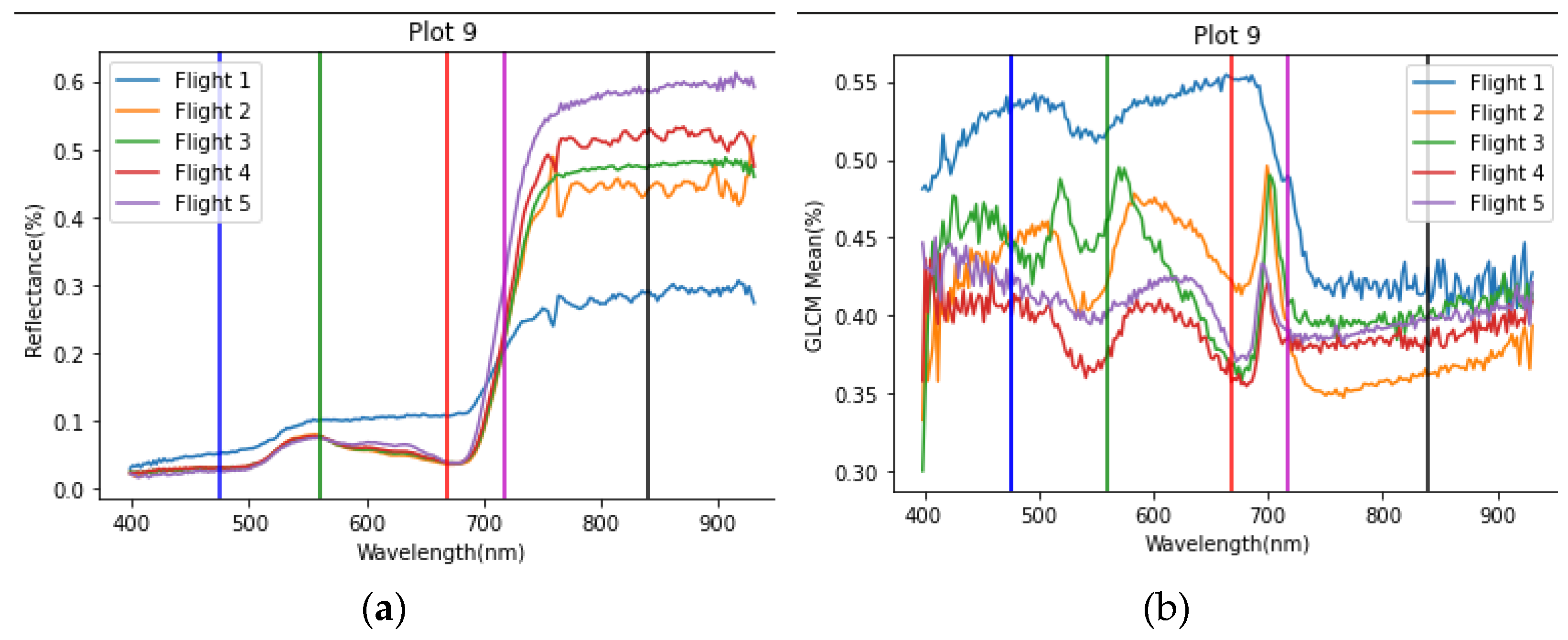
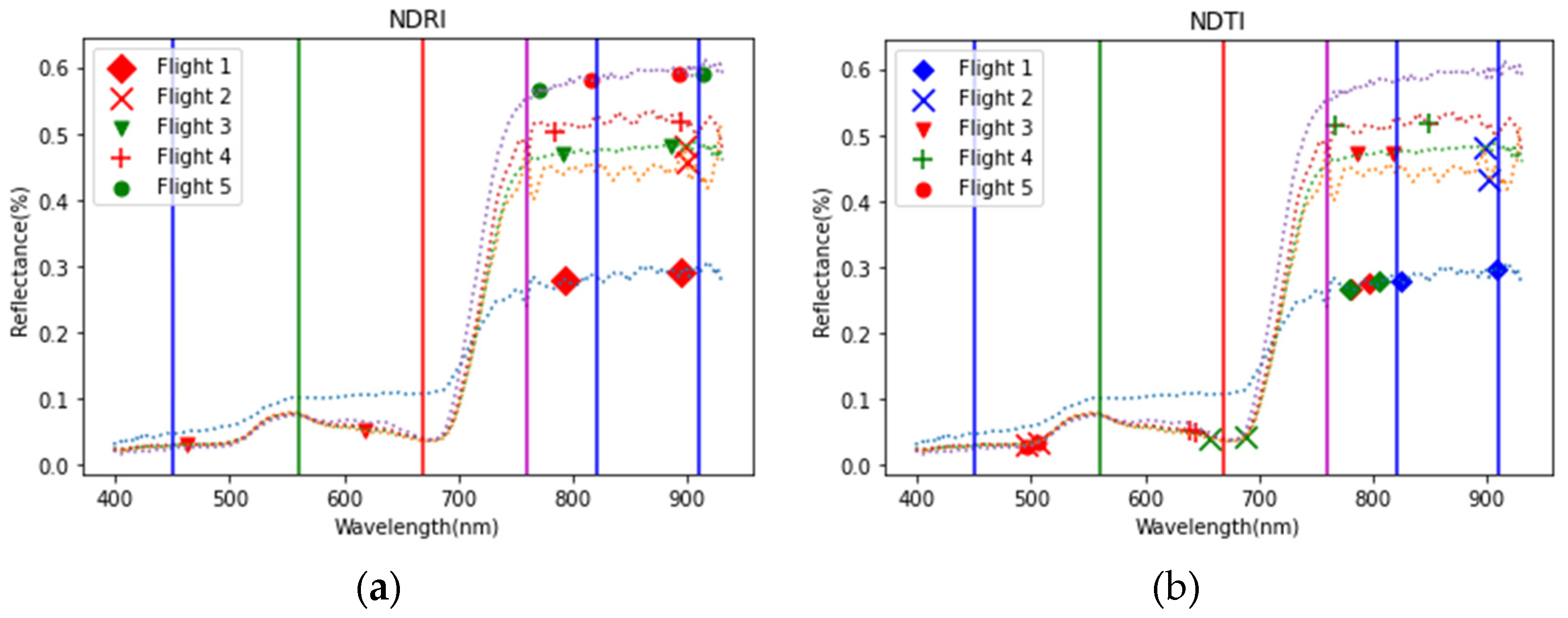
2.4.1. Reflectance Features
2.4.2. Texture Features
2.5. Model Evaluation and Feature Search
2.5.1. Single Feature Approach
2.5.2. Double Feature Approach
2.5.3. Modified Stepwise Regression
3. Results
3.1. Single Features
3.2. Double Features
3.3. Multiple Features
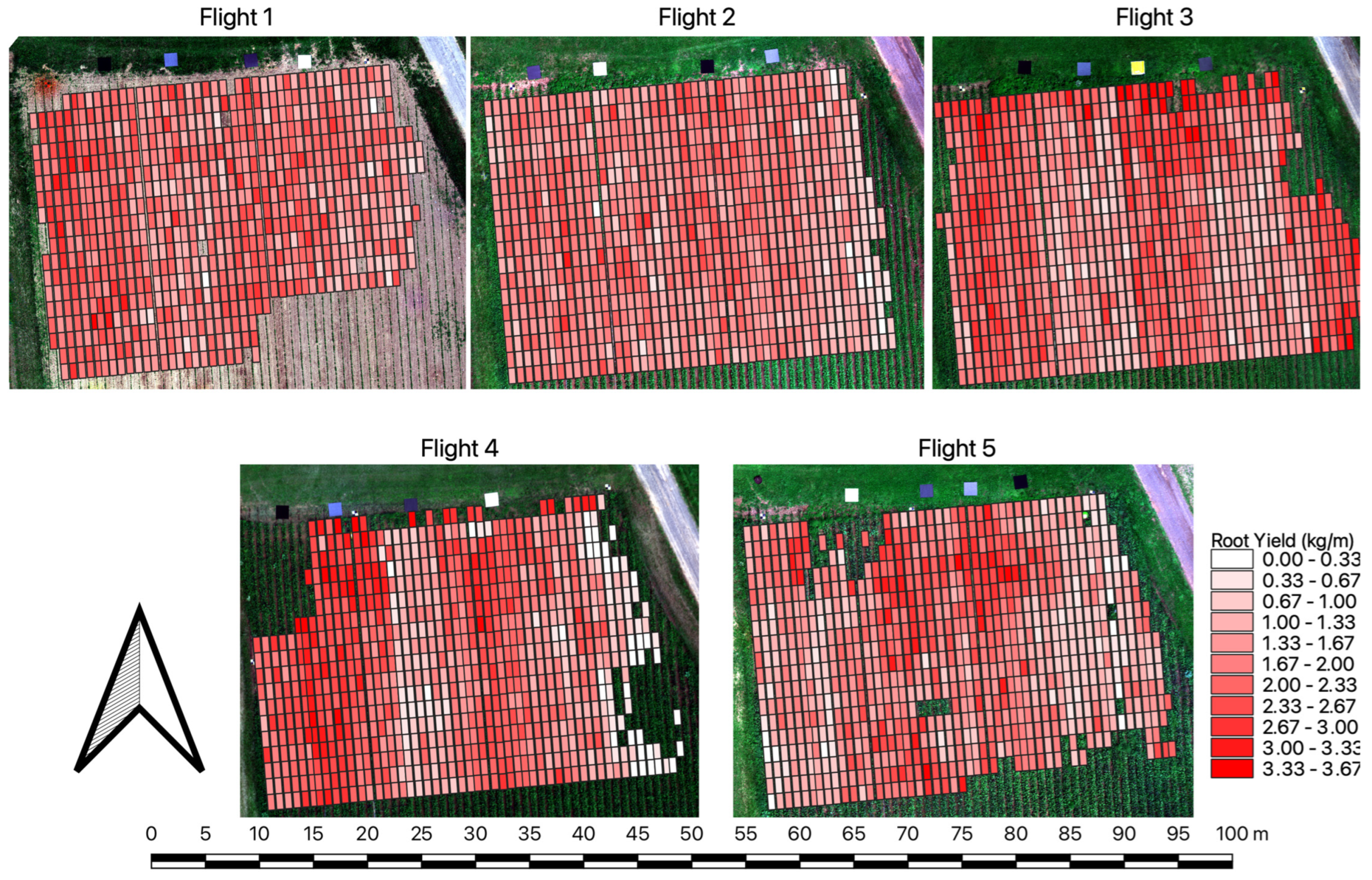
3.4. Extrapolating Yield Modeling Results to the Field Scale
4. Discussion
4.1. Significance of Obtained Features
4.2. Model Performance
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A. Top Features from Each Feature Selection Method
| Flight 1 | Flight 2 | Flight 3 | Flight 4 | Flight 5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| wavelength pair (nm) | R2 | wavelength pair (nm) | R2 | wavelength pair (nm) | R2 | wavelength pair (nm) | R2 | wavelength pair (nm) | R2 |
| [830.4, 765.9] | 0.61 | [897.2, 895.0] | 0.61 | [883.8, 832.6] | 0.73 | [872.7, 799.2] | 0.70 | [834.9, 770.3] | 0.68 |
| [494.3, 485.4] | 0.60 | [899.4, 897.2] | 0.59 | [903.9, 897.2] | 0.70 | [881.6, 808.2] | 0.67 | [892.7, 814.8] | 0.67 |
| [895.0, 765.9] | 0.54 | [801.5, 757.0] | 0.54 | [903.9, 852.7] | 0.68 | [895.0, 785.9] | 0.66 | [883.8, 794.8] | 0.66 |
| [810.4, 736.9] | 0.53 | [897.2, 890.5] | 0.54 | [874.9, 788.1] | 0.68 | [895.0, 783.7] | 0.66 | [926.1, 823.7] | 0.65 |
| [797.0 765.9] | 0.51 | [801.5, 761.4] | 0.52 | [618.9, 463.1] | 0.67 | [888.3, 790.3] | 0.65 | [895.0, 823.7] | 0.65 |
| [785.9, 765.9] | 0.50 | [897.2, 892.7] | 0.51 | [886.1, 832.6] | 0.67 | [888.3, 832.6] | 0.64 | [886.1, 770.3] | 0.64 |
| [826.0, 765.9] | 0.50 | [832.6, 812.6] | 0.51 | [897.2, 788.1] | 0.67 | [877.2, 785.9] | 0.63 | [910.5, 770.3] | 0.63 |
| [821.5, 759.2] | 0.49 | [834.9, 812.6] | 0.50 | [892.7, 788.1] | 0.66 | [846.0, 785.9] | 0.63 | [912.8, 770.3] | 0.63 |
| [765.9, 734.7] | 0.49 | [897.2, 826.0] | 0.49 | [485.4, 454.2] | 0.66 | [892.7, 808.2] | 0.63 | [926.1, 777.0] | 0.63 |
| [895.0, 792.6] | 0.49 | [803.7, 761.4] | 0.49 | [886.1, 790.3] | 0.66 | [921.7, 834.9] | 0.63 | [923.9, 821.5] | 0.61 |
| Flight 1 | Flight 2 | Flight 3 | Flight 4 | Flight 5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| wavelength pair (nm) | R2 | wavelength pair (nm) | R2 | wavelength pair (nm) | R2 | wavelength pair (nm) | R2 | wavelength pair (nm) | R2 |
| [850.4, 781.4] | 0.65 | [901.6, 897.2] | 0.36 | [469.8, 452.0] | 0.63 | [558.8, 541.0] | 0.50 | [610.0, 607.8] | 0.52 |
| [805.9, 781.4] | 0.59 | [590.0, 581.1] | 0.33 | [701.3, 612.3] | 0.59 | [874.9, 763.6] | 0.49 | [681.3, 679.0] | 0.33 |
| [908.3, 823.7] | 0.57 | [587.8, 583.3] | 0.33 | [890.5, 765.9] | 0.58 | [643.4, 639.0] | 0.49 | [618.9, 607.8] | 0.29 |
| [850.4, 785.9] | 0.52 | [687.9, 654.6] | 0.32 | [817.1, 785.9] | 0.58 | [808.2, 765.9] | 0.48 | [478.7, 469.8] | 0.29 |
| [850.4, 748.0] | 0.51 | [590.0, 583.3] | 0.31 | [699.1, 612.3] | 0.56 | [870.5, 763.6] | 0.47 | [614.5, 607.8] | 0.29 |
| [850.4, 743.6] | 0.49 | [590.0, 576.7] | 0.31 | [590.0, 576.7] | 0.53 | [874.9, 777.0] | 0.47 | [505.4, 496.5] | 0.29 |
| [805.9, 779.2] | 0.47 | [590.0, 585.6] | 0.30 | [701.3, 607.8] | 0.53 | [874.9, 765.9] | 0.46 | [420.8, 414.2] | 0.28 |
| [805.9, 748.0] | 0.47 | [687.9, 656.8] | 0.28 | [890.5, 768.1] | 0.53 | [848.2, 765.9] | 0.45 | [505.4, 487.6] | 0.26 |
| [850.4, 759.2] | 0.46 | [505.4, 496.5] | 0.27 | [518.8, 507.6] | 0.52 | [857.1, 792.6] | 0.44 | [681.3, 674.6] | 0.26 |
| [797.0, 781.4] | 0.44 | [912.8, 910.5] | 0.26 | [701.3, 603.4] | 0.51 | [874.9, 812.6] | 0.43 | [708.0, 398.6] | 0.25 |
References
- Clifford, T.; Howatson, G.; West, D.J.; Stevenson, E.J. The Potential Benefits of Red Beetroot Supplementation in Health and Disease. Nutrients 2015, 7, 2801–2822. [Google Scholar] [CrossRef] [PubMed]
