Detection of Maize Crop Phenology Using Planet Fusion
Abstract
1. Introduction
- Compare various template selection strategies and propose the use of multiple field observations during the detection of the phenological stage.
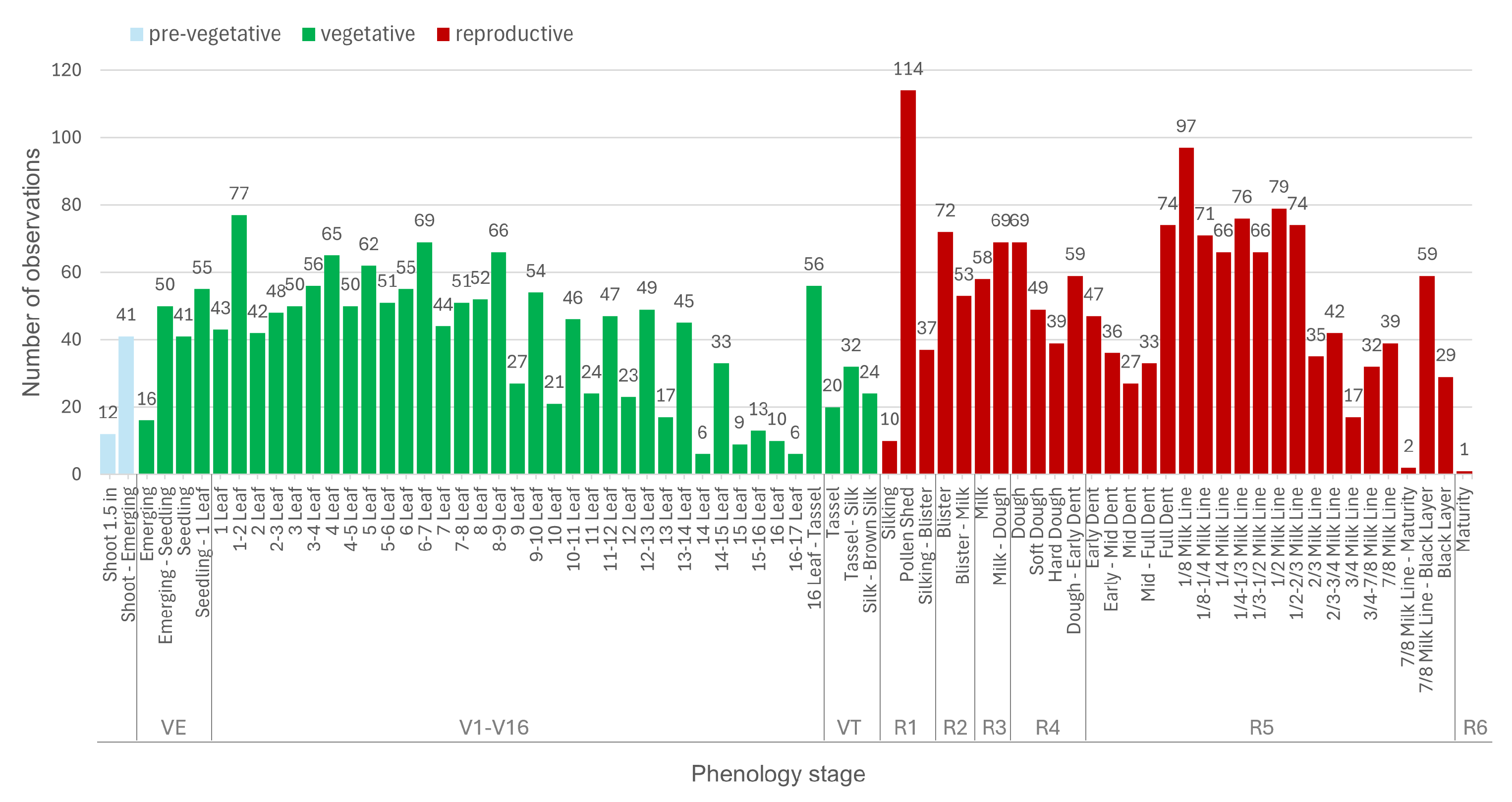
- Identify and delineate 70 micro-stages of maize growth using a comprehensive dataset comprising over 200 fields and 3200 observations.
- Explore the effectiveness of different vegetation indices and image bands for accurate growth stage identification.
- Evaluate the performance of algorithms using near-daily-temporal-resolution harmonized data obtained from two datasets which combine different sensors: Planet Fusion (PF) and Harmonized Landsat Sentinel-2 (HLS).
- Assess the generalizability and effectiveness of the proposed method in a second, distinct geographical region, thus expanding the scope of applicability beyond the initial study areas.
2. Datasets and Preprocessing
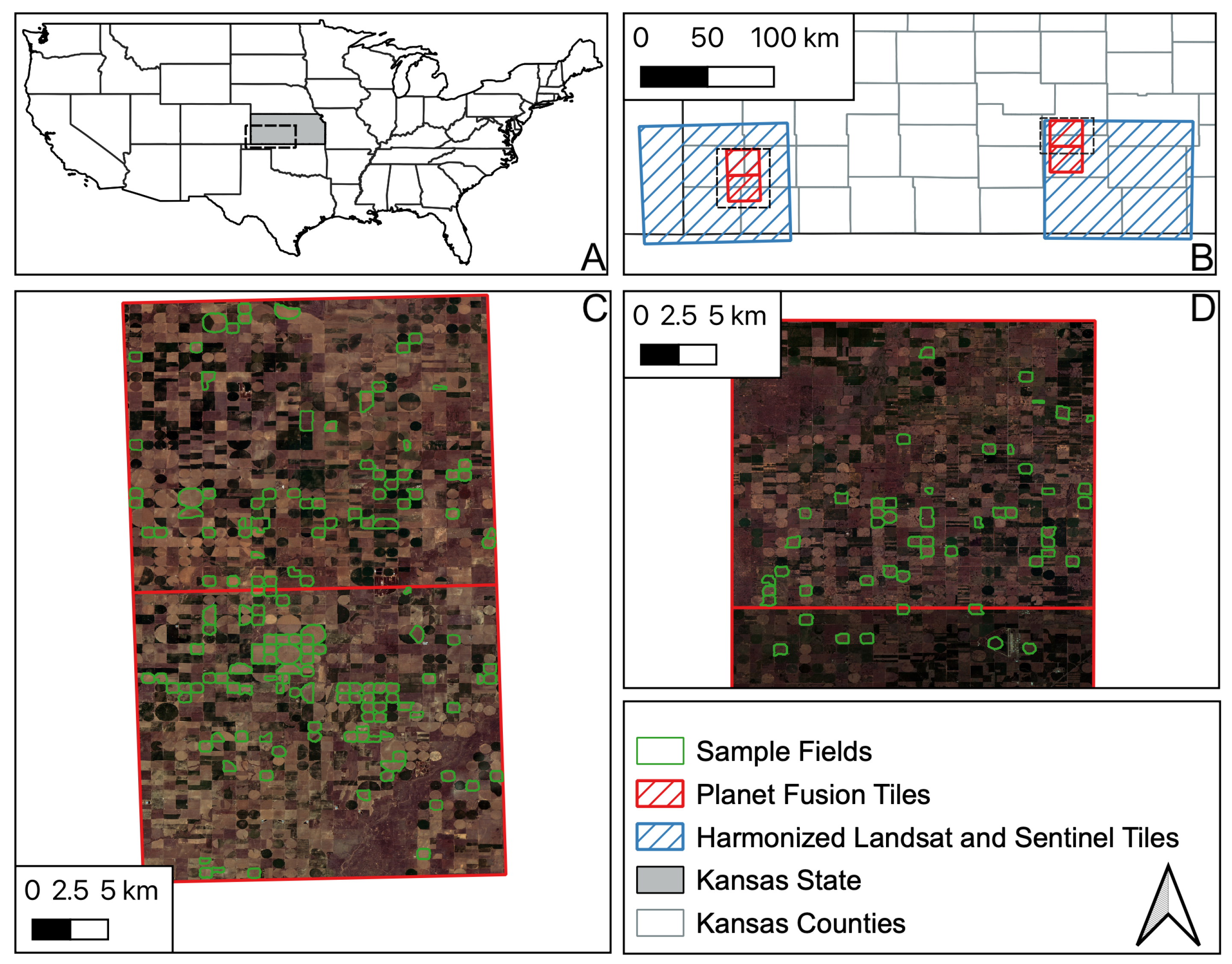
2.1. Remote Sensing Dataset
2.1.1. Planet Fusion (PF)
2.1.2. Harmonized Landsat and Sentinel-2 (HLS)
2.2. Maize Phenology Observation Datasets
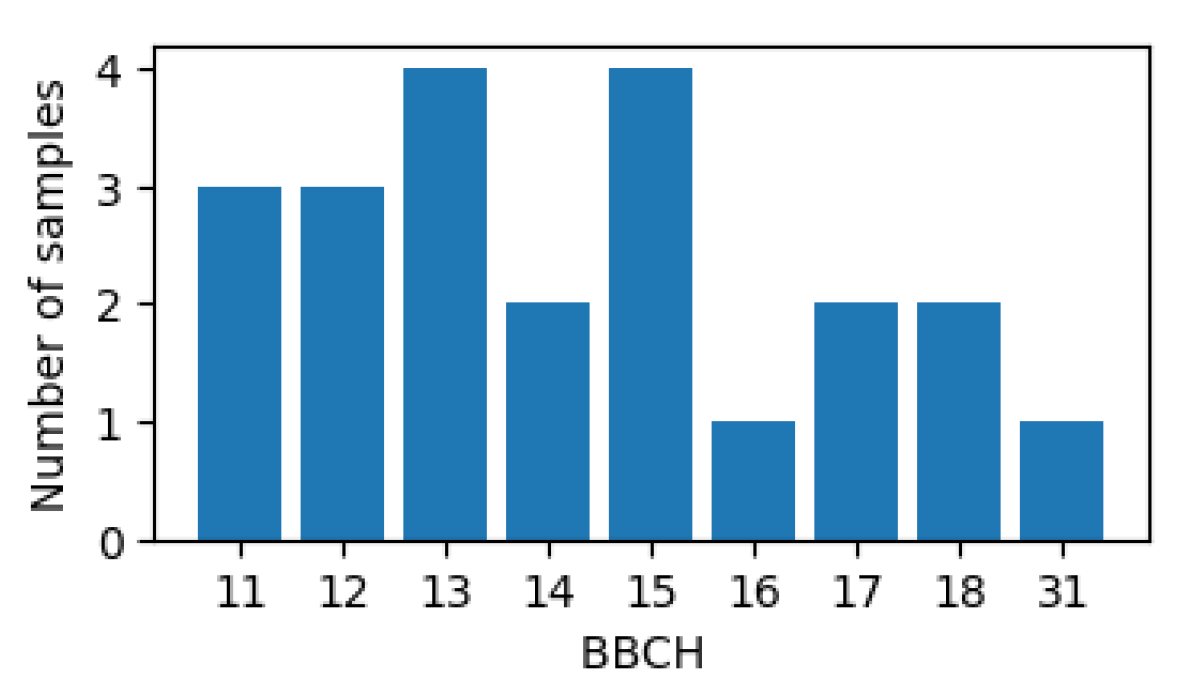
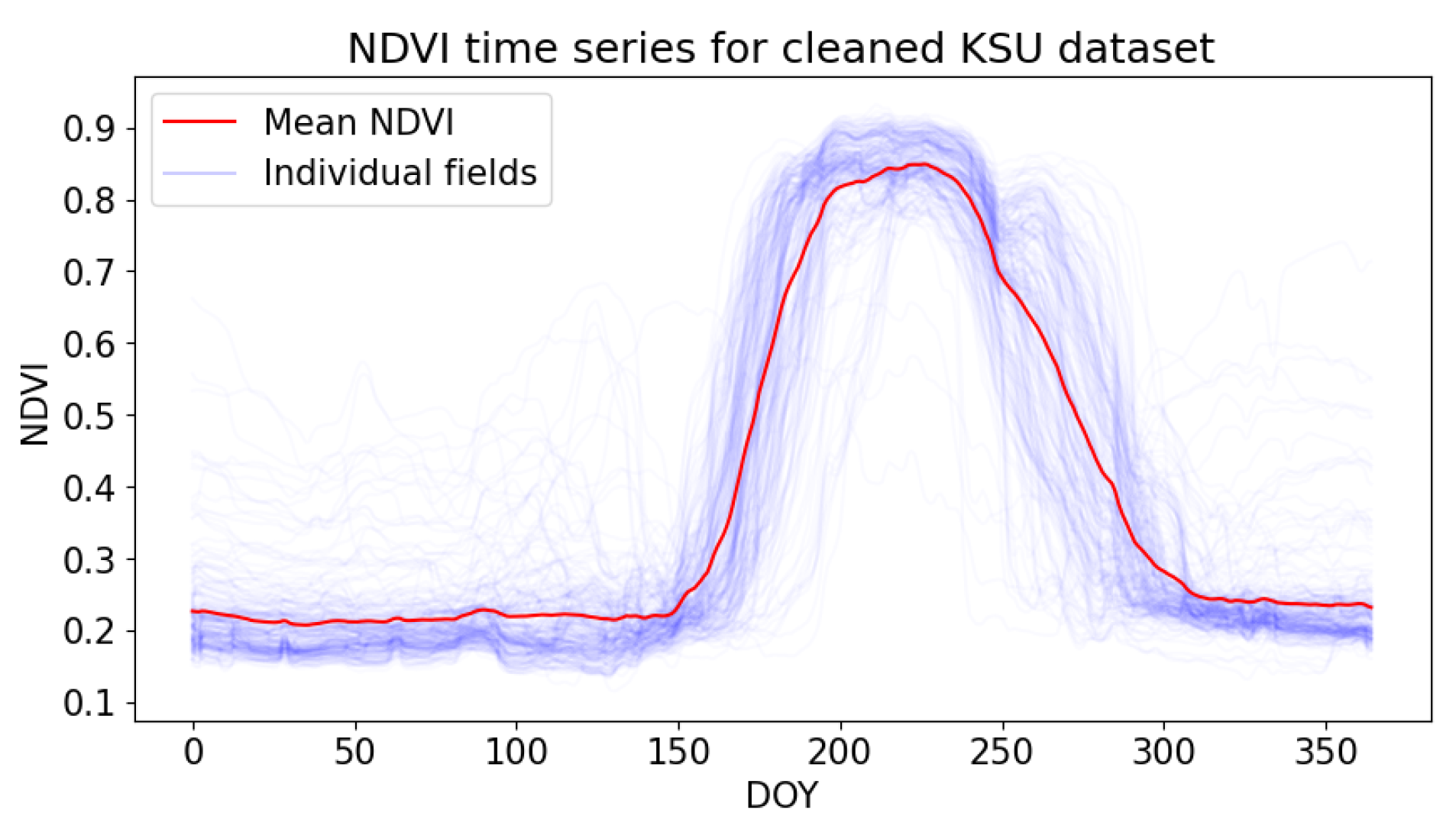
2.2.1. Kansas Dataset
2.2.2. PIAF Dataset
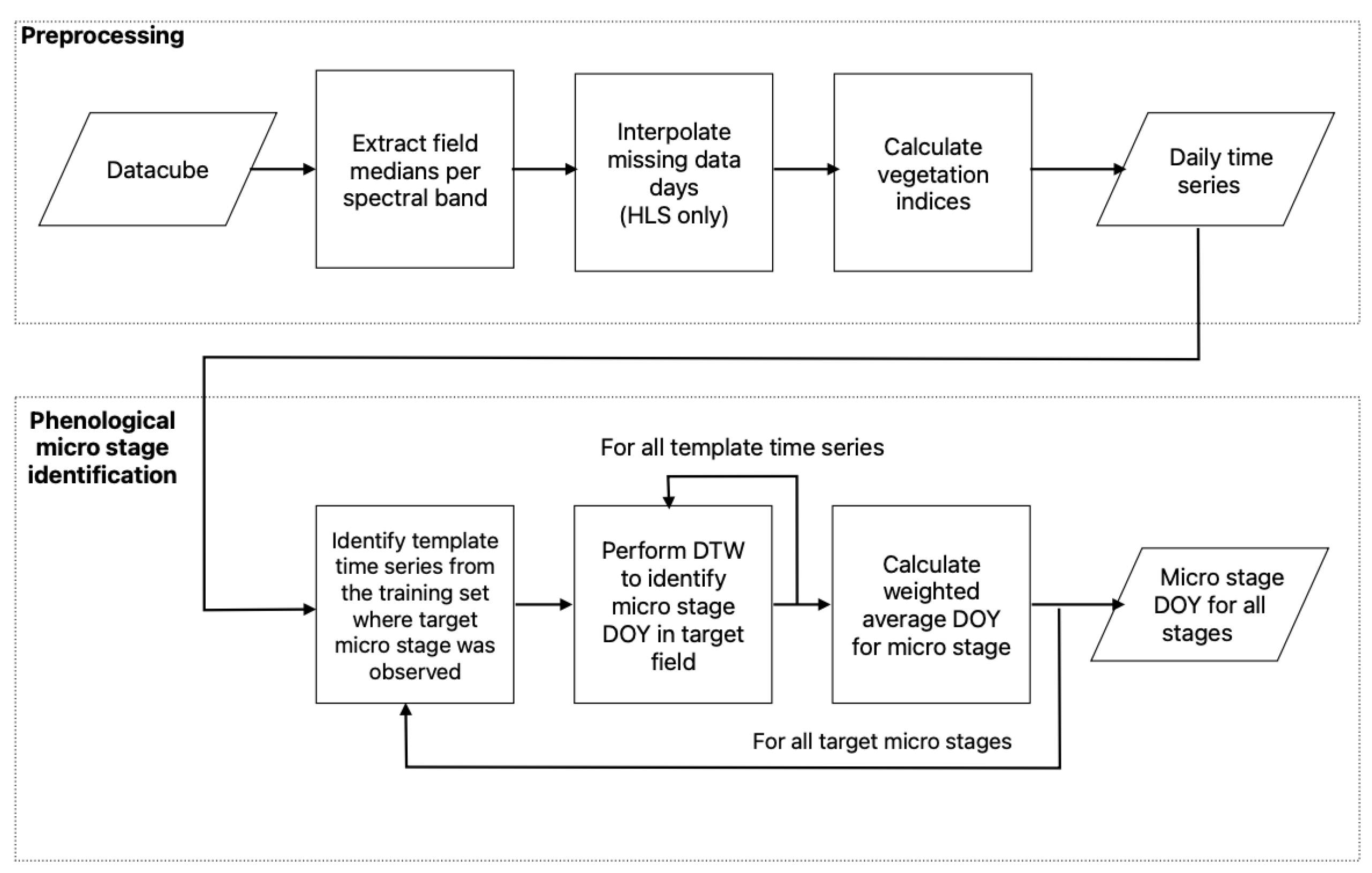
3. Methodology
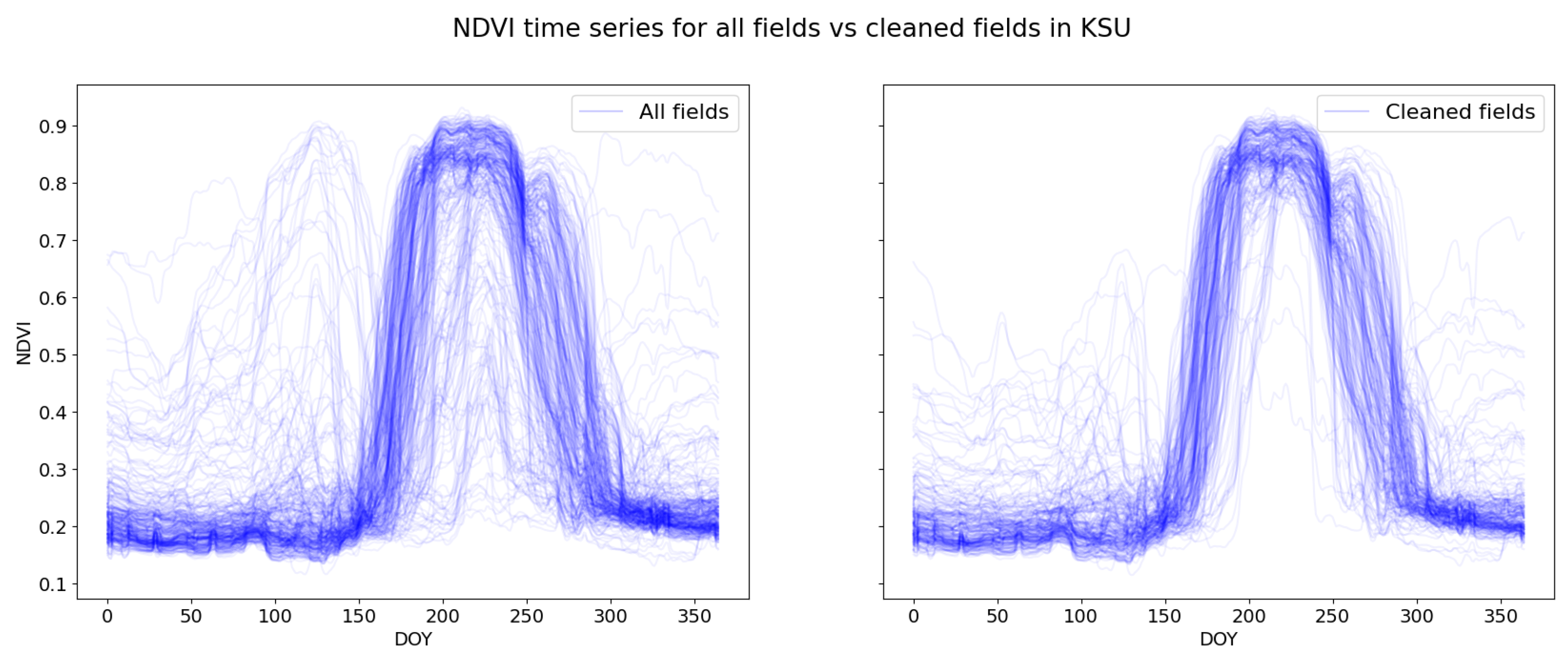
3.1. Preparation of the Time Series
3.2. Dynamic Time Warping with Weighted Average
4. Experiments
4.1. Performance Metrics
4.2. Comparison of DTW Methods
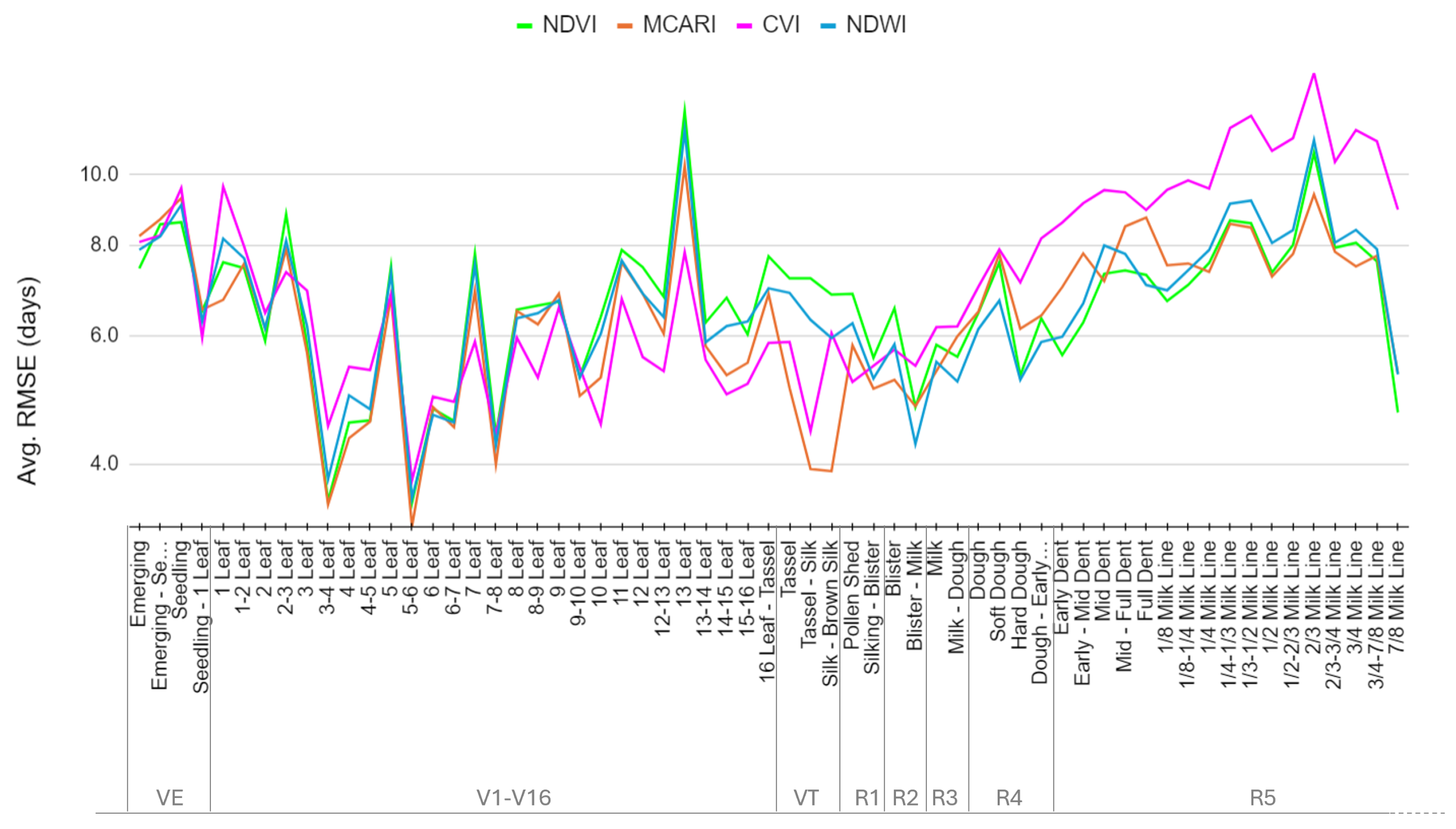
4.3. Performance of Spectral Indices and Bands
4.4. Comparison of Planet Fusion and HLS
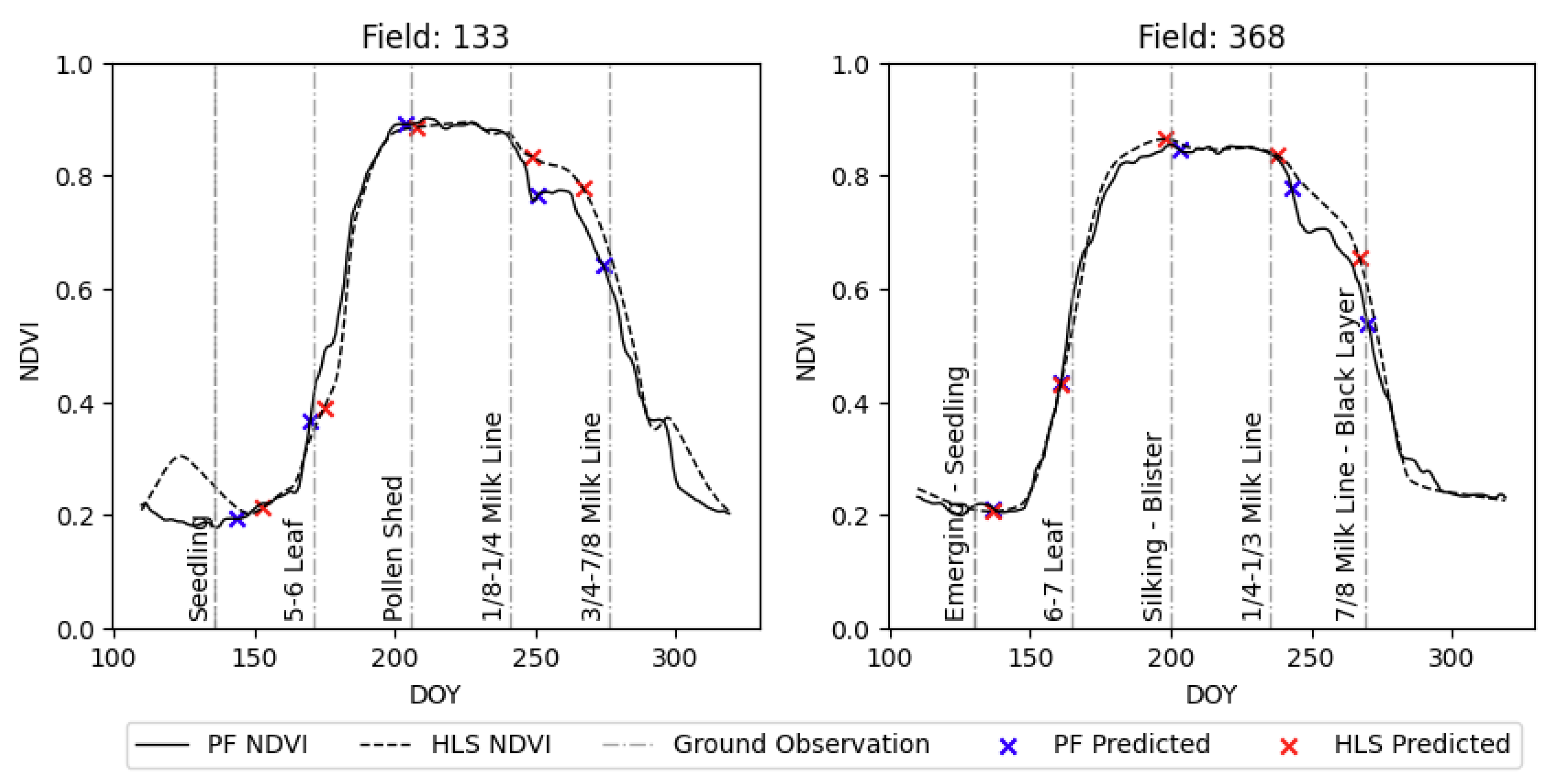
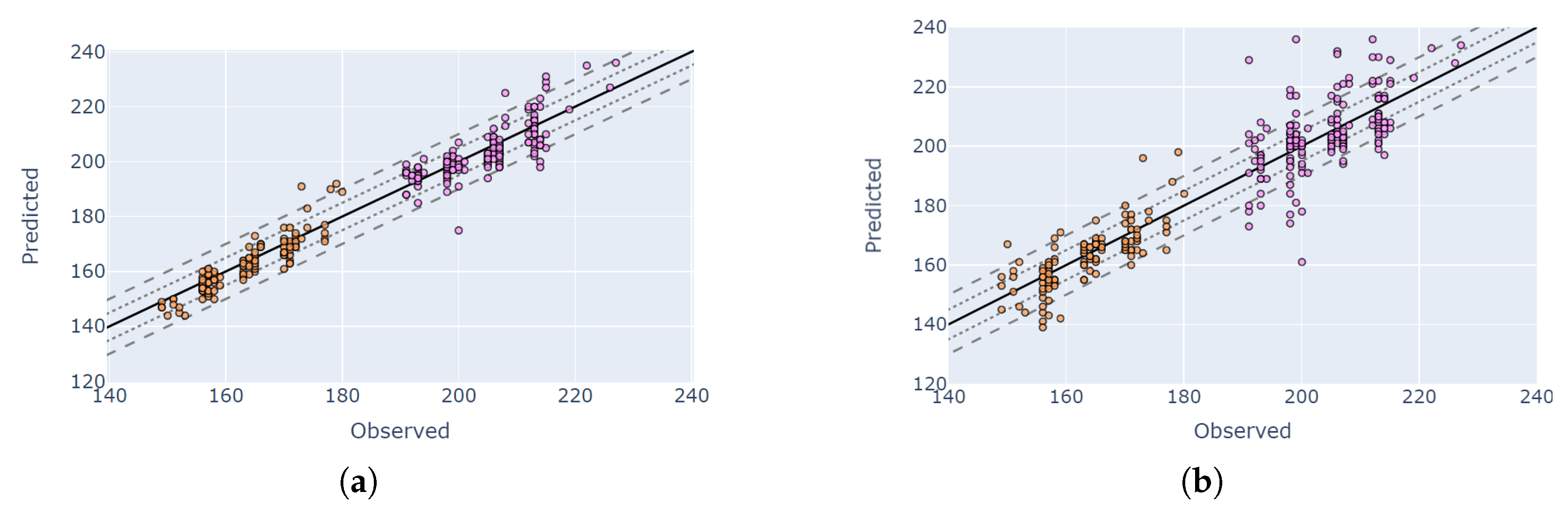
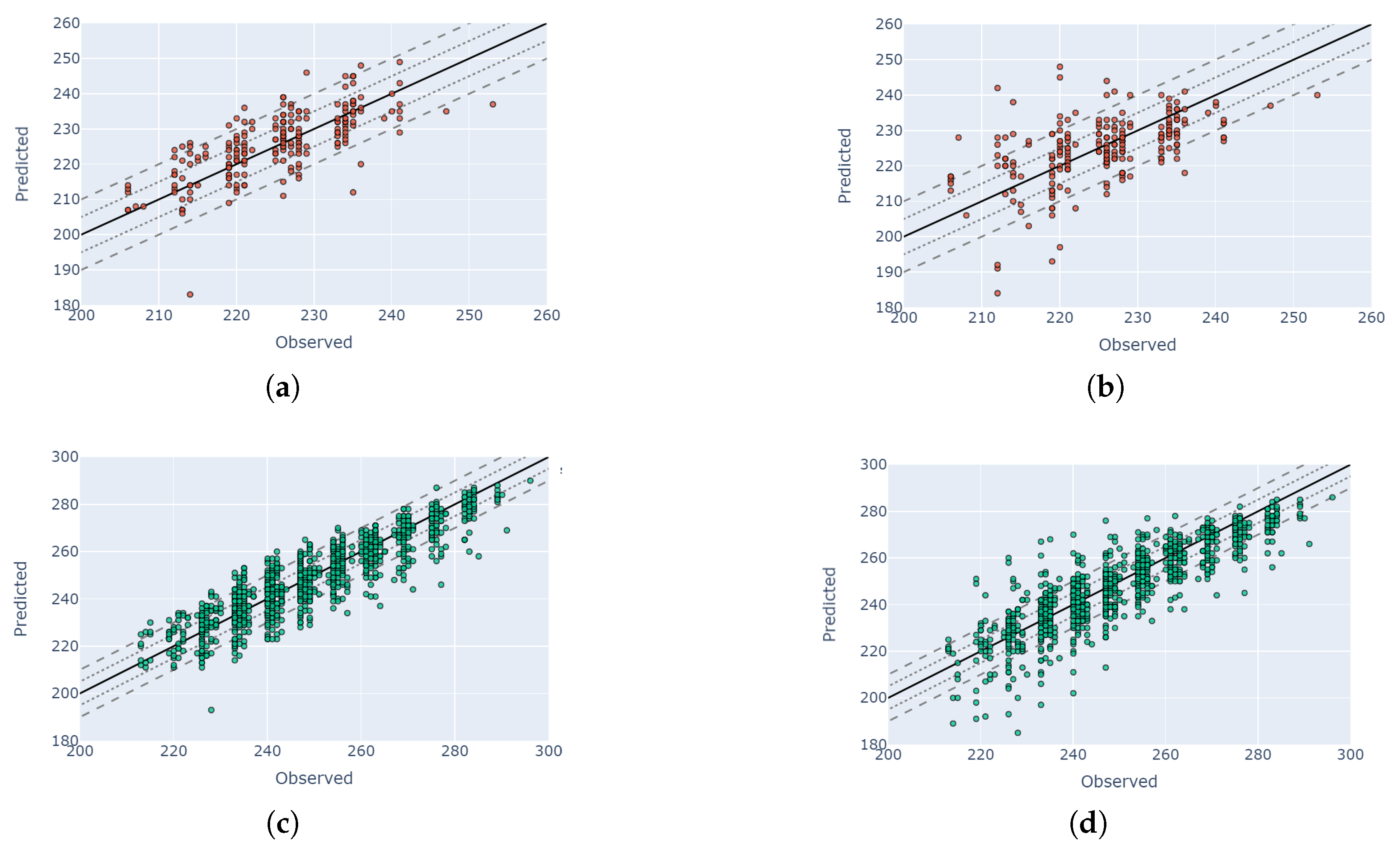
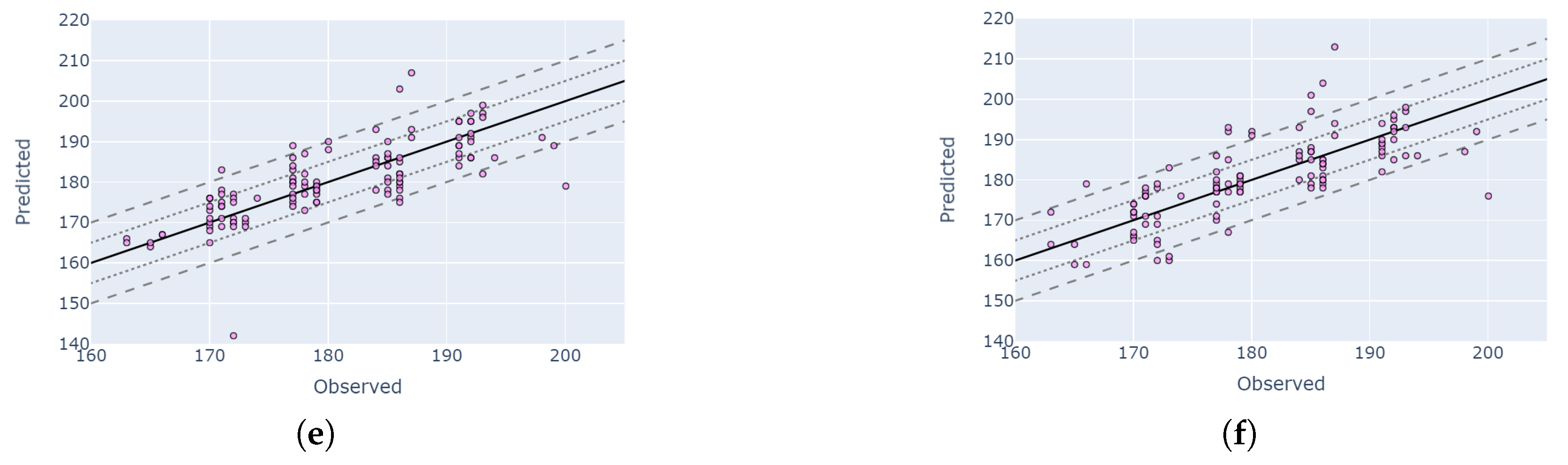
4.5. Visual Validation for PIAF Fields
5. Discussion and Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Stage | NDVI | MCARI | EVI | EVI2 | CVI | kNDVI | NDWI | Band G | Band R | Band NIR |
|---|---|---|---|---|---|---|---|---|---|---|
| VE: Emerging | 7.4 | 8.2 | 8.1 | 7.4 | 8.1 | 7.3 | 7.9 | 8.6 | 7.7 | 10.8 |
| VE: Emerging—Seedling | 8.5 | 8.7 | 7.3 | 8.5 | 8.3 | 8.6 | 8.2 | 5.9 | 5.9 | 8.2 |
| VE: Seedling | 8.6 | 9.3 | 7.9 | 8.5 | 9.6 | 8.4 | 9.1 | 8.0 | 7.9 | 8.5 |
| VE: Seedling—1-leaf | 6.5 | 6.5 | 5.8 | 6.5 | 6.0 | 6.6 | 6.3 | 6.6 | 6.4 | 8.7 |
| V1: 1-leaf | 7.6 | 6.7 | 6.8 | 7.5 | 9.6 | 7.5 | 8.2 | 8.2 | 7.7 | 12.2 |
| V1: 1–2-leaf | 7.4 | 7.6 | 7.1 | 7.5 | 8.0 | 7.5 | 7.7 | 6.5 | 6.6 | 9.9 |
| V2: 2-leaf | 5.9 | 6.2 | 5.5 | 5.9 | 6.5 | 6.0 | 6.1 | 5.4 | 5.0 | 8.4 |
| V2: 2–3-leaf | 8.8 | 7.9 | 8.9 | 8.8 | 7.3 | 8.8 | 8.1 | 9.2 | 9.7 | 9.3 |
| V3: 3-leaf | 5.8 | 5.7 | 5.6 | 5.8 | 6.9 | 5.9 | 6.2 | 6.3 | 6.1 | 7.8 |
| V3: 3–4-leaf | 3.6 | 3.5 | 3.4 | 3.5 | 4.5 | 3.6 | 3.8 | 4.3 | 4.2 | 7.6 |
| V4: 4-leaf | 4.6 | 4.3 | 4.2 | 4.6 | 5.4 | 4.6 | 5.0 | 4.9 | 4.5 | 7.8 |
| V4: 4–5-leaf | 4.6 | 4.6 | 4.6 | 4.6 | 5.4 | 4.6 | 4.8 | 5.9 | 5.5 | 7.4 |
| V5: 5-leaf | 7.5 | 6.8 | 7.7 | 7.5 | 6.9 | 7.5 | 7.3 | 8.0 | 8.3 | 8.4 |
| V5: 5–6-leaf | 3.5 | 3.3 | 3.3 | 3.5 | 3.8 | 3.5 | 3.6 | 4.0 | 4.0 | 7.9 |
| V6: 6-leaf | 4.8 | 4.8 | 4.9 | 4.8 | 5.0 | 4.8 | 4.7 | 5.2 | 5.1 | 7.6 |
| V6: 6–7-leaf | 4.6 | 4.5 | 4.6 | 4.5 | 4.9 | 4.5 | 4.6 | 5.1 | 4.9 | 6.0 |
| V7: 7-leaf | 7.8 | 6.9 | 8.1 | 7.8 | 5.9 | 7.9 | 7.5 | 8.6 | 8.7 | 7.6 |
| V7: 7–8-leaf | 4.4 | 4.0 | 4.5 | 4.2 | 4.4 | 4.3 | 4.2 | 4.8 | 4.7 | 5.0 |
| V8: 8-leaf | 6.5 | 6.5 | 6.6 | 6.5 | 6.0 | 6.5 | 6.3 | 6.5 | 6.6 | 7.8 |
