Quantifying the Variation in Reflectance Spectra of Metrosideros polymorpha Canopies across Environmental Gradients
Abstract
:1. Introduction
- In a tropical/sub-tropical environment, do canopy spectra reflect the environmental filtering, and thus evolutionary divergence, of canopy properties in a single species?
- How does the spectral variability of M. polymorpha capture the environmental filtering of traits across these environmental gradients in the context of the LES?
- What is the relative performance of models using canopy spectra versus leaf trait indices estimated from reflectance data in differentiating the effects of environmental filtering on a single species?
2. Materials and Methods
2.1. Study Sites
2.2. Data Processing
2.3. Analysis
3. Results
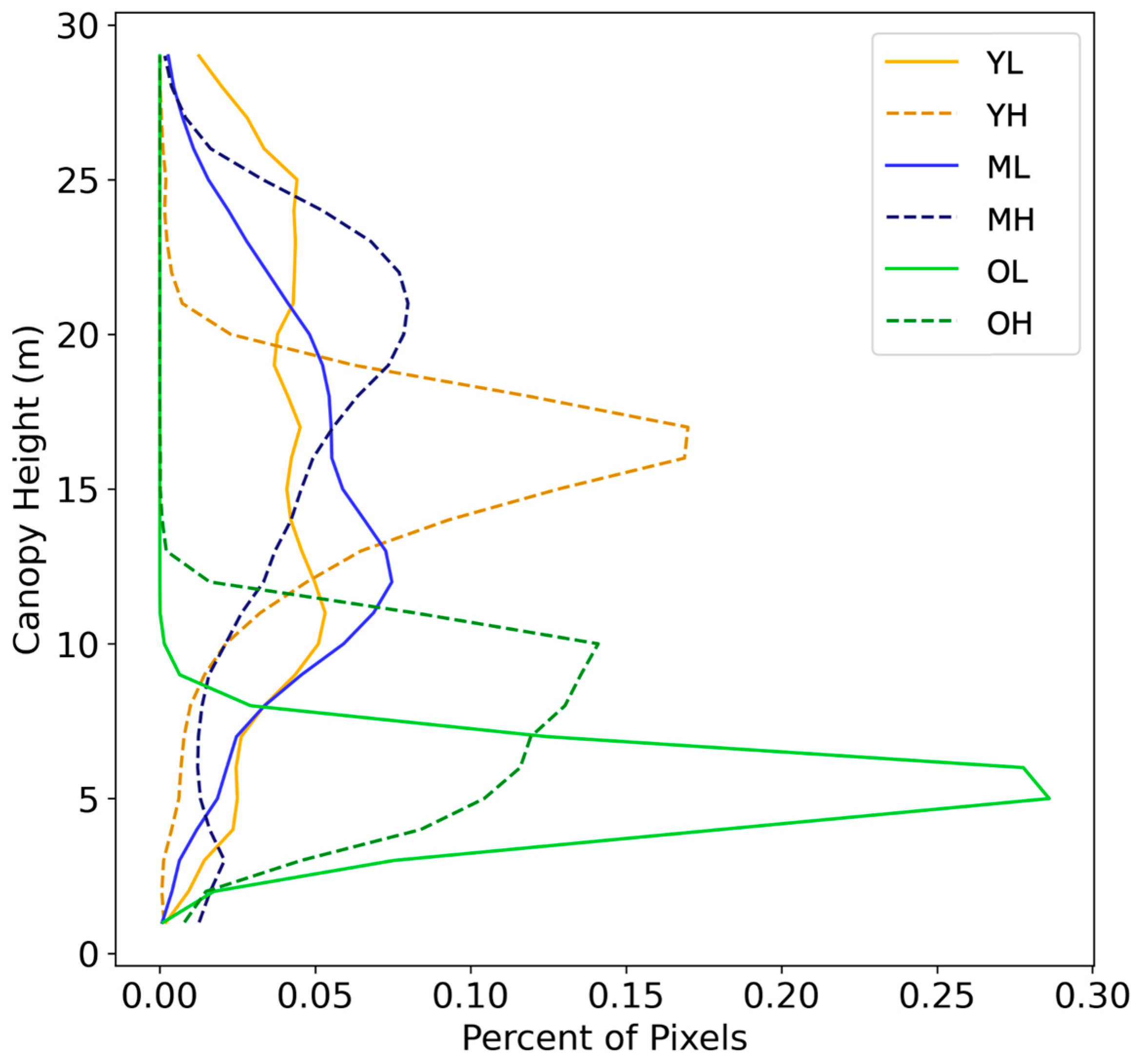
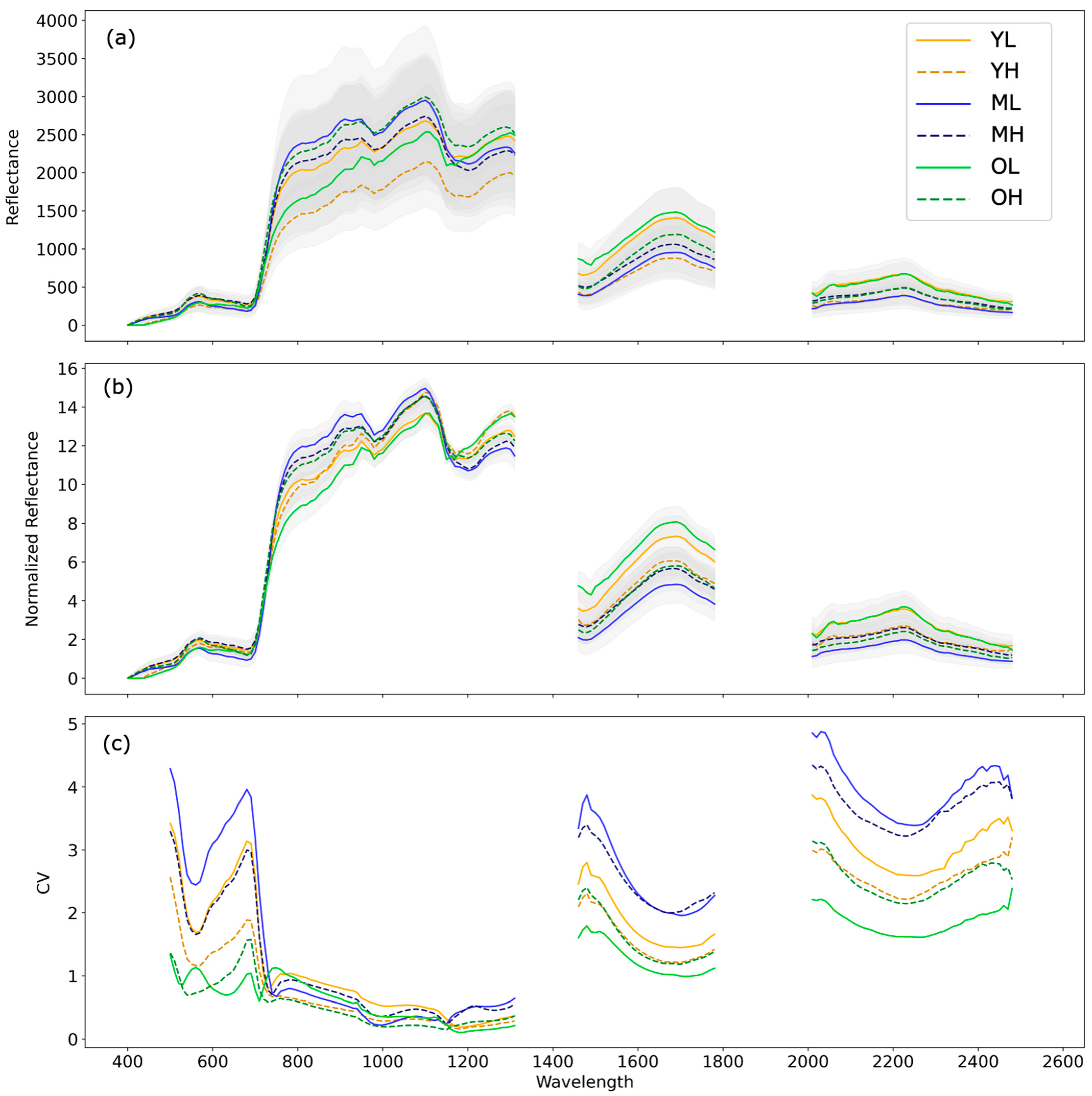
3.1. Inter-Site Variation
3.2. Intra-Site Variation
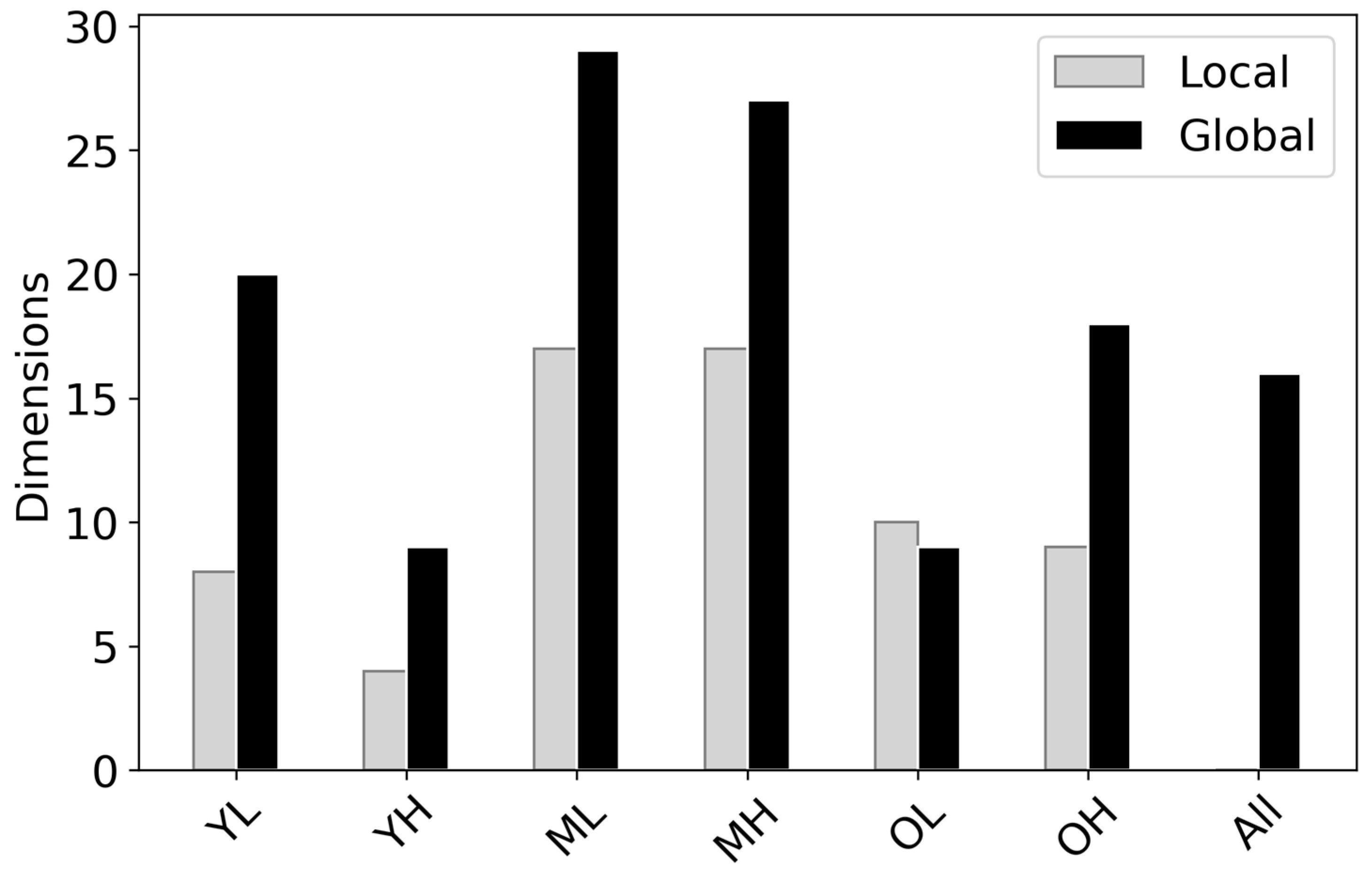
3.3. Canopy Traits
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Weiher, E.; Clarke, G.D.P.; Keddy, P.A. Community Assembly Rules, Morphological Dispersion, and the Coexistence of Plant Species. Oikos 1998, 81, 309–322. [Google Scholar] [CrossRef]
- Keddy, P.A. Assembly and response rules: Two goals for predictive community ecology. J. Veg. Sci. 1992, 3, 157–164. [Google Scholar] [CrossRef] [Green Version]
- Macarthur, R.; Levins, R. The Limiting Similarity, Convergence, and Divergence of Coexisting Species. Am. Nat. 1967, 101, 377–385. [Google Scholar] [CrossRef]
- Morin, X.; Lechowicz, M. Contemporary perspectives on the niche that can improve models of species range shifts under climate change. Biol. Lett. 2003, 4, 573–576. [Google Scholar] [CrossRef] [Green Version]
- Buzzard, V.; Hulshof, C.M.; Birt, T.; Violle, C.; Enquist, B.J. Re-growing a tropical dry forest: Functional plant trait composition and community assembly during succession. Funct. Ecol. 2016, 30, 1006–1013. [Google Scholar] [CrossRef] [Green Version]
- Richardson, D.M.; Pyšek, P.; Rejmánek, M.; Barbour, M.G.; Panetta, F.D.; West, C.J. Naturalization and invasion of alien plants: Concepts and definitions. Divers. Distrib. 2000, 6, 93–107. [Google Scholar] [CrossRef]
- Swenson, N.G.; Enquist, B.J.; Pither, J.; Kerkhoff, A.J.; Boyle, B.; Weiser, M.D.; Elser, J.J.; Fagan, W.F.; Forero-Montaña, J.; Fyllas, N.; et al. The biogeography and filtering of woody plant functional diversity in North and South America. Glob. Ecol. Biogeogr. 2012, 21, 798–808. [Google Scholar] [CrossRef]
- Kraft, N.J.B.; Adler, P.B.; Godoy, O.; James, E.C.; Fuller, S.; Levine, J.M. Community assembly, coexistence and the environmental filtering metaphor. Funct. Ecol. 2015, 29, 592–599. [Google Scholar] [CrossRef]