- Tanumihardjo, S.A.; Suri, D.; Simon, P.; Goldman, I.L. Vegetables of Temperate Climates: Carrot, Parsnip, and Beetroot. In Encyclopedia of Food and Health; Caballero, B., Finglas, P.M., Toldrá, F., Eds.; Academic Press: Oxford, UK, 2016; pp. 387–392. ISBN 978-0-12-384953-3. [Google Scholar]
- Pedreño, M.A.; Escribano, J. Studying the Oxidation and the Antiradical Activity of Betalain from Beetroot. J. Biol. Educ. 2000, 35, 49–51. [Google Scholar] [CrossRef]
- Gengatharan, A.; Dykes, G.A.; Choo, W.S. Betalains: Natural Plant Pigments with Potential Application in Functional Foods. LWT Food Sci. Technol. 2015, 64, 645–649. [Google Scholar] [CrossRef]
- Toth, C.; Jóźków, G. Remote Sensing Platforms and Sensors: A Survey. ISPRS J. Photogramm. Remote Sens. 2016, 115, 22–36. [Google Scholar] [CrossRef]
- Zhang, C.; Kovacs, J.M. The Application of Small Unmanned Aerial Systems for Precision Agriculture: A Review. Precis. Agric. 2012, 13, 693–712. [Google Scholar] [CrossRef]
- Chancia, R.; Bates, T.; Vanden Heuvel, J.; van Aardt, J. Assessing Grapevine Nutrient Status from Unmanned Aerial System (UAS) Hyperspectral Imagery. Remote Sens. 2021, 13, 4489. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Li, D.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Combining Unmanned Aerial Vehicle (UAV)-Based Multispectral Imagery and Ground-Based Hyperspectral Data for Plant Nitrogen Concentration Estimation in Rice. Front. Plant Sci. 2018, 9, 936. [Google Scholar] [CrossRef]
- Ahmad, A.; Aggarwal, V.; Saraswat, D.; El Gamal, A.; Johal, G.S. GeoDLS: A Deep Learning-Based Corn Disease Tracking and Location System Using RTK Geolocated UAS Imagery. Remote Sens. 2022, 14, 4140. [Google Scholar] [CrossRef]
- Oh, S.; Lee, D.-Y.; Gongora-Canul, C.; Ashapure, A.; Carpenter, J.; Cruz, A.P.; Fernandez-Campos, M.; Lane, B.Z.; Telenko, D.E.P.; Jung, J.; et al. Tar Spot Disease Quantification Using Unmanned Aircraft Systems (UAS) Data. Remote Sens. 2021, 13, 2567. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Zhou, M.; Li, D.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Improved Estimation of Rice Aboveground Biomass Combining Textural and Spectral Analysis of UAV Imagery. Precis. Agric. 2019, 20, 611–629. [Google Scholar] [CrossRef]
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Long, H.; Yue, J.; Li, Z.; Yang, G.; Yang, X.; Fan, L. Estimation of Crop Growth Parameters Using UAV-Based Hyperspectral Remote Sensing Data. Sensors 2020, 20, 1296. [Google Scholar] [CrossRef] [PubMed]
- Simpson, J.E.; Holman, F.H.; Nieto, H.; El-Madany, T.S.; Migliavacca, M.; Martin, M.P.; Burchard-Levine, V.; Cararra, A.; Blöcher, S.; Fiener, P.; et al. UAS-Based High Resolution Mapping of Evapotranspiration in a Mediterranean Tree-Grass Ecosystem. Agric. For. Meteorol. 2022, 321, 108981. [Google Scholar] [CrossRef]
- de Oca, A.M.; Flores, G. A UAS Equipped with a Thermal Imaging System with Temperature Calibration for Crop Water Stress Index Computation. In Proceedings of the 2021 International Conference on Unmanned Aircraft Systems (ICUAS), Athens, Greece, 15–18 June 2021; pp. 714–720. [Google Scholar]
- Singh, V.; Rana, A.; Bishop, M.; Filippi, A.M.; Cope, D.; Rajan, N.; Bagavathiannan, M. Chapter Three—Unmanned Aircraft Systems for Precision Weed Detection and Management: Prospects and Challenges. In Advances in Agronomy; Sparks, D.L., Ed.; Academic Press: Cambridge, MA, USA, 2020; Volume 159, pp. 93–134. [Google Scholar]
- Yu, N.; Li, L.; Schmitz, N.; Tian, L.F.; Greenberg, J.A.; Diers, B.W. Development of Methods to Improve Soybean Yield Estimation and Predict Plant Maturity with an Unmanned Aerial Vehicle Based Platform. Remote Sens. Environ. 2016, 187, 91–101. [Google Scholar] [CrossRef]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting Grain Yield in Rice Using Multi-Temporal Vegetation Indices from UAV-Based Multispectral and Digital Imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Hassanzadeh, A.; Zhang, F.; van Aardt, J.; Murphy, S.P.; Pethybridge, S.J. Broadacre Crop Yield Estimation Using Imaging Spectroscopy from Unmanned Aerial Systems (UAS): A Field-Based Case Study with Snap Bean. Remote Sens. 2021, 13, 3241. [Google Scholar] [CrossRef]