| V8: 8–9-leaf | 6.6 | 6.2 | 7.0 | 6.6 | 5.3 | 6.6 | 6.5 | 7.1 | 7.3 | 6.8 |
| V9: 9-leaf | 6.7 | 6.9 | 6.9 | 6.8 | 6.6 | 6.8 | 6.7 | 6.8 | 6.8 | 6.8 |
| V9: 9–10-leaf | 5.3 | 5.0 | 5.4 | 5.2 | 5.4 | 5.1 | 5.3 | 5.7 | 5.6 | 5.1 |
| V10: 10-leaf | 6.4 | 5.3 | 6.5 | 6.3 | 4.5 | 6.4 | 6.0 | 5.6 | 6.2 | 4.7 |
| V11: 10–11-leaf | 7.9 | 6.7 | 8.4 | 7.8 | 5.7 | 7.8 | 7.3 | 8.5 | 8.7 | 6.2 |
| V11: 11-leaf | 7.9 | 7.6 | 8.3 | 7.8 | 6.7 | 7.9 | 7.6 | 7.2 | 7.7 | 8.9 |
| V12: 11–12-leaf | 6.9 | 6.3 | 6.9 | 6.9 | 5.9 | 6.8 | 6.7 | 6.6 | 6.9 | 5.3 |
| V12: 12-leaf | 7.5 | 6.9 | 7.9 | 7.4 | 5.6 | 7.5 | 6.9 | 6.7 | 7.2 | 6.5 |
| V12: 12–13-leaf | 6.8 | 6.0 | 6.9 | 6.8 | 5.4 | 6.8 | 6.4 | 6.4 | 6.7 | 5.8 |
| V13: 13-leaf | 12.2 | 10.3 | 13.4 | 12.1 | 7.8 | 12.4 | 11.5 | 12.7 | 13.5 | 7.0 |
| V13: 13–14-leaf | 6.3 | 5.8 | 6.2 | 6.2 | 5.6 | 6.2 | 5.9 | 6.4 | 6.4 | 5.3 |
| V14: 14–15-leaf | 6.8 | 5.3 | 7.2 | 6.8 | 5.0 | 6.8 | 6.2 | 6.4 | 6.4 | 5.3 |
| V15: 15–16-leaf | 6.0 | 5.5 | 6.1 | 6.0 | 5.2 | 6.2 | 6.3 | 5.5 | 5.7 | 5.4 |
| V16: 16-leaf—Tassel | 7.7 | 6.9 | 8.8 | 7.7 | 5.9 | 7.6 | 7.0 | 8.5 | 9.2 | 6.3 |
| VT: Tassel | 7.2 | 5.1 | 6.9 | 7.2 | 5.9 | 7.1 | 6.9 | 7.1 | 6.6 | 6.0 |
| VT: Tassel—Silk | 7.2 | 3.9 | 7.1 | 7.2 | 4.4 | 7.2 | 6.3 | 6.0 | 6.9 | 4.3 |
| VT: Silk—Brown Silk | 6.8 | 3.9 | 6.9 | 6.8 | 6.1 | 6.7 | 6.0 | 7.6 | 8.1 | 6.1 |
| Stage | NDVI | MCARI | EVI | EVI2 | CVI | kNDVI | NDWI | Band G | Band R | Band NIR |
|---|---|---|---|---|---|---|---|---|---|---|
| VE: Emerging | 4.0 | 6.0 | 5.5 | 4.0 | 5.5 | 4.0 | 5.0 | 6.5 | 5.0 | 7.5 |
| VE: Emerging—Seedling | 5.0 | 4.0 | 5.0 | 5.5 | 6.0 | 6.0 | 5.5 | 4.0 | 4.5 | 4.0 |
| VE: Seedling | 6.0 | 6.0 | 6.0 | 6.0 | 8.0 | 6.0 | 7.0 | 6.0 | 5.0 | 4.0 |
| VE: Seedling—1-leaf | 4.0 | 5.0 | 4.0 | 4.0 | 4.0 | 4.0 | 5.0 | 5.0 | 5.0 | 7.0 |
| V1: 1-leaf | 6.0 | 4.0 | 5.0 | 6.0 | 5.0 | 6.0 | 5.0 | 3.0 | 4.0 | 5.0 |
| V1: 1–2-leaf | 5.0 | 4.0 | 4.0 | 5.0 | 5.0 | 5.0 | 5.0 | 3.0 | 4.0 | 6.0 |
| V2: 2-leaf | 3.5 | 3.0 | 3.0 | 3.5 | 3.5 | 3.5 | 3.5 | 3.0 | 2.5 | 5.0 |
| V2: 2–3-leaf | 3.0 | 3.0 | 2.0 | 3.0 | 2.0 | 2.5 | 3.0 | 3.0 | 3.0 | 5.0 |
| V3: 3-leaf | 3.5 | 3.0 | 3.0 | 3.0 | 4.0 | 3.5 | 4.0 | 3.5 | 3.0 | 5.0 |
| V3: 3–4-leaf | 2.0 | 2.0 | 2.0 | 2.0 | 2.5 | 2.0 | 2.5 | 3.0 | 2.0 | 3.0 |
| V4: 4-leaf | 3.0 | 3.0 | 3.0 | 3.0 | 4.0 | 3.0 | 4.0 | 3.0 | 3.0 | 4.0 |
| V4: 4–5-leaf | 3.0 | 2.5 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 5.0 |
| V5: 5-leaf | 3.0 | 2.0 | 3.0 | 3.0 | 3.5 | 3.0 | 3.0 | 3.0 | 3.0 | 5.0 |
| V5: 5–6-leaf | 3.0 | 2.0 | 2.0 | 3.0 | 3.0 | 3.0 | 2.0 | 3.0 | 3.0 | 4.0 |
| V6: 6-leaf | 2.0 | 2.0 | 2.0 | 2.0 | 3.0 | 2.0 | 2.0 | 2.0 | 2.0 | 4.0 |
| V6: 6–7-leaf | 3.0 | 2.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 3.0 | 4.0 |
| V7: 7-leaf | 3.0 | 3.0 | 3.0 | 3.0 | 3.5 | 3.0 | 3.0 | 2.0 | 2.0 | 4.0 |
| V7: 7–8-leaf | 3.0 | 2.0 | 3.0 | 3.0 | 3.0 | 3.0 | 2.0 | 3.0 | 3.0 | 3.0 |
| V8: 8-leaf | 4.0 | 4.0 | 4.0 | 4.0 | 3.5 | 4.0 | 4.0 | 4.0 | 4.0 | 3.0 |
| V8: 8–9-leaf | 3.0 | 3.5 | 3.0 | 3.0 | 3.5 | 3.0 | 3.0 | 3.0 | 3.0 | 3.5 |
| V9: 9-leaf | 3.0 | 3.0 | 2.0 | 3.0 | 4.0 | 3.0 | 3.0 | 3.0 | 3.0 | 4.0 |
| V9: 9–10-leaf | 4.0 | 3.0 | 4.0 | 4.0 | 3.5 | 4.0 | 4.0 | 4.0 | 4.0 | 3.0 |
| V10: 10-leaf | 3.0 | 3.0 | 4.0 | 3.0 | 3.0 | 3.0 | 3.0 | 4.0 | 4.0 | 4.0 |
| V11: 11-leaf | 5.5 | 6.0 | 5.5 | 5.5 | 4.0 | 5.5 | 5.0 | 4.0 | 5.0 | 6.0 |
| V12: 12-leaf | 5.0 | 4.0 | 5.0 | 5.0 | 5.0 | 5.0 | 4.0 | 5.0 | 5.0 | 5.0 |