- Anderegg, L.D.L.; Berner, L.T.; Badgley, G.; Sethi, M.L.; Law, B.E.; HilleRisLambers, J. Within-species patterns challenge our understanding of the leaf economics spectrum. Ecol. Lett. 2018, 21, 734–744. [Google Scholar] [CrossRef]
- Schneider, F.D.; Morsdorf, F.; Schmid, B.; Petchey, O.L.; Hueni, A.; Schimel, D.S.; Schaepman, M.E. Mapping functional diversity from remotely sensed morphological and physiological forest traits. Nat. Commun. 2017, 8, 1441. [Google Scholar] [CrossRef] [Green Version]
- Albert, C.H.; Thuiller, W.; Yoccoz, N.G.; Soudant, A.; Boucher, F.; Saccone, P.; Lavorel, S. Intraspecific functional variability: Extent, structure and sources of variation. J. Ecol. 2010, 98, 604–613. [Google Scholar] [CrossRef]
- Wright, I.J.; Reich, P.B.; Westoby, M.; Ackerly, D.D.; Baruch, Z.; Bongers, F.; Cavender-Bares, J.; Chapin, T.; Cornelissen, J.H.C.; Diemer, M.; et al. The worldwide leaf economics spectrum. Nature 2004, 428, 821–827. [Google Scholar] [CrossRef]
- Read, Q.D.; Moorhead, L.C.; Swenson, N.G.; Bailey, J.K.; Sanders, N.J. Convergent effects of elevation on functional leaf traits within and among species. Funct. Ecol. 2014, 28, 37–45. [Google Scholar] [CrossRef]
- Midolo, G.; De Frenne, P.; Hölzel, N.; Wellstein, C. Global patterns of intraspecific leaf trait responses to elevation. Glob. Chang. Biol. 2019, 25, 2485–2498. [Google Scholar] [CrossRef] [PubMed]
- Wright, J.P.; Sutton-Grier, A. Does the leaf economic spectrum hold within local species pools across varying environmental conditions? Funct. Ecol. 2012, 26, 1390–1398. [Google Scholar] [CrossRef]
- Martin, R.E.; Asner, G.P.; Sack, L. Genetic variation in leaf pigment, optical and photosynthetic function among diverse phenotypes of Metrosideros polymorpha grown in a common garden. Oecologia 2007, 151, 387–400. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying forest canopy traits: Imaging spectroscopy versus field survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Draper, F.C.; Baraloto, C.; Brodrick, P.G.; Phillips, O.L.; Martinez, R.V.; Coronado, E.N.H.; Baker, T.R.; Gómez, R.Z.; Guerra, C.A.A.; Flores, M.; et al. Imaging spectroscopy predicts variable distance decay across contrasting Amazonian tree communities. J. Ecol. 2019, 107, 696–710. [Google Scholar] [CrossRef]
- Kalacska, M.; Sanchez-Azofeifa, G.A.; Rivard, B.; Caelli, T.; White, H.P.; Calvo-Alvarado, J.C. Ecological fingerprinting of ecosystem succession: Estimating secondary tropical dry forest structure and diversity using imaging spectroscopy. Remote Sens. Environ. 2007, 108, 82–96. [Google Scholar] [CrossRef]
- DuBois, S.; Desai, A.R.; Singh, A.; Serbin, S.P.; Goulden, M.L.; Baldocchi, D.D.; Ma, S.; Oechel, W.C.; Wharton, S.; Kruger, E.L.; et al. Using imaging spectroscopy to detect variation in terrestrial ecosystem productivity across a water-stressed landscape. Ecol. Appl. 2018, 28, 1313–1324. [Google Scholar] [CrossRef]