- Eismann, M. Hyperspectral Remote Sensing; Society of Photo-Optical Instrumentation Engineers: Bellingham, WA, USA, 2012. [Google Scholar]
- Georganos, S.; Grippa, T.; Vanhuysse, S.; Lennert, M.; Shimoni, M.; Kalogirou, S.; Wolff, E. Less Is More: Optimizing Classification Performance through Feature Selection in a Very-High-Resolution Remote Sensing Object-Based Urban Application. GISci. Remote Sens. 2018, 55, 221–242. [Google Scholar] [CrossRef]
- Khodr, J.; Younes, R. Dimensionality Reduction on Hyperspectral Images: A Comparative Review Based on Artificial Datas. In Proceedings of the 2011 4th International Congress on Image and Signal Processing, Shanghai, China, 15–17 October 2011; Volume 4, pp. 1875–1883. [Google Scholar]
- Kokaly, R.F.; Clark, R.N. Spectroscopic Determination of Leaf Biochemistry Using Band-Depth Analysis of Absorption Features and Stepwise Multiple Linear Regression. Remote Sens. Environ. 1999, 67, 267–287. [Google Scholar] [CrossRef]
- Hassanzadeh, A.; van Aardt, J.; Murphy, S.P.; Pethybridge, S.J. Yield Modeling of Snap Bean Based on Hyperspectral Sensing: A Greenhouse Study. JARS 2020, 14, 024519. [Google Scholar] [CrossRef]
- Raj, R.; Walker, J.P.; Pingale, R.; Banoth, B.N.; Jagarlapudi, A. Leaf Nitrogen Content Estimation Using Top-of-Canopy Airborne Hyperspectral Data. Int. J. Appl. Earth Obs. Geoinf. 2021, 104, 102584. [Google Scholar] [CrossRef]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I. Textural Features for Image Classification. IEEE Trans. Syst. Man Cybern. 1973, SMC-3, 610–621. [Google Scholar] [CrossRef]
- He, D.-C.; Wang, L. Texture Unit, Texture Spectrum, and Texture Analysis. IEEE Trans. Geosci. Remote Sens. 1990, 28, 509–512. [Google Scholar]
- Unser, M. Texture Classification and Segmentation Using Wavelet Frames. IEEE Trans. Image Process. 1995, 4, 1549–1560. [Google Scholar] [CrossRef] [PubMed]
- Riou, R.; Seyler, F. Texture Analysis of Tropical Rain Forest Infrared Satellite Images. Photogramm. Eng. Remote Sens. 1997, 63, 515–521. [Google Scholar]
- Podest, E.; Saatchi, S. Application of Multiscale Texture in Classifying JERS-1 Radar Data over Tropical Vegetation. Int. J. Remote Sens. 2002, 23, 1487–1506. [Google Scholar] [CrossRef]
- Nyoungui, A.N.; Tonye, E.; Akono, A. Evaluation of Speckle Filtering and Texture Analysis Methods for Land Cover Classification from SAR Images. Int. J. Remote Sens. 2002, 23, 1895–1925. [Google Scholar] [CrossRef]
- Marceau, D.J.; Howarth, P.J.; Dubois, J.M.M.; Gratton, D.J. Evaluation of the Grey-Level Co-Occurrence Matrix Method for Land-Cover Classification Using SPOT Imagery. IEEE Trans. Geosci. Remote Sens. 1990, 28, 513–519. [Google Scholar] [CrossRef]
- Franklin, S.E.; Hall, R.J.; Moskal, L.M.; Maudie, A.J.; Lavigne, M.B. Incorporating Texture into Classification of Forest Species Composition from Airborne Multispectral Images. Int. J. Remote Sens. 2000, 21, 61–79. [Google Scholar] [CrossRef]
- Augusteijn, M.F.; Clemens, L.E.; Shaw, K.A. Performance Evaluation of Texture Measures for Ground Cover Identification in Satellite Images by Means of a Neural Network Classifier. IEEE Trans. Geosci. Remote Sens. 1995, 33, 616–626. [Google Scholar] [CrossRef]
- Franklin, S.E.; Peddle, D.R. Spectral Texture for Improved Class Discrimination in Complex Terrain. Int. J. Remote Sens. 1989, 10, 1437–1443. [Google Scholar] [CrossRef]
- Lu, D.; Batistella, M. Exploring TM Image Texture and Its Relationships with Biomass Estimation in Rondônia, Brazilian Amazon. Acta Amaz. 2005, 35, 249–257. [Google Scholar] [CrossRef]
- Sarker, L.R.; Nichol, J.E. Improved Forest Biomass Estimates Using ALOS AVNIR-2 Texture Indices. Remote Sens. Environ. 2011, 115, 968–977. [Google Scholar] [CrossRef]