| V12: 12–13-leaf | 5.0 | 4.0 | 5.0 | 5.0 | 4.0 | 5.0 | 5.0 | 5.0 | 5.0 | 4.0 |
| V13: 13-leaf | 6.0 | 6.0 | 5.0 | 5.0 | 6.0 | 5.0 | 6.0 | 6.0 | 6.0 | 5.0 |
| V13: 13–14-leaf | 5.0 | 5.0 | 5.0 | 5.0 | 4.0 | 5.0 | 5.0 | 6.0 | 6.0 | 4.0 |
| V14: 14–15-leaf | 4.0 | 3.0 | 5.0 | 4.0 | 3.0 | 4.0 | 4.0 | 5.0 | 4.0 | 3.0 |
| V15: 15–16-leaf | 5.0 | 6.0 | 4.0 | 5.0 | 3.0 | 5.0 | 4.0 | 3.0 | 4.0 | 3.0 |
| V16: 16-leaf—Tassel | 5.5 | 5.0 | 5.0 | 5.0 | 4.0 | 5.0 | 5.0 | 5.0 | 6.0 | 5.0 |
| VT: Tassel | 6.0 | 4.0 | 5.5 | 6.0 | 4.5 | 6.0 | 6.0 | 5.5 | 5.0 | 5.0 |
| VT: Tassel—Silk | 5.0 | 3.0 | 4.5 | 5.0 | 3.0 | 5.0 | 5.0 | 5.0 | 6.0 | 3.0 |
| VT: Silk—Brown Silk | 5.5 | 3.0 | 5.0 | 5.5 | 5.0 | 5.5 | 4.0 | 8.0 | 8.0 | 2.5 |
| Stage | PF NDVI | PF MCARI | EVI | EVI2 | CVI | kNDVI | NDWI | Band G | Band R | Band NIR |
|---|---|---|---|---|---|---|---|---|---|---|
| R1: Pollen Shed | 6.9 | 5.8 | 7.4 | 6.8 | 5.2 | 6.9 | 6.2 | 7.3 | 7.7 | 6.1 |
| R1: Silking—Blister | 5.6 | 5.1 | 6.2 | 5.6 | 5.5 | 5.6 | 5.2 | 6.1 | 5.6 | 5.9 |
| R2: Blister | 6.6 | 5.2 | 7.1 | 6.5 | 5.7 | 6.5 | 5.8 | 6.6 | 6.8 | 7.2 |
| R2: Blister—Milk | 4.8 | 4.8 | 5.6 | 4.7 | 5.5 | 4.8 | 4.3 | 4.2 | 4.6 | 6.0 |
| R3: Milk | 5.8 | 5.4 | 6.5 | 5.8 | 6.2 | 5.7 | 5.5 | 6.8 | 6.3 | 7.3 |
| R3: Milk—Dough | 5.6 | 6.0 | 6.4 | 5.6 | 6.2 | 5.6 | 5.2 | 5.5 | 5.5 | 7.5 |
| R4: Dough | 6.5 | 6.5 | 7.8 | 6.5 | 7.0 | 6.5 | 6.1 | 6.1 | 6.1 | 7.7 |
| R4: Soft Dough | 7.6 | 7.8 | 10.2 | 7.6 | 7.9 | 7.6 | 6.7 | 8.9 | 9.1 | 8.6 |
| R4: Hard Dough | 5.3 | 6.1 | 6.5 | 5.4 | 7.1 | 5.3 | 5.2 | 6.5 | 6.3 | 7.0 |
| R4: Dough—Early Dent | 6.3 | 6.4 | 7.4 | 6.3 | 8.2 | 6.4 | 5.9 | 6.4 | 6.2 | 8.3 |
| R5: Early Dent | 5.6 | 7.0 | 7.2 | 5.6 | 8.6 | 5.6 | 6.0 | 5.9 | 6.0 | 8.6 |
| R5: Early—Mid Dent | 6.3 | 7.8 | 8.0 | 6.3 | 9.1 | 6.3 | 6.7 | 6.7 | 6.7 | 9.1 |
| R5: Mid Dent | 7.3 | 7.1 | 6.7 | 7.4 | 9.5 | 7.2 | 8.0 | 7.4 | 6.6 | 7.4 |
| R5: Mid—Full Dent | 7.4 | 8.5 | 7.3 | 7.3 | 9.5 | 7.4 | 7.8 | 7.1 | 6.2 | 10.6 |
| R5: Full Dent | 7.3 | 8.7 | 9.3 | 7.2 | 8.9 | 7.3 | 7.1 | 7.2 | 6.9 | 9.6 |
| R5: 1/8 Milk Line | 6.7 | 7.5 | 7.8 | 6.7 | 9.5 | 6.7 | 6.9 | 7.5 | 7.1 | 8.8 |
| R5: 1/8–1/4 Milk Line | 7.1 | 7.6 | 8.2 | 7.0 | 9.8 | 7.1 | 7.4 | 8.3 | 7.5 | 9.3 |
| R5: 1/4 Milk Line | 7.6 | 7.4 | 8.2 | 7.5 | 9.6 | 7.5 | 7.9 | 7.9 | 7.5 | 8.0 |
| R5: 1/4–1/3 Milk Line | 8.7 | 8.6 | 8.9 | 8.6 | 11.6 | 8.6 | 9.1 | 10.0 | 8.7 | 10.1 |
| R5: 1/3–1/2 Milk Line | 8.6 | 8.5 | 9.1 | 8.6 | 12.1 | 8.5 | 9.2 | 8.5 | 8.6 | 8.6 |
| R5: 1/2 Milk Line | 7.3 | 7.2 | 7.4 | 7.3 | 10.8 | 7.3 | 8.1 | 8.4 | 7.7 | 8.1 |
| R5: 1/2–2/3 Milk Line | 8.0 | 7.8 | 8.2 | 7.9 | 11.2 | 8.0 | 8.4 | 8.0 | 8.2 | 9.2 |
| R5: 2/3 Milk Line | 10.7 | 9.4 | 11.0 | 10.6 | 13.8 | 10.6 | 11.2 | 10.6 | 10.8 | 10.3 |
| R5: 2/3–3/4 Milk Line | 7.9 | 7.8 | 8.4 | 7.9 | 10.4 | 7.8 | 8.1 | 7.6 | 7.9 | 9.3 |
| R5: 3/4 Milk Line | 8.1 | 7.5 | 7.7 | 8.1 | 11.5 | 8.1 | 8.4 | 12.0 | 9.0 | 10.3 |
| R5: 3/4–7/8 Milk Line | 7.6 | 7.7 | 8.0 | 7.5 | 11.1 | 7.5 | 7.9 | 10.2 | 8.2 | 9.1 |
| R5: 7/8 Milk Line | 4.7 | 5.3 | 5.0 | 4.7 | 9.0 | 4.7 | 5.3 | 8.2 | 5.6 | 7.1 |
| R5: 7/8 Milk L.—Black L. | 8.7 | 9.3 | 8.9 | 8.7 | 12.1 | 8.7 | 9.1 | 9.2 | 8.5 | 12.4 |
| R5: Black Layer | 8.3 | 7.8 | 8.2 | 8.2 | 12.0 | 8.1 | 9.1 | 10.8 | 7.9 | 8.5 |
| Stage | PF NDVI | PF MCARI | EVI | EVI2 | CVI | kNDVI | NDWI | Band G | Band R | Band NIR |
|---|---|---|---|---|---|---|---|---|---|---|
| R1: Pollen Shed | 5.0 | 3.0 | 4.0 | 5.0 | 4.0 | 4.5 | 5.0 | 5.0 | 5.0 | 4.0 |
| R1: Silking—Blister | 5.0 | 2.0 | 4.0 | 5.0 | 4.0 | 5.0 | 4.0 | 4.0 | 5.0 | 4.0 |