- Kokaly, R.F.; Asner, G.P.; Ollinger, S.V.; Martin, M.E.; Wessman, C.A. Characterizing canopy biochemistry from imaging spectroscopy and its application to ecosystem studies. Remote Sens. Environ. 2009, 113, S78–S91. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.; Carranza, L.; Martinez, P.; Houcheime, M.; Sinca, F.; Weiss, P. Spectroscopy of canopy chemicals in humid tropical forests. Remote Sens. Environ. 2011, 115, 3587–3598. [Google Scholar] [CrossRef]
- Asner, G.P.; Knapp, D.E.; Anderson, C.B.; Martin, R.E.; Vaughn, N. Large-scale climatic and geophysical controls on the leaf economics spectrum. Proc. Natl. Acad. Sci. USA 2016, 113, E4043–E4051. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Chlus, A.; Geygan, R.; Ye, Z.; Zheng, T.; Singh, A.; Couture, J.J.; Cavender-Bares, J.; Kruger, E.L.; Townsend, P.A. Foliar functional traits from imaging spectroscopy across biomes in eastern North America. New Phytol. 2020, 228, 494–511. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Serbin, S.P.; McNeil, B.E.; Kingdon, C.C.; Townsend, P.A. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecol. Appl. 2015, 25, 2180–2197. [Google Scholar] [CrossRef] [Green Version]
- Asner, G.P.; Martin, R.E. Convergent elevation trends in canopy chemical traits of tropical forests. Glob. Chang. Biol. 2016, 22, 2216–2227. [Google Scholar] [CrossRef]
- Féret, J.-B.; Asner, G.P. Mapping tropical forest canopy diversity using high-fidelity imaging spectroscopy. Ecol. Appl. 2014, 24, 1289–1296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vitousek, P.M. Nutrient Cycling and Limitation: Hawai‘i as a Model System; Princeton University Press: Princeton, NJ, USA, 2004. [Google Scholar]
- Morrison, K.R.; Stacy, E.A. Intraspecific divergence and evolution of a life-history trade-off along a successional gradient in Hawaii’s Metrosideros polymorpha. J. Evol. Biol. 2014, 27, 1192–1204. [Google Scholar] [CrossRef] [PubMed]
- Stacy, E.A.; Johansen, J.B.; Sakishima, T.; Price, D.K.; Pillon, Y. Incipient radiation within the dominant Hawaiian tree Metrosideros polymorpha. Heredity 2014, 113, 334–342. [Google Scholar] [CrossRef] [Green Version]
- Vitousek, P.M.; Aplet, G.; Turner, D.; Lockwood, J.J. The Mauna Loa environmental matrix: Foliar and soil nutrients. Oecologia 1992, 89, 372–382. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Chadwick, O.; Crews, T.E.; Fownes, J.; Hendricks, D.M.; Herbert, D. Soil and ecosystem development across the Hawaiian Islands. GSA Today 1997, 7, 1–8. [Google Scholar]
- Herbert, D.A.; Fownes, J.H. Forest Productivity and Efficiency of Resource Use across a Chronosequence of Tropical Montane Soils. Ecosystems 1999, 2, 242–254. [Google Scholar] [CrossRef]