- Eckert, S. Improved Forest Biomass and Carbon Estimations Using Texture Measures from WorldView-2 Satellite Data. Remote Sens. 2012, 4, 810–829. [Google Scholar] [CrossRef]
- Kelsey, K.C.; Neff, J.C. Estimates of Aboveground Biomass from Texture Analysis of Landsat Imagery. Remote Sens. 2014, 6, 6407–6422. [Google Scholar] [CrossRef]
- Féret, J.-B.; Gitelson, A.A.; Noble, S.D.; Jacquemoud, S. PROSPECT-D: Towards Modeling Leaf Optical Properties through a Complete Lifecycle. Remote Sens. Environ. 2017, 193, 204–215. [Google Scholar] [CrossRef]
- Zheng, C.; Sun, D.-W.; Zheng, L. Recent Applications of Image Texture for Evaluation of Food Qualities—A Review. Trends Food Sci. Technol. 2006, 17, 113–128. [Google Scholar] [CrossRef]
- Xie, C.; He, Y. Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves. Sensors 2016, 16, 676. [Google Scholar] [CrossRef]
- Zhu, F.; Zhang, D.; He, Y.; Liu, F.; Sun, D.-W. Application of Visible and Near Infrared Hyperspectral Imaging to Differentiate Between Fresh and Frozen–Thawed Fish Fillets. Food Bioprocess Technol. 2013, 6, 2931–2937. [Google Scholar] [CrossRef]
- Guo, A.; Huang, W.; Ye, H.; Dong, Y.; Ma, H.; Ren, Y.; Ruan, C. Identification of Wheat Yellow Rust Using Spectral and Texture Features of Hyperspectral Images. Remote Sens. 2020, 12, 1419. [Google Scholar] [CrossRef]
- Jia, B.; Wang, W.; Yoon, S.-C.; Zhuang, H.; Li, Y.-F. Using a Combination of Spectral and Textural Data to Measure Water-Holding Capacity in Fresh Chicken Breast Fillets. Appl. Sci. 2018, 8, 343. [Google Scholar] [CrossRef]
- Yang, Y.; Wang, W.; Zhuang, H.; Yoon, S.-C.; Jiang, H. Fusion of Spectra and Texture Data of Hyperspectral Imaging for the Prediction of the Water-Holding Capacity of Fresh Chicken Breast Filets. Appl. Sci. 2018, 8, 640. [Google Scholar] [CrossRef]
- Olson, D.; Chatterjee, A.; Franzen, D.W. Can We Select Sugarbeet Harvesting Dates Using Drone-Based Vegetation Indices? Agron. J. 2019, 111, 2619–2624. [Google Scholar] [CrossRef]
- Al-Gaadi, K.A.; Hassaballa, A.A.; Tola, E.; Kayad, A.G.; Madugundu, R.; Alblewi, B.; Assiri, F. Prediction of Potato Crop Yield Using Precision Agriculture Techniques. PLoS ONE 2016, 11, e0162219. [Google Scholar] [CrossRef] [PubMed]
- Luo, S.; He, Y.; Li, Q.; Jiao, W.; Zhu, Y.; Zhao, X. Nondestructive Estimation of Potato Yield Using Relative Variables Derived from Multi-Period LAI and Hyperspectral Data Based on Weighted Growth Stage. Plant Methods 2020, 16, 150. [Google Scholar] [CrossRef] [PubMed]
- Li, B.; Xu, X.; Zhang, L.; Han, J.; Bian, C.; Li, G.; Liu, J.; Jin, L. Above-Ground Biomass Estimation and Yield Prediction in Potato by Using UAV-Based RGB and Hyperspectral Imaging. ISPRS J. Photogramm. Remote Sens. 2020, 162, 161–172. [Google Scholar] [CrossRef]
- Li, D.; Miao, Y.; Gupta, S.K.; Rosen, C.J.; Yuan, F.; Wang, C.; Wang, L.; Huang, Y. Improving Potato Yield Prediction by Combining Cultivar Information and UAV Remote Sensing Data Using Machine Learning. Remote Sens. 2021, 13, 3322. [Google Scholar] [CrossRef]
- Suarez, L.A.; Robson, A.; McPhee, J.; O’Halloran, J.; van Sprang, C. Accuracy of Carrot Yield Forecasting Using Proximal Hyperspectral and Satellite Multispectral Data. Precis. Agric. 2020, 21, 1304–1326. [Google Scholar] [CrossRef]
- Olson, D.; Chatterjee, A.; Franzen, D.W.; Day, S.S. Relationship of Drone-Based Vegetation Indices with Corn and Sugarbeet Yields. Agron. J. 2019, 111, 2545–2557. [Google Scholar] [CrossRef]
- Chancia, R.; van Aardt, J.; Pethybridge, S.; Cross, D.; Henderson, J. Predicting Table Beet Root Yield with Multispectral UAS Imagery. Remote Sens. 2021, 13, 2180. [Google Scholar] [CrossRef]
- RedEdge-M User Manual (PDF)—Legacy. Available online: https://support.micasense.com/hc/en-us/articles/115003537673-RedEdge-M-User-Manual-PDF-Legacy (accessed on 21 July 2022).