| R2: Blister | 5.0 | 4.0 | 4.0 | 4.5 | 3.0 | 4.5 | 4.0 | 4.5 | 4.0 | 5.0 |
| R2: Blister—Milk | 3.0 | 3.0 | 3.0 | 3.0 | 4.0 | 3.0 | 3.0 | 3.0 | 3.0 | 4.0 |
| R3: Milk | 3.0 | 4.0 | 4.5 | 3.0 | 4.0 | 3.0 | 3.0 | 4.0 | 3.5 | 5.0 |
| R3: Milk—Dough | 4.0 | 5.0 | 4.0 | 4.0 | 4.0 | 4.0 | 4.0 | 3.0 | 3.0 | 5.0 |
| R4: Dough | 4.0 | 4.0 | 6.0 | 4.0 | 4.0 | 4.0 | 4.0 | 3.0 | 4.0 | 6.0 |
| R4: Soft Dough | 4.0 | 4.0 | 6.0 | 4.0 | 5.0 | 4.0 | 4.0 | 4.0 | 5.0 | 6.0 |
| R4: Hard Dough | 3.0 | 5.0 | 4.0 | 3.0 | 5.0 | 3.0 | 3.0 | 4.0 | 3.0 | 5.0 |
| R4: Dough—Early Dent | 4.0 | 6.0 | 6.0 | 4.0 | 5.0 | 4.0 | 3.0 | 3.0 | 4.0 | 7.0 |
| R5: Early Dent | 4.0 | 6.0 | 5.0 | 5.0 | 6.0 | 5.0 | 5.0 | 4.0 | 4.0 | 7.0 |
| R5: Early–Mid Dent | 4.5 | 6.0 | 5.0 | 4.5 | 6.5 | 4.5 | 5.0 | 4.0 | 5.0 | 7.0 |
| R5: Mid Dent | 5.0 | 4.0 | 5.0 | 5.0 | 8.0 | 4.0 | 6.0 | 4.0 | 4.0 | 6.0 |
| R5: Mid–Full Dent | 4.0 | 6.0 | 6.0 | 4.0 | 6.0 | 4.0 | 4.0 | 4.0 | 4.0 | 7.0 |
| R5: Full Dent | 5.0 | 6.0 | 6.0 | 5.0 | 7.5 | 5.0 | 6.0 | 5.0 | 6.0 | 7.0 |
| R5: 1/8 Milk Line | 5.0 | 5.0 | 7.0 | 5.0 | 6.0 | 5.0 | 4.0 | 4.0 | 5.0 | 6.0 |
| R5: 1/8–1/4 Milk Line | 4.0 | 5.0 | 6.0 | 4.0 | 6.0 | 4.0 | 4.0 | 6.0 | 5.0 | 6.0 |
| R5: 1/4 Milk Line | 4.0 | 5.0 | 5.0 | 4.0 | 5.0 | 4.0 | 4.0 | 5.0 | 5.0 | 5.0 |
| R5: 1/4–1/3 Milk Line | 6.0 | 6.5 | 6.0 | 6.0 | 7.0 | 6.0 | 6.0 | 6.5 | 7.0 | 7.0 |
| R5: 1/3–1/2 Milk Line | 5.0 | 5.0 | 5.5 | 5.0 | 6.0 | 5.0 | 5.0 | 6.0 | 5.0 | 6.0 |
| R5: 1/2 Milk Line | 5.0 | 5.0 | 5.0 | 5.0 | 7.0 | 5.0 | 5.0 | 5.0 | 5.0 | 5.0 |
| R5: 1/2–2/3 Milk Line | 5.0 | 4.0 | 5.0 | 5.0 | 6.0 | 5.0 | 5.0 | 5.0 | 6.0 | 5.0 |
| R5: 2/3 Milk Line | 6.0 | 4.0 | 6.0 | 6.0 | 5.0 | 6.0 | 5.0 | 6.0 | 5.0 | 7.0 |
| R5: 2/3–3/4 Milk Line | 3.0 | 4.0 | 4.5 | 3.0 | 6.0 | 3.0 | 4.0 | 4.5 | 5.0 | 6.0 |
| R5: 3/4 Milk Line | 4.0 | 5.0 | 4.0 | 4.0 | 5.0 | 4.0 | 5.0 | 7.0 | 3.0 | 6.0 |
| R5: 3/4–7/8 Milk Line | 4.5 | 4.5 | 5.0 | 4.5 | 6.5 | 4.5 | 5.0 | 8.5 | 4.0 | 8.0 |
| R5: 7/8 Milk Line | 3.0 | 3.0 | 4.0 | 3.0 | 5.0 | 3.0 | 4.0 | 6.0 | 3.0 | 4.0 |
| R5: 7/8 Milk L.—Black L. | 5.0 | 5.0 | 4.0 | 5.0 | 7.0 | 5.0 | 5.0 | 6.0 | 4.0 | 7.0 |
| R5: Black Layer | 6.0 | 5.0 | 5.0 | 6.0 | 7.0 | 6.0 | 7.0 | 6.0 | 5.0 | 7.0 |
| Stage | PF NDVI | PF MCARI | HLS NDVI | HLS MCARI |
|---|---|---|---|---|
| VE: Emerging | 7.4 | 8.2 | 14.0 | 9.5 |
| VE: Emerging—Seedling | 8.5 | 8.7 | 10.1 | 11.6 |
| VE: Seedling | 8.6 | 9.3 | 10.8 | 13.2 |
| VE: Seedling—1-leaf | 6.5 | 6.5 | 11.2 | 9.7 |
| V1: 1-leaf | 7.6 | 6.7 | 7.5 | 11.6 |
| V1: 1–2-leaf | 7.4 | 7.6 | 9.2 | 9.9 |
| V2: 2-leaf | 5.9 | 6.2 | 8.2 | 11.7 |
| V2: 2–3-leaf | 8.8 | 7.9 | 8.6 | 9.5 |
| V3: 3-leaf | 5.8 | 5.7 | 9.4 | 8.8 |
| V3: 3–4-leaf | 3.6 | 3.5 | 7.4 | 7.4 |
| V4: 4-leaf | 4.6 | 4.3 | 6.9 | 10.5 |
| V4: 4–5-leaf | 4.6 | 4.6 | 6.2 | 6.0 |
| V5: 5-leaf | 7.5 | 6.8 | 7.9 | 9.9 |
| V5: 5–6-leaf | 3.5 | 3.3 | 4.4 | 5.7 |
| V6: 6-leaf | 4.8 | 4.8 | 5.9 | 7.0 |
| V6: 6–7-leaf | 4.6 | 4.5 | 6.6 | 8.0 |
| V7: 7-leaf | 7.8 | 6.9 | 7.3 | 7.2 |
| V7: 7–8-leaf | 4.4 | 4.0 | 6.1 | 5.4 |
| V8: 8-leaf | 6.5 | 6.5 | 7.1 | 7.3 |
| V8: 8–9-leaf | 6.6 | 6.2 | 9.0 | 7.9 |
| V9: 9-leaf | 6.7 | 6.9 | 6.8 | 6.5 |
| V9: 9–10-leaf | 5.3 | 5.0 | 8.1 | 6.9 |
| V10: 10-leaf | 6.4 | 5.3 | 7.1 | 14.4 |
| V10: 10–11-leaf | 7.9 | 6.7 | 11.0 | 9.3 |
| V11: 11-leaf | 7.9 | 7.6 | 8.6 | 8.3 |
| V11: 11–12-leaf | 6.9 | 6.3 | 9.6 | 8.8 |
| V12: 12-leaf | 7.5 | 6.9 | 7.7 | 8.0 |
| V12: 12–13-leaf | 6.8 | 6.0 | 9.2 | 9.7 |