- James, S.A.; Puttock, C.F.; Cordell, S.; Adams, R.P. Morphological and genetic variation within Metrosideros polymorpha (Myrtaceae) on Hawai‘i. N. Z. J. Bot. 2004, 42, 263–270. [Google Scholar] [CrossRef] [Green Version]
- Izuno, A.; Kitayama, K.; Onoda, Y.; Tsujii, Y.; Hatakeyama, M.; Nagano, A.J.; Honjo, M.N.; Shimizu-Inatsugi, R.; Kudoh, H.; Shimizu, K.K.; et al. The population genomic signature of environmental association and gene flow in an ecologically divergent tree species Metrosideros polymorpha (Myrtaceae). Mol. Ecol. 2017, 26, 1515–1532. [Google Scholar] [CrossRef] [Green Version]
- Stacy, E.A.; Johansen, J.B.; Sakishima, T.; Price, D.K. Genetic analysis of an ephemeral intraspecific hybrid zone in the hypervariable tree, Metrosideros polymorpha, on Hawai‘i Island. Heredity 2016, 117, 173–183. [Google Scholar] [CrossRef] [Green Version]
- Joel, G.; Aplet, G.; Vitousek, P.M. Leaf Morphology Along Environmental Gradients in Hawaiian Metrosideros polymorpha. Biotropica 1994, 26, 17–22. [Google Scholar] [CrossRef]
- Martin, R.E.; Asner, G.P. Leaf Chemical and Optical Properties of Metrosideros polymorpha across Environmental Gradients in Hawaii. Biotropica 2009, 41, 292–301. [Google Scholar] [CrossRef]
- Cordell, S.; Goldstein, G.; Mueller-Dombois, D.; Webb, D.; Vitousek, P.M. Physiological and morphological variation in Metrosideros polymorpha, a dominant Hawaiian tree species, along an altitudinal gradient: The role of phenotypic plasticity. Oecologia 1998, 113, 188–196. [Google Scholar] [CrossRef] [PubMed]
- Fisher, J.B.; Goldstein, G.; Jones, T.J.; Cordell, S. Wood vessel diameter is related to elevation and genotype in the Hawaiian tree Metrosideros polymorpha (Myrtaceae). Am. J. Bot. 2007, 94, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Tsujii, Y.; Onoda, Y.; Izuno, A.; Isagi, Y.; Kitayama, K. A quantitative analysis of phenotypic variations of Metrosideros polymorpha within and across populations along environmental gradients on Mauna Loa, Hawaii. Oecologia 2016, 180, 1049–1059. [Google Scholar] [CrossRef]
- Cordell, S.; Goldstein, G.; Meinzer, F.C.; Handley, L.L. Allocation of nitrogen and carbon in leaves of Metrosideros polymorpha regulates carboxylation capacity and δ13C along an altitudinal gradient. Funct. Ecol. 1999, 13, 811–818. [Google Scholar] [CrossRef]
- Choi, J.Y.; Purugganan, M.; Stacy, E.A. Divergent Selection and Primary Gene Flow Shape Incipient Speciation of a Riparian Tree on Hawaii Island. Mol. Biol. Evol. 2020, 37, 695–710. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitayama, K.; Pattison, R.; Cordell, S.; Webb, D.; Mueller-dombois, D. Ecological and Genetic Implications of Foliar Polymorphism in Metrosideros polymorphaGaud. (Myrtaceae) in a Habitat Matrix on Mauna Loa, Hawaii. Ann. Bot. 1997, 80, 491–497. [Google Scholar] [CrossRef] [Green Version]