- Hyperspectral and Operational Software. Available online: https://www.headwallphotonics.com/products/software (accessed on 24 July 2022).
- PIX4Dmapper: Professional Photogrammetry Software for Drone Mapping. Available online: https://www.pix4d.com/product/pix4dmapper-photogrammetry-software (accessed on 24 July 2022).
- Atmospheric Correction. Available online: https://www.l3harrisgeospatial.com/docs/atmosphericcorrection.html#empirical_line_calibration (accessed on 24 July 2022).
- Rasti, B.; Scheunders, P.; Ghamisi, P.; Licciardi, G.; Chanussot, J. Noise Reduction in Hyperspectral Imagery: Overview and Application. Remote Sens. 2018, 10, 482. [Google Scholar] [CrossRef]
- Chen, G.; Qian, S.-E. Denoising of Hyperspectral Imagery Using Principal Component Analysis and Wavelet Shrinkage. IEEE Trans. Geosci. Remote Sens. 2011, 49, 973–980. [Google Scholar] [CrossRef]
- Velicer, W.F. Determining the Number of Components from the Matrix of Partial Correlations. Psychometrika 1976, 41, 321–327. [Google Scholar] [CrossRef]
- Chen, G.Y.; Zhu, W.-P. Signal Denoising Using Neighbouring Dual-Tree Complex Wavelet Coefficients. In Proceedings of the 2009 Canadian Conference on Electrical and Computer Engineering, St. John’s, NL, Canada, 3–6 May 2009; pp. 565–568. [Google Scholar]
- Kruse, F.A.; Lefkoff, A.B.; Boardman, J.W.; Heidebrecht, K.B.; Shapiro, A.T.; Barloon, P.J.; Goetz, A.F.H. The Spectral Image Processing System (SIPS)—Interactive Visualization and Analysis of Imaging Spectrometer Data. Remote Sens. Environ. 1993, 44, 145–163. [Google Scholar] [CrossRef]
- Huete, A.R.; Jackson, R.D.; Post, D.F. Spectral Response of a Plant Canopy with Different Soil Backgrounds. Remote Sens. Environ. 1985, 17, 37–53. [Google Scholar] [CrossRef]
- Colwell, J.E. Vegetation Canopy Reflectance. Remote Sens. Environ. 1974, 3, 175–183. [Google Scholar] [CrossRef]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef]
- Townshend, J.R.G.; Goff, T.E.; Tucker, C.J. Multitemporal Dimensionality of Images of Normalized Difference Vegetation Index at Continental Scales. IEEE Trans. Geosci. Remote Sens. 1985, GE-23, 888–895. [Google Scholar] [CrossRef]
- Zhu, Z.; Bi, J.; Pan, Y.; Ganguly, S.; Anav, A.; Xu, L.; Samanta, A.; Piao, S.; Nemani, R.R.; Myneni, R.B. Global Data Sets of Vegetation Leaf Area Index (LAI)3g and Fraction of Photosynthetically Active Radiation (FPAR)3g Derived from Global Inventory Modeling and Mapping Studies (GIMMS) Normalized Difference Vegetation Index (NDVI3g) for the Period 1981 to 2011. Remote Sens. 2013, 5, 927–948. [Google Scholar] [CrossRef]
- Schaefer, M.T.; Lamb, D.W. A Combination of Plant NDVI and LiDAR Measurements Improve the Estimation of Pasture Biomass in Tall Fescue (Festuca Arundinacea Var. Fletcher). Remote Sens. 2016, 8, 109. [Google Scholar] [CrossRef]
- Hassan, M.A.; Yang, M.; Rasheed, A.; Yang, G.; Reynolds, M.; Xia, X.; Xiao, Y.; He, Z. A Rapid Monitoring of NDVI across the Wheat Growth Cycle for Grain Yield Prediction Using a Multi-Spectral UAV Platform. Plant Sci. 2019, 282, 95–103. [Google Scholar] [CrossRef]
- Sun, Y.; Ren, H.; Zhang, T.; Zhang, C.; Qin, Q. Crop Leaf Area Index Retrieval Based on Inverted Difference Vegetation Index and NDVI. IEEE Geosci. Remote Sens. Lett. 2018, 15, 1662–1666. [Google Scholar] [CrossRef]
- Li, F.; Miao, Y.; Feng, G.; Yuan, F.; Yue, S.; Gao, X.; Liu, Y.; Liu, B.; Ustin, S.L.; Chen, X. Improving Estimation of Summer Maize Nitrogen Status with Red Edge-Based Spectral Vegetation Indices. Field Crops Res. 2014, 157, 111–123. [Google Scholar] [CrossRef]
- Tan, C.-W.; Zhang, P.-P.; Zhou, X.-X.; Wang, Z.-X.; Xu, Z.-Q.; Mao, W.; Li, W.-X.; Huo, Z.-Y.; Guo, W.-S.; Yun, F. Quantitative Monitoring of Leaf Area Index in Wheat of Different Plant Types by Integrating NDVI and Beer-Lambert Law. Sci. Rep. 2020, 10, 929. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, N.; Saito, Y.; Maki, M.; Homma, K. Simulation of Reflectance and Vegetation Indices for Unmanned Aerial Vehicle (UAV) Monitoring of Paddy Fields. Remote Sens. 2019, 11, 2119. [Google Scholar] [CrossRef]