| V13: 13-leaf | 12.2 | 10.3 | 13.2 | 13.5 |
| V13: 13–14-leaf | 6.3 | 5.8 | 8.9 | 7.7 |
| V14: 14–15-leaf | 6.8 | 5.3 | 10.2 | 8.7 |
| V15: 15–16-leaf | 6.0 | 5.5 | 12.3 | 11.2 |
| V16: 16-leaf—Tassel | 7.7 | 6.9 | 12.1 | 10.3 |
| VT: Tassel | 7.2 | 5.1 | 10.7 | 8.9 |
| VT: Tassel—Silk | 7.2 | 3.9 | 9.9 | 9.8 |
| VT: Silk—Brown Silk | 6.8 | 3.9 | 6.4 | 4.5 |
| Stage | PF NDVI | PF MCARI | HLS NDVI | HLS MCARI |
|---|---|---|---|---|
| R1: Pollen Shed | 6.9 | 5.8 | 10.4 | 9.4 |
| R1: Silking—Blister | 5.6 | 5.1 | 11.4 | 7.9 |
| R2: Blister | 6.6 | 5.2 | 9.2 | 9.1 |
| R2: Blister—Milk | 4.8 | 4.8 | 10.4 | 7.9 |
| R3: Milk | 5.8 | 5.4 | 11.0 | 11.2 |
| R3: Milk—Dough | 5.6 | 6.0 | 9.1 | 9.3 |
| R4: Dough | 6.5 | 6.5 | 9.9 | 8.2 |
| R4: Soft Dough | 7.6 | 7.8 | 10.0 | 8.1 |
| R4: Hard Dough | 5.3 | 6.1 | 9.7 | 11.6 |
| R4: Dough—Early Dent | 6.3 | 6.4 | 8.0 | 7.8 |
| R5: Early Dent | 5.6 | 7.0 | 9.0 | 8.1 |
| R5: Early–Mid Dent | 6.3 | 7.8 | 7.5 | 7.9 |
| R5: Mid Dent | 7.3 | 7.1 | 8.7 | 12.8 |
| R5: Mid–Full Dent | 7.4 | 8.5 | 10.9 | 10.3 |
| R5: Full Dent | 7.3 | 8.7 | 10.1 | 9.7 |
| R5: 1/8 Milk Line | 6.7 | 7.5 | 9.7 | 8.9 |
| R5: 1/8-1/4 Milk Line | 7.1 | 7.6 | 9.3 | 9.3 |
| R5: 1/4 Milk Line | 7.6 | 7.4 | 10.0 | 9.3 |
| R5: 1/4-1/3 Milk Line | 8.7 | 8.6 | 9.6 | 8.5 |
| R5: 1/3-1/2 Milk Line | 8.6 | 8.5 | 10.2 | 9.5 |
| R5: 1/2 Milk Line | 7.3 | 7.2 | 7.7 | 7.6 |
| R5: 1/2-2/3 Milk Line | 8.0 | 7.8 | 10.3 | 9.5 |
| R5: 2/3 Milk Line | 10.7 | 9.4 | 10.6 | 10.3 |
| R5: 2/3-3/4 Milk Line | 7.9 | 7.8 | 7.9 | 10.2 |
| R5: 3/4 Milk Line | 8.1 | 7.5 | 7.2 | 8.3 |
| R5: 3/4-7/8 Milk Line | 7.6 | 7.7 | 10.6 | 9.2 |
| R5: 7/8 Milk Line | 4.7 | 5.3 | 6.0 | 6.7 |
| R5: 7/8 Milk Line—B. Layer | 8.7 | 9.3 | 9.1 | 10.6 |
| R5: Black Layer | 8.3 | 7.8 | 11.4 | 7.7 |
| Average (all stages) | 6.9 | 6.5 | 9.0 | 9.1 |
References
- Zeng, L.; Wardlow, B.D.; Xiang, D.; Hu, S.; Li, D. A review of vegetation phenological metrics extraction using time-series, multispectral satellite data. Remote Sens. Environ. 2020, 237, 111511. [Google Scholar] [CrossRef]
- Gao, F.; Zhang, X. Mapping Crop Phenology in Near Real-Time Using Satellite Remote Sensing: Challenges and Opportunities. J. Remote Sens. 2021, 2021, 8379391. [Google Scholar] [CrossRef]
- Pipia, L.; Belda, S.; Franch, B.; Verrelst, J. Trends in Satellite Sensors and Image Time Series Processing Methods for Crop Phenology Monitoring. In Information and Communication Technologies for Agriculture—Theme I: Sensors; Springer International Publishing: Cham, Switzerland, 2022; pp. 199–231. [Google Scholar] [CrossRef]
- Helman, D. Land surface phenology: What do we really ‘see’ from space? Sci. Total Environ. 2018, 618, 665–673. [Google Scholar] [CrossRef] [PubMed]
- Pan, Z.; Huang, J.; Zhou, Q.; Wang, L.; Cheng, Y.; Zhang, H.; Blackburn, G.A.; Yan, J.; Liu, J. Mapping crop phenology using NDVI time-series derived from HJ-1 A/B data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 188–197. [Google Scholar] [CrossRef]
- Jonas Schreier, G.G.; Dubovyk, O. Crop-specific phenomapping by fusing Landsat and Sentinel data with MODIS time series. Eur. J. Remote Sens. 2021, 54, 47–58. [Google Scholar] [CrossRef]
- Lu, L.; Wang, C.; Gao, H.; Li, Q. Detecting winter wheat phenology with SPOT-VEGETATION data in the North China Plain. Geocarto Int. 2014, 29, 244–255. [Google Scholar] [CrossRef]
- Rodigheri, G.; Sanches, I.D.; Richetti, J.; Tsukahara, R.Y.; Lawes, R.; Bendini, H.d.N.; Adami, M. Estimating Crop Sowing and Harvesting Dates Using Satellite Vegetation Index: A Comparative Analysis. Remote Sens. 2023, 15, 5366. [Google Scholar] [CrossRef]