- Stacy, E.A.; Sakishima, T. Phylogeography of the highly dispersible landscape-dominant woody species complex, Metrosideros, in Hawaii. J. Biogeogr. 2019, 46, 2215–2231. [Google Scholar] [CrossRef]
- Blonder, B.; Graae, B.J.; Greer, B.; Haagsma, M.; Helsen, K.; Kapás, R.E.; Pai, H.; Rieksta, J.; Sapena, D.; Still, C.J.; et al. Remote sensing of ploidy level in quaking aspen (Populus tremuloides Michx.). J. Ecol. 2020, 108, 175–188. [Google Scholar] [CrossRef] [Green Version]
- Cavender-Bares, J.; Meireles, J.E.; Couture, J.J.; Kaproth, M.A.; Kingdon, C.C.; Singh, A.; Serbin, S.P.; Center, A.; Zuniga, E.; Pilz, G.; et al. Associations of Leaf Spectra with Genetic and Phylogenetic Variation in Oaks: Prospects for Remote Detection of Biodiversity. Remote Sens. 2016, 8, 221. [Google Scholar] [CrossRef] [Green Version]
- Madritch, M.D.; Kingdon, C.C.; Singh, A.; Mock, K.E.; Lindroth, R.L.; Townsend, P.A. Imaging spectroscopy links aspen genotype with below-ground processes at landscape scales. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 194. [Google Scholar] [CrossRef]
- Sherrod, D.R.; Sinton, J.M.; Watkins, S.E.; Brunt, K.M. Geologic Map of the State of Hawai‘i: U.S. Geologica Survey Open-File Report. 2007; p. 1089. Available online: http://pubs.usgs.gov/of/2007/1089/ (accessed on 4 January 2023).
- Asner, G.P.; Knapp, D.E.; Boardman, J.; Green, R.O.; Kennedy-Bowdoin, T.; Eastwood, M.; Martin, R.E.; Anderson, C.; Field, C.B. Carnegie Airborne Observatory-2: Increasing science data dimensionality via high-fidelity multi-sensor fusion. Remote Sens. Environ. 2012, 124, 454–465. [Google Scholar] [CrossRef]
- Schaepman-Strub, G.; Schaepman, M.; Martonchik, J.; Schaaf, C. Whats in a Satellite Albedo Product? IEEE Int. Symp. Geosci. Remote Sens. 2006, 6, 2848–2852. [Google Scholar]
- Miller, C.J. Performance assessment of ACORN atmospheric correction algorithm. In Proceedings of the Algorithms and Technologies for Multispectral, Hyperspectral, and Ultraspectral Imagery VIII, SPIE 2002, Melbourne, Australia, 16–18 December 2002; Volume 4725, pp. 438–449. [Google Scholar]
- Asner, G.P.; Knapp, D.E.; Kennedy-Bowdoin, T.; Jones, M.O.; Martin, R.E.; Boardman, J.W.; Field, C.B. Carnegie Airborne Observatory: In-flight fusion of hyperspectral imaging and waveform light detection and ranging for three-dimensional studies of ecosystems. J. Appl. Remote Sens. 2007, 1, 013536. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E.; Schmidtlein, S. Brightness-normalized Partial Least Squares Regression for hyperspectral data. J. Quant. Spectrosc. Radiat. Transf. 2010, 111, 1947–1957. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Macler, B.A.; Plummer, S.E.; Peterson, D.L. Reflectance spectroscopy of fresh whole leaves for the estimation of chemical concentration. Remote Sens. Environ. 1992, 39, 153–166. [Google Scholar] [CrossRef]