- Chen, S.; She, D.; Zhang, L.; Guo, M.; Liu, X. Spatial Downscaling Methods of Soil Moisture Based on Multisource Remote Sensing Data and Its Application. Water 2019, 11, 1401. [Google Scholar] [CrossRef]
- Marshall, M.; Thenkabail, P.; Biggs, T.; Post, K. Hyperspectral Narrowband and Multispectral Broadband Indices for Remote Sensing of Crop Evapotranspiration and Its Components (Transpiration and Soil Evaporation). Agric. For. Meteorol. 2016, 218–219, 122–134. [Google Scholar] [CrossRef]
- Thenkabail, P.S.; Smith, R.B.; De Pauw, E. Hyperspectral Vegetation Indices and Their Relationships with Agricultural Crop Characteristics. Remote Sens. Environ. 2000, 71, 158–182. [Google Scholar] [CrossRef]
- Koppe, W.; Li, F.; Gnyp, M.; Miao, Y.; Jia, L.; Chen, X.; Zhang, F.; Bareth, G. Evaluating Multispectral and Hyperspectral Satellite Remote Sensing Data for Estimating Winter Wheat Growth Parameters at Regional Scale in the North China Plain. Photogramm. Fernerkund. Geoinf. 2010, 2010, 167–178. [Google Scholar] [CrossRef]
- Zheng, H.; Ma, J.; Zhou, M.; Li, D.; Yao, X.; Cao, W.; Zhu, Y.; Cheng, T. Enhancing the Nitrogen Signals of Rice Canopies across Critical Growth Stages through the Integration of Textural and Spectral Information from Unmanned Aerial Vehicle (UAV) Multispectral Imagery. Remote Sens. 2020, 12, 957. [Google Scholar] [CrossRef]
- Yue, J.; Yang, G.; Tian, Q.; Feng, H.; Xu, K.; Zhou, C. Estimate of Winter-Wheat above-Ground Biomass Based on UAV Ultrahigh-Ground-Resolution Image Textures and Vegetation Indices. ISPRS J. Photogramm. Remote Sens. 2019, 150, 226–244. [Google Scholar] [CrossRef]
- Wong, T.-T. Performance Evaluation of Classification Algorithms by K-Fold and Leave-One-out Cross Validation. Pattern Recognit. 2015, 48, 2839–2846. [Google Scholar] [CrossRef]
- Smith, G. Step Away from Stepwise. J. Big Data 2018, 5, 32. [Google Scholar] [CrossRef]
- Grossman, Y.L.; Ustin, S.L.; Jacquemoud, S.; Sanderson, E.W.; Schmuck, G.; Verdebout, J. Critique of Stepwise Multiple Linear Regression for the Extraction of Leaf Biochemistry Information from Leaf Reflectance Data. Remote Sens. Environ. 1996, 56, 182–193. [Google Scholar] [CrossRef]
- Whittingham, M.J.; Stephens, P.A.; Bradbury, R.B.; Freckleton, R.P. Why Do We Still Use Stepwise Modelling in Ecology and Behaviour? J. Anim. Ecol. 2006, 75, 1182–1189. [Google Scholar] [CrossRef] [PubMed]
- Hegyi, G.; Garamszegi, L.Z. Using Information Theory as a Substitute for Stepwise Regression in Ecology and Behavior. Behav. Ecol. Sociobiol. 2011, 65, 69–76. [Google Scholar] [CrossRef]
- Karagiannopoulos, M.; Anyfantis, D.; Kotsiantis, S.; Pintelas, P. Feature Selection for Regression Problems. Educational Software Development Laboratory, Department of Mathematics, University of Patras: Patras, Greece, 2007; pp. 20–22. [Google Scholar]
- Jović, A.; Brkić, K.; Bogunović, N. A Review of Feature Selection Methods with Applications. In Proceedings of the 2015 38th International Convention on Information and Communication Technology, Electronics and Microelectronics (MIPRO), Opatija, Croatia, 25–29 May 2015; pp. 1200–1205. [Google Scholar]
- Craney, T.A.; Surles, J.G. Model-Dependent Variance Inflation Factor Cutoff Values. Qual. Eng. 2002, 14, 391–403. [Google Scholar] [CrossRef]
- Stine, R.A. Graphical Interpretation of Variance Inflation Factors. Am. Stat. 1995, 49, 53–56. [Google Scholar] [CrossRef]
- Gao, B.-C.; Montes, M.J.; Davis, C.O. Refinement of Wavelength Calibrations of Hyperspectral Imaging Data Using a Spectrum-Matching Technique. Remote Sens. Environ. 2004, 90, 424–433. [Google Scholar] [CrossRef]
- Wolfe, W.L. Introduction to Imaging Spectrometers; SPIE Press: Bellingham, WA, USA, 1997; ISBN 978-0-8194-2260-6. [Google Scholar]