- Xu, X.; Conrad, C.; Doktor, D. Optimising Phenological Metrics Extraction for Different Crop Types in Germany Using the Moderate Resolution Imaging Spectrometer (MODIS). Remote Sens. 2017, 9, 254. [Google Scholar] [CrossRef]
- Yang, Y.; Tao, B.; Liang, L.; Huang, Y.; Matocha, C.; Lee, C.D.; Sama, M.; Masri, B.E.; Ren, W. Detecting Recent Crop Phenology Dynamics in Corn and Soybean Cropping Systems of Kentucky. Remote Sens. 2021, 13, 1615. [Google Scholar] [CrossRef]
- d’Andrimont, R.; Taymans, M.; Lemoine, G.; Ceglar, A.; Yordanov, M.; van der Velde, M. Detecting flowering phenology in oil seed rape parcels with Sentinel-1 and -2 time series. Remote Sens. Environ. 2020, 239, 111660. [Google Scholar] [CrossRef]
- Harfenmeister, K.; Itzerott, S.; Weltzien, C.; Spengler, D. Detecting Phenological Development of Winter Wheat and Winter Barley Using Time Series of Sentinel-1 and Sentinel-2. Remote Sens. 2021, 13, 5036. [Google Scholar] [CrossRef]
- Zhou, M.; Ma, X.; Wang, K.; Cheng, T.; Tian, Y.; Wang, J.; Zhu, Y.; Hu, Y.; Niu, Q.; Gui, L.; et al. Detection of phenology using an improved shape model on time-series vegetation index in wheat. Comput. Electron. Agric. 2020, 173, 105398. [Google Scholar] [CrossRef]
- Gao, F.; Anderson, M.; Daughtry, C.; Karnieli, A.; Hively, D.; Kustas, W. A within-season approach for detecting early growth stages in corn and soybean using high temporal and spatial resolution imagery. Remote Sens. Environ. 2020, 242, 111752. [Google Scholar] [CrossRef]
- Gao, F.; Anderson, M.C.; Hively, W.D. Detecting Cover Crop End-Of-Season Using VENµS and Sentinel-2 Satellite Imagery. Remote Sens. 2020, 12, 3524. [Google Scholar] [CrossRef]
- Zhang, C.; Xie, Z.; Shang, J.; Liu, J.; Dong, T.; Tang, M.; Feng, S.; Cai, H. Detecting winter canola (Brassica napus) phenological stages using an improved shape-model method based on time-series UAV spectral data. Crop J. 2022, 10, 1353–1362. [Google Scholar] [CrossRef]
- Chen, S.; Yi, Q.; Wang, F.; Zheng, J.; Li, J. Improving the matching degree between remotely sensed phenological dates and physiological growing stages of soybean by a dynamic offset-adjustment strategy. Sci. Total Environ. 2024, 906, 167783. [Google Scholar] [CrossRef] [PubMed]
- Houborg, R.; McCabe, M.F. Daily Retrieval of NDVI and LAI at 3 m Resolution via the Fusion of CubeSat, Landsat, and MODIS Data. Remote Sens. 2018, 10, 890. [Google Scholar] [CrossRef]
- Shen, Y.; Zhang, X.; Yang, Z.; Ye, Y.; Wang, J.; Gao, S.; Liu, Y.; Wang, W.; Tran, K.H.; Ju, J. Developing an operational algorithm for near-real-time monitoring of crop progress at field scales by fusing harmonized Landsat and Sentinel-2 time series with geostationary satellite observations. Remote Sens. Environ. 2023, 296, 113729. [Google Scholar] [CrossRef]
- Nieto, L.; Houborg, R.; Zajdband, A.; Jumpasut, A.; Prasad, P.V.; Olson, B.J.; Ciampitti, I.A. Impact of high-cadence earth observation in maize crop phenology classification. Remote Sens. 2022, 14, 469. [Google Scholar] [CrossRef]
- Meroni, M.; d’Andrimont, R.; Vrieling, A.; Fasbender, D.; Lemoine, G.; Rembold, F.; Seguini, L.; Verhegghen, A. Comparing land surface phenology of major European crops as derived from SAR and multispectral data of Sentinel-1 and -2. Remote Sens. Environ. 2021, 253, 112232. [Google Scholar] [CrossRef]