- Ustin, S.L.; Roberts, D.A.; Gamon, J.A.; Asner, G.P.; Green, R.O. Using Imaging Spectroscopy to Study Ecosystem Processes and Properties. BioScience 2004, 54, 523–534. [Google Scholar] [CrossRef]
- Wang, R.; Gamon, J.A.; Cavender-Bares, J.; Townsend, P.A.; Zygielbaum, A.I. The spatial sensitivity of the spectral diversity–biodiversity relationship: An experimental test in a prairie grassland. Ecol. Appl. 2018, 28, 541–556. [Google Scholar] [CrossRef] [Green Version]
- Thompson, D.R.; Boardman, J.W.; Eastwood, M.L.; Green, R.O. A large airborne survey of Earth’s visible-infrared spectral dimensionality. Opt. Express 2017, 25, 9186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boardman, J.W.; Green, R.O. Exploring the spectral variability of the Earth as measured by AVIRIS in 1999. Summaries of the ninth annual JPL airborne geosciences workshop. Jet Propuls. Lab. Spec. Publ. 2000, 18, 10. [Google Scholar]
- Green, R.O.; Boardman, J.W. Exploration of the relationship between information content and signal-to-noise ratio and spatial resolution in AVIRIS spectral data. Spectrum 2000, 7, 8. [Google Scholar]
- Dai, J.; Vaughn, N.R.; Seeley, M.; Heckler, J.; Thompson, D.R.; Asner, G.P. Spectral dimensionality of imaging spectroscopy data over diverse landscapes and spatial resolutions. J. Appl. Remote Sens. 2022, 16, 044518. [Google Scholar] [CrossRef]
- Baldeck, C.A.; Colgan, M.S.; Féret, J.-B.; Levick, S.R.; Martin, R.E.; Asner, G.P. Landscape-scale variation in plant community composition of an African savanna from airborne species mapping. Ecol. Appl. 2014, 24, 84–93. [Google Scholar] [CrossRef] [PubMed]
- Baldeck, C.A.; Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E.; Kellner, J.R.; Wright, S.J. Operational Tree Species Mapping in a Diverse Tropical Forest with Airborne Imaging Spectroscopy. PLoS ONE 2015, 10, e0118403. [Google Scholar] [CrossRef] [PubMed]
- Balzotti, C.S.; Asner, G.P.; Adkins, E.D.; Parsons, E.W. Spatial drivers of composition and connectivity across endangered tropical dry forests. J. Appl. Ecol. 2020, 57, 1593–1604. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Gomez-Chova, L.; Calpe-Maravilla, J.; Martin-Guerrero, J.D.; Soria-Olivas, E.; Alonso-Chorda, L.; Moreno, J. Robust support vector method for hyperspectral data classification and knowledge discovery. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1530–1542. [Google Scholar] [CrossRef]
- Feret, J.-B.; Asner, G.P. Tree Species Discrimination in Tropical Forests Using Airborne Imaging Spectroscopy. IEEE Trans. Geosci. Remote Sens. 2013, 51, 73–84. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef] [Green Version]
- Pedregosa, F.; Varoquaux, G.; Gramfort, A.; Michel, V.; Thirion, B.; Grisel, O.; Blondel, M.; Prettenhofer, P.; Weiss, R.; Dubourg, V.; et al. Scikit-learn: Machine Learning in Python. J. Mach. Learn. Res. 2011, 12, 2825–2830. [Google Scholar]