- Ceclu, L.; Nistor, O.-V. Red Beetroot: Composition and Health Effects—A Review. J. Nutr. Med. Diet Care 2020, 6, 1–9. [Google Scholar] [CrossRef]
- Blackburn, G.A. Quantifying Chlorophylls and Caroteniods at Leaf and Canopy Scales: An Evaluation of Some Hyperspectral Approaches. Remote Sens. Environ. 1998, 66, 274–285. [Google Scholar] [CrossRef]
- Stroppiana, D.; Boschetti, M.; Brivio, P.A.; Bocchi, S. Plant Nitrogen Concentration in Paddy Rice from Field Canopy Hyperspectral Radiometry. Field Crops Res. 2009, 111, 119–129. [Google Scholar] [CrossRef]
- Wei, M.C.F.; Maldaner, L.F.; Ottoni, P.M.N.; Molin, J.P. Carrot Yield Mapping: A Precision Agriculture Approach Based on Machine Learning. AI 2020, 1, 229–241. [Google Scholar] [CrossRef]


| Date | Milestone | Growth Stages |
|---|---|---|
| 20-May | Crop planted | Germination |
| 16-Jun | 1st Flight | Leaf development (crop emergence) |
| 7-Jul | 2nd Flight | Leaf development (more than 9 leaves unfolded) |
| 15-Jul | 3rd Flight | Rosette growth |
| 20-Jul | 4th Flight | Rosette growth (cont’d) |
| 2-Aug | 5th Flight | Harvest-ready |
| 5-Aug | Harvest | Harvest-ready |
| Flight 1 | ||||
|---|---|---|---|---|
| Features | R2 | R2adj | RMSE (%) | VIF |
| d [895.0, 792.6] t [908.3, 823.7] | 0.77 | 0.74 | 16.13 | (1.241, 1.241) |
| d [826.0, 765.9] t [805.9, 779.2] | 0.76 | 0.73 | 16.23 | (1.195, 1.195) |
| d [494.3, 485.4] t [908.3, 823.7] | 0.76 | 0.73 | 16.32 | (1.496, 1.496) |
| d [895.0, 765.9] t [908.3, 823.7] | 0.75 | 0.72 | 16.66 | (1.334, 1.334) |
| d [494.3, 485.4] t [797.0, 781.4] | 0.75 | 0.72 | 16.73 | (1.324, 1.324) |
| Flight 2 | ||||
| Features | R2 | R2adj | RMSE (%) | VIF |
| d [897.2, 895.0] t [901.6, 897.2] | 0.73 | 0.69 | 17.37 | (1.281, 1.281) |
| d [801.5, 761.4] t [687.9, 656.8] | 0.71 | 0.67 | 18.16 | (1.103, 1.103) |
| d [899.4, 897.2] t [505.4, 496.5] | 0.71 | 0.67 | 18.27 | (1.118, 1.118) |
| d [899.4, 897.2] t [687.9, 654.6] | 0.69 | 0.65 | 18.66 | (1.264, 1.264) |
| d [803.7, 761.4] t [587.8, 583.3] | 0.67 | 0.63 | 19.03 | (1.097, 1.097) |
| Flight 3 | ||||
| Features | R2 | R2adj | RMSE (%) | VIF |
| d [897.2, 788.1] t [469.8, 452.0] | 0.82 | 0.80 | 14.23 | (1.651, 1.651) |
| d [886.1, 790.3] t [817.1, 785.9] | 0.81 | 0.79 | 14.54 | (1.555, 1.555) |
| d [874.9, 788.1] t [469.8, 452.0] | 0.81 | 0.78 | 14.72 | (1.77, 1.77) |
| d [903.9, 897.2] t [469.8, 452.0] | 0.80 | 0.78 | 14.97 | (1.766, 1.766) |
| d [886.1, 790.3] t [469.8, 452.0] | 0.79 | 0.76 | 15.32 | (1.756, 1.756) |
| Flight 4 | ||||
| Features | R2 | R2adj | RMSE (%) | VIF |
| d [895.0, 785.9] t [848.2, 765.9] | 0.85 | 0.83 | 13.20 | (1.198, 1.198) |
| d [895.0, 785.9] t [808.2, 765.9] | 0.83 | 0.81 | 13.70 | (1.226, 1.226) |
| d [895.0, 783.7] t [848.2, 765.9] | 0.83 | 0.81 | 13.90 | (1.252, 1.252) |
| d [895.0, 785.9] t [874.9, 763.6] | 0.82 | 0.80 | 14.15 | (1.341, 1.341) |
| d [895.0, 783.7] t [857.1, 792.6] | 0.82 | 0.80 | 14.23 | (1.264, 1.264) |
| Flight 5 | ||||
| Features | R2 | R2adj | RMSE (%) | VIF |
| d [892.7, 814.8] t [505.4, 496.5] | 0.82 | 0.79 | 14.41 | (1.108, 1.108) |
| d [883.8, 794.8] t [614.5, 607.8] | 0.79 | 0.77 | 15.19 | (1.178, 1.178) |
| d [910.5, 770.3] t [505.4, 496.5] | 0.78 | 0.75 | 15.71 | (1.146, 1.146) |
| d [886.1, 770.3] t [505.4, 496.5] | 0.76 | 0.73 | 16.43 | (1.186, 1.186) |
| d [883.8, 794.8] t [610.0, 607.8] | 0.76 | 0.73 | 16.46 | (1.694, 1.694) |
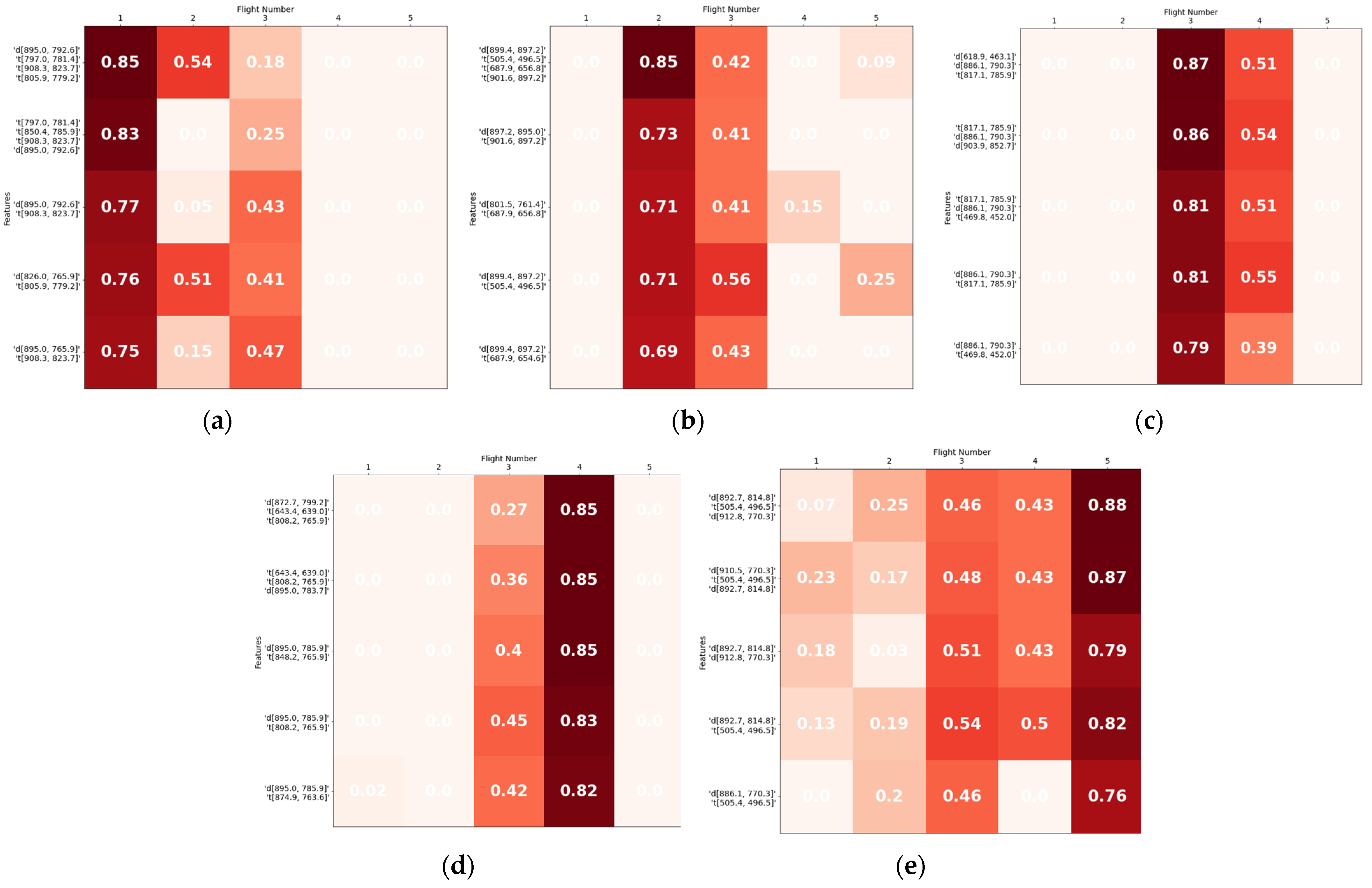
| Flight 1 | |||||
|---|---|---|---|---|---|
| features | R2 | R2adj | RMSE (%) | VIF | p-values |
| ['d [895.0, 792.6]', 't [797.0, 781.4]', 't [908.3, 823.7]', 't [805.9, 779.2]'] | 0.85 | 0.81 | 12.93 | [1.463, 1.632 2.136, 1.879] | [0.004, 0.027 0.019, 0.042] |
| ['t [797.0, 781.4]', 'd [895.0, 792.6]', 't [850.4, 743.6]', 't [908.3, 823.7]'] | 0.84 | 0.79 | 13.59 | [1.648, 1.387 1.944, 2.205] | [0.043, 0.002 0.048, 0.026] |