- Lobert, F.; Löw, J.; Schwieder, M.; Gocht, A.; Schlund, M.; Hostert, P.; Erasmi, S. A deep learning approach for deriving winter wheat phenology from optical and SAR time series at field level. Remote Sens. Environ. 2023, 298, 113800. [Google Scholar] [CrossRef]
- Jiang, C.; Guan, K.; Huang, Y.; Jong, M. A vehicle imaging approach to acquire ground truth data for upscaling to satellite data: A case study for estimating harvesting dates. Remote Sens. Environ. 2024, 300, 113894. [Google Scholar] [CrossRef]
- Baumann, M.; Ozdogan, M.; Richardson, A.D.; Radeloff, V.C. Phenology from Landsat when data is scarce: Using MODIS and Dynamic Time-Warping to combine multi-year Landsat imagery to derive annual phenology curves. Int. J. Appl. Earth Obs. Geoinf. 2017, 54, 72–83. [Google Scholar] [CrossRef]
- Giorgino, T. Computing and Visualizing Dynamic Time Warping Alignments in R: The dtw Package. J. Stat. Softw. 2009, 31, 1–24. [Google Scholar] [CrossRef]
- Petitjean, F.; Inglada, J.; Gançarski, P. Satellite image time series analysis under time warping. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3081–3095. [Google Scholar] [CrossRef]
- Belgiu, M.; Csillik, O. Sentinel-2 cropland mapping using pixel-based and object-based time-weighted dynamic time warping analysis. Remote Sens. Environ. 2018, 204, 509–523. [Google Scholar] [CrossRef]
- Zhao, F.; Yang, G.; Yang, X.; Cen, H.; Zhu, Y.; Han, S.; Yang, H.; He, Y.; Zhao, C. Determination of key phenological phases of winter wheat based on the time-weighted dynamic time warping algorithm and MODIS time-series data. Remote Sens. 2021, 13, 1836. [Google Scholar] [CrossRef]
- Ye, J.; Bao, W.; Liao, C.; Chen, D.; Hu, H. Corn phenology detection using the derivative dynamic time warping method and sentinel-2 time series. Remote Sens. 2023, 15, 3456. [Google Scholar] [CrossRef]
- Team, P.F. Planet Fusion Monitoring Technical Specification, Version 1.3.0; Technical Report; Planet Labs: San Francisco, CA, USA, 2024. [Google Scholar]
- Houborg, R.; McCabe, M.F. A Cubesat enabled Spatio-Temporal Enhancement Method (CESTEM) utilizing Planet, Landsat and MODIS data. Remote Sens. Environ. 2018, 209, 211–226. [Google Scholar] [CrossRef]
- Claverie, M.; Ju, J.; Masek, J.G.; Dungan, J.L.; Vermote, E.F.; Roger, J.C.; Skakun, S.V.; Justice, C. The Harmonized Landsat and Sentinel-2 surface reflectance data set. Remote Sens. Environ. 2018, 219, 145–161. [Google Scholar] [CrossRef]
- EO Research Team. eo-learn (v1.5.5). Zenodo. Available online: https://zenodo.org/records/12166103 (accessed on 14 June 2024).
- Qiu, S.; Zhu, Z.; He, B. Fmask 4.0: Improved cloud and cloud shadow detection in Landsats 4–8 and Sentinel-2 imagery. Remote Sens. Environ. 2019, 231, 111205. [Google Scholar] [CrossRef]
- Akima, H. A new method of interpolation and smooth curve fitting based on local procedures. J. ACM (JACM) 1970, 17, 589–602. [Google Scholar] [CrossRef]
- Tang, Z.; Amatulli, G.; Pellikka, P.K.E.; Heiskanen, J. Spectral Temporal Information for Missing Data Reconstruction (STIMDR) of Landsat Reflectance Time Series. Remote Sens. 2022, 14, 172. [Google Scholar] [CrossRef]
- Yan, L.; Roy, D. Automated crop field extraction from multi-temporal Web Enabled Landsat Data. Remote Sens. Environ. 2014, 144, 42–64. [Google Scholar] [CrossRef]
- Ciampitti, I.A.; Elmore, R.W.; Lauer, J. Corn growth and development. Dent 2011, 5, 1–24. [Google Scholar]
- Harrell, D.M.; Wilhelm, W.W.; McMaster, G.S. Scales 2: Computer program to convert among developmental stage scales for corn and small grains. Agron. J. 1998, 90, 235–238. [Google Scholar] [CrossRef]