- Porder, S.; Asner, G.P.; Vitousek, P.M. Ground-based and remotely sensed nutrient availability across a tropical landscape. Proc. Natl. Acad. Sci. USA 2005, 102, 10909–10912. [Google Scholar] [CrossRef] [Green Version]
- Balzotti, C.S.; Asner, G.P. Biotic and Abiotic Controls Over Canopy Function and Structure in Humid Hawaiian Forests. Ecosystems 2018, 21, 331–348. [Google Scholar] [CrossRef]
- Corn, C.A.; Hiesey, W.M. Altitudinal Variation in Hawaiian Metrosideros. Am. J. Bot. 1973, 60, 991–1002. [Google Scholar] [CrossRef]
- Drake, D.R.; Mueller-Dombois, D. Population Development of Rain Forest Trees on a Chronosequence of Hawaiian Lava Flows. Ecology 1993, 74, 1012–1019. [Google Scholar] [CrossRef]
- Stemmermann, L. Ecological studies of Hawaiian Metrosideros in a successional context. Pac. Sci. 1983, 37, 361–373. [Google Scholar]
- Poorter, H.; Niinemets, Ü.; Poorter, L.; Wright, I.J.; Villar, R. Causes and consequences of variation in leaf mass per area (LMA): A meta-analysis. New Phytol. 2009, 182, 565–588. [Google Scholar] [CrossRef]
- Westoby, M.; Falster, D.; Moles, A.T.; Vesk, P.; Wright, I.J. Plant Ecological Strategies: Some Leading Dimensions of Variation Between Species. Annu. Rev. Ecol. Syst. 2002, 33, 125–159. [Google Scholar] [CrossRef] [Green Version]
- Wright, S.J.; Kitajima, K.; Kraft, N.J.B.; Reich, P.B.; Wright, I.J.; Bunker, D.E.; Condit, R.; Dalling, J.W.; Davies, S.J.; Díaz, S.; et al. Functional traits and the growth–mortality trade-off in tropical trees. Ecology 2010, 91, 3664–3674. [Google Scholar] [CrossRef] [PubMed]
- Crews, T.E.; Kitayama, K.; Fownes, J.H.; Riley, R.H.; Herbert, D.A.; Mueller-Dombois, D.; Vitousek, P.M. Changes in Soil Phosphorus Fractions and Ecosystem Dynamics across a Long Chronosequence in Hawaii. Ecology 1995, 76, 1407–1424. [Google Scholar] [CrossRef]
- Vitousek, P.M.; Matson, P.A.; Turner, D.R. Elevational and age gradients in hawaiian montane rainforest: Foliar and soil nutrients. Oecologia 1988, 77, 565–570. [Google Scholar] [CrossRef]
- Hoof, J.; Sack, L.; Webb, D.T.; Nilsen, E.T. Contrasting Structure and Function of Pubescent and Glabrous Varieties of Hawaiian Metrosideros polymorpha (Myrtaceae) at High Elevation. Biotropica 2008, 40, 113–118. [Google Scholar] [CrossRef]
- Kitayama, K.; Mueller-Dombois, D. Vegetation changes along gradients of long-term soil development in the Hawaiian montane rainforest zone. Vegetatio 1995, 120, 1–20. [Google Scholar] [CrossRef]
- Stacy, E.; Johnson, M. Floral Variation across Three Varieties of the Landscape-Dominant Tree Metrosideros polymorpha (Myrtaceae): Insights from a Hawaii Island Common Garden. Int. J. Plant Sci. 2021, 182, 46–58. [Google Scholar] [CrossRef]