| ['t [797.0, 781.4]', 't [850.4, 785.9]', 't [908.3, 823.7]', 'd [895.0, 792.6]'] | 0.83 | 0.78 | 13.74 | [1.640, 1.959 1.938, 1.743] | [0.021, 0.046 0.010, 0.019] |
| ['d [494.3, 485.4]', 't [797.0, 781.4]', 't [908.3, 823.7]'] | 0.82 | 0.78 | 14.28 | [1.589, 1.595 1.803] | [0.003, 0.041, 0.012] |
| Flight 2 | |||||
| Features | R2 | R2adj | RMSE (%) | VIF | p-values |
| ['d [899.4, 897.2]', 't [505.4, 496.5]', 't [687.9, 656.8]', 't [901.6, 897.2]'] | 0.85 | 0.81 | 12.93 | [1.813, 1.219 1.361, 1.586] | [0.006, 0.006 0.014, 0.019] |
| Flight 3 | |||||
| Features | R2 | R2adj | RMSE (%) | VIF | p-values |
| ['d [618.9, 463.1]', 'd [886.1, 790.3]', 't [817.1, 785.9]'] | 0.87 | 0.85 | 11.85 | [2.266, 1.839, 2.078] | [0.011, 0.002, 0.020] |
| ['t [817.1, 785.9]', 'd [886.1, 790.3]', 'd [903.9, 852.7]'] | 0.86 | 0.83 | 12.63 | [2.070, 1.955, 2.457] | [0.021, 0.004, 0.020] |
| ['d [903.9, 852.7]', 'd [886.1, 790.3]', 't [469.8, 452.0]'] | 0.84 | 0.81 | 13.46 | [2.405, 2.082, 2.289] | [0.022, 0.008, 0.023] |
| ['t [817.1, 785.9]', 'd [886.1, 832.6]', 'd [618.9, 463.1]'] | 0.84 | 0.80 | 13.45 | [2.149, 2.044, 2.310] | [0.038, 0.006, 0.019] |
| ['d [886.1, 790.3]', 't [469.8, 452.0]', 'd [618.9, 463.1]'] | 0.83 | 0.80 | 13.69 | [1.962, 2.379, 2.299] | [0.004, 0.027, 0.033] |
| ['t [469.8, 452.0]', 'd [886.1, 790.3]', 'd [892.7, 788.1]'] | 0.83 | 0.79 | 13.92 | [2.026, 2.284, 2.293] | [0.008, 0.024, 0.024] |
| ['t [817.1, 785.9]', 'd [886.1, 790.3]', 't [469.8, 452.0]'] | 0.81 | 0.77 | 14.51 | [2.276, 1.852, 2.569] | [0.045, 0.003, 0.049] |
| ['t [817.1, 785.9]', 'd [886.1, 832.6]', 'd [874.9, 788.1]'] | 0.81 | 0.77 | 14.60 | [2.074, 2.370, 2.700] | [0.032, 0.022, 0.037] |
| Flight 4 | |||||
| Features | R2 | R2adj | RMSE (%) | VIF | p-values |
| ['t [643.4, 639.0]', 't [848.2, 765.9]', 'd [895.0, 783.7]'] | 0.90 | 0.87 | 10.81 | [1.666, 1.332, 1.672] | [0.007, 0.001, 0.000] |
| ['d [872.7, 799.2]', 't [643.4, 639.0]', 't [808.2, 765.9]'] | 0.85 | 0.81 | 13.15 | [1.951, 1.603, 1.624] | [0.003, 0.008, 0.025] |
| ['t [643.4, 639.0]', 't [808.2, 765.9]', 'd [895.0, 783.7]'] | 0.85 | 0.81 | 13.20 | [1.676, 1.413, 1.713] | [0.012, 0.003, 0.001] |
| Flight 5 | |||||
| Features | R2 | R2adj | RMSE (%) | VIF | p-values |
| ['d [892.7, 814.8]', 't [505.4, 496.5]', 'd [912.8, 770.3]'] | 0.88 | 0.85 | 11.80 | [1.680, 1.446, 2.184] | [0.000, 0.015, 0.016] |
| ['d [910.5, 770.3]', 't [505.4, 496.5]', 'd [892.7, 814.8]'] | 0.87 | 0.84 | 12.14 | [2.929, 1.148, 2.833] | [0.036, 0.002, 0.009] |
| ['d [926.1, 823.7]', 'd [883.8, 794.8]'] | 0.81 | 0.78 | 14.60 | [1.975, 1.975] | [0.004, 0.004] |
| ['d [883.8, 794.8]', 'd [926.1, 777.0]'] | 0.80 | 0.77 | 15.08 | [1.884, 1.884] | [0.003, 0.008] |
| ['d [892.7, 814.8]', 'd [912.8, 770.3]'] | 0.79 | 0.77 | 15.27 | [1.674, 1.674] | [0.001, 0.003] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Saif, M.S.; Chancia, R.; Pethybridge, S.; Murphy, S.P.; Hassanzadeh, A.; van Aardt, J. Forecasting Table Beet Root Yield Using Spectral and Textural Features from Hyperspectral UAS Imagery. Remote Sens. 2023, 15, 794. https://doi.org/10.3390/rs15030794
Saif MS, Chancia R, Pethybridge S, Murphy SP, Hassanzadeh A, van Aardt J. Forecasting Table Beet Root Yield Using Spectral and Textural Features from Hyperspectral UAS Imagery. Remote Sensing. 2023; 15(3):794. https://doi.org/10.3390/rs15030794
Chicago/Turabian StyleSaif, Mohammad S., Robert Chancia, Sarah Pethybridge, Sean P. Murphy, Amirhossein Hassanzadeh, and Jan van Aardt. 2023. "Forecasting Table Beet Root Yield Using Spectral and Textural Features from Hyperspectral UAS Imagery" Remote Sensing 15, no. 3: 794. https://doi.org/10.3390/rs15030794
APA StyleSaif, M. S., Chancia, R., Pethybridge, S., Murphy, S. P., Hassanzadeh, A., & van Aardt, J. (2023). Forecasting Table Beet Root Yield Using Spectral and Textural Features from Hyperspectral UAS Imagery. Remote Sensing, 15(3), 794. https://doi.org/10.3390/rs15030794