- Ustin, S.L.; Riaño, D.; Hunt, E.R. Estimating canopy water content from spectroscopy. Isr. J. Plant Sci. 2012, 60, 9–23. [Google Scholar] [CrossRef] [Green Version]
- Townsend, P.A.; Foster, J.R.; Chastain, R.A.; Currie, W.S. Application of imaging spectroscopy to mapping canopy nitrogen in the forests of the central Appalachian Mountains using Hyperion and AVIRIS. IEEE Trans. Geosci. Remote Sens. 2003, 41, 1347–1354. [Google Scholar] [CrossRef]



| Site | Description (Substrate Age, Elevation) | Area (ha) | Substrate Age Range (Years) | Mean Elevation (m) | TCH Max (m) | Mean NDVI | Mean LMA (g/m2) | Mean % N |
|---|---|---|---|---|---|---|---|---|
| YL | Young, Low | 9 | 200–750 | 200 | 34 | 0.75 | 199.5 | 1.03 |
| YH | Young, High | 11 | <200 | 1200 | 32 | 0.77 | 271.4 | 0.73 |
| ML | Medium, Low | 53 | 5000–11,000 | 750 | 36 | 0.85 | 198.1 | 1.80 |
| MH | Medium, High | 49 | 5000–64,000 | 1500 | 34 | 0.75 | 290.6 | 1.33 |
| OL | Old, Low | 8 | 260,000–500,000 | 900 | 10 | 0.85 | 196.2 | 0.54 |
| OH | Old, High | 10 | 260,000–500,000 | 1100 | 16 | 0.87 | 248.0 | 0.85 |
| Chemistry | Abbreviation | Units |
|---|---|---|
| Nitrogen | N | % |
| Leaf mass per area | LMA | g m−2 |
| Chlorophyll | Chl | mg g−1 |
| Carotenoids | Car | mg g−1 |
| Phosphorus | P | % |
| Iron | Fe | μg g−1 |
| Calcium | Ca | % |
| Total Carbon | Tot C | % |
| Soluble Carbon | Sol C | % |
| Method | Purpose |
|---|---|
| Mean Brightness-Normalized Reflectance |
|
| Coefficient of Variation |
|
| Principal Component Analysis (PCA) |
|
| Intrinsic Spectral Dimensionality |
|
| Euclidean Distance |
|
| Support Vector Machine (SVM) |
|
| Predicted | |||||||
|---|---|---|---|---|---|---|---|
| YL | YH | ML | MH | OL | OH | ||
| Actual | YL | 99.8 | 0 | 0 | 0.1 | 0 | 0 |
| YH | 0 | 99.9 | 0 | 0 | 0 | 0 | |
| ML | 0.2 | 0 | 99.6 | 0.1 | 0 | 0.2 | |
| MH | 0.1 | 0 | 0.1 | 99.7 | 0 | 0.1 | |
| OL | 0 | 0 | 0 | 0 | 100 | 0 | |
| OH | 0 | 0 | 0.3 | 0.2 | 0 | 99.6 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seeley, M.M.; Martin, R.E.; Vaughn, N.R.; Thompson, D.R.; Dai, J.; Asner, G.P. Quantifying the Variation in Reflectance Spectra of Metrosideros polymorpha Canopies across Environmental Gradients. Remote Sens. 2023, 15, 1614. https://doi.org/10.3390/rs15061614
Seeley MM, Martin RE, Vaughn NR, Thompson DR, Dai J, Asner GP. Quantifying the Variation in Reflectance Spectra of Metrosideros polymorpha Canopies across Environmental Gradients. Remote Sensing. 2023; 15(6):1614. https://doi.org/10.3390/rs15061614
Chicago/Turabian StyleSeeley, Megan M., Roberta E. Martin, Nicholas R. Vaughn, David R. Thompson, Jie Dai, and Gregory P. Asner. 2023. "Quantifying the Variation in Reflectance Spectra of Metrosideros polymorpha Canopies across Environmental Gradients" Remote Sensing 15, no. 6: 1614. https://doi.org/10.3390/rs15061614