- proPlant GmbH. Products-PIAF. Available online: https://proplant.de/produkte/ (accessed on 14 June 2004).
- Huete, A.; Liu, H.; Batchily, K.; Van Leeuwen, W. A comparison of vegetation indices over a global set of TM images for EOS-MODIS. Remote Sens. Environ. 1997, 59, 440–451. [Google Scholar] [CrossRef]
- Jiang, Z.; Huete, A.R.; Didan, K.; Miura, T. Development of a two-band enhanced vegetation index without a blue band. Remote Sens. Environ. 2008, 112, 3833–3845. [Google Scholar] [CrossRef]
- Camps-Valls, G.; Campos-Taberner, M.; Moreno-Martínez, Á.; Walther, S.; Duveiller, G.; Cescatti, A.; Mahecha, M.D.; Muñoz-Marí, J.; García-Haro, F.J.; Guanter, L.; et al. A unified vegetation index for quantifying the terrestrial biosphere. Sci. Adv. 2021, 7, eabc7447. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Vincini, M.; Frazzi, E. Comparing narrow and broad-band vegetation indices to estimate leaf chlorophyll content in planophile crop canopies. Precis. Agric. 2011, 12, 334–344. [Google Scholar] [CrossRef]
- McFeeters, S.K. The use of the Normalized Difference Water Index (NDWI) in the delineation of open water features. Int. J. Remote Sens. 1996, 17, 1425–1432. [Google Scholar] [CrossRef]
- Mori, A.; Uchida, S.; Kurazume, R.; Taniguchi, R.I.; Hasegawa, T.; Sakoe, H. Early recognition and prediction of gestures. In Proceedings of the 18th International Conference on Pattern Recognition (ICPR’06), Hong Kong, China, 20–24 August 2006; IEEE: New York, NY, USA, 2006; Volume 3, pp. 560–563. [Google Scholar]
- Itakura, F. Minimum prediction residual principle applied to speech recognition. IEEE Trans. Acoust. Speech Signal Process. 1975, 23, 67–72. [Google Scholar] [CrossRef]
- Sakamoto, T.; Wardlow, B.D.; Gitelson, A.A.; Verma, S.B.; Suyker, A.E.; Arkebauer, T.J. A two-step filtering approach for detecting maize and soybean phenology with time-series MODIS data. Remote Sens. Environ. 2010, 114, 2146–2159. [Google Scholar] [CrossRef]
- Clark, J.D.; Fernández, F.G.; Camberato, J.J.; Carter, P.R.; Ferguson, R.B.; Franzen, D.W.; Kitchen, N.R.; Laboski, C.A.; Nafziger, E.D.; Sawyer, J.E.; et al. Weather and soil in the US Midwest influence the effectiveness of single-and split-nitrogen applications in corn production. Agron. J. 2020, 112, 5288–5299. [Google Scholar] [CrossRef]
- Carrera, C.S.; Savin, R.; Slafer, G.A. Critical period for yield determination across grain crops. Trends Plant Sci. 2023, 29, 329–342. [Google Scholar] [CrossRef]
- Saravia, D.; Salazar, W.; Valqui-Valqui, L.; Quille-Mamani, J.; Porras-Jorge, R.; Corredor, F.A.; Barboza, E.; Vásquez, H.V.; Casas Diaz, A.V.; Arbizu, C.I. Yield Predictions of Four Hybrids of Maize (Zea mays) Using Multispectral Images Obtained UAV Coast Peru. Agronomy 2022, 12, 2630. [Google Scholar] [CrossRef]
- Viña, A.; Gitelson, A.; Rundquist, D.; Keydan, G.; Leavitt, B.; Schepers, J. Monitoring maize (Zea mays L.) Phenol. Remote Sensing. Agron. J. 2004, 96, 1139–1147. [Google Scholar] [CrossRef]
- Huang, X.; Liu, J.; Zhu, W.; Atzberger, C.; Liu, Q. The Optimal Threshold and Vegetation Index Time Series for Retrieving Crop Phenology Based on a Modified Dynamic Threshold Method. Remote Sens. 2019, 11, 2725. [Google Scholar] [CrossRef]
- Brewer, K.; Clulow, A.; Sibanda, M.; Gokool, S.; Naiken, V.; Mabhaudhi, T. Predicting the Chlorophyll Content of Maize over Phenotyping as a Proxy for Crop Health in Smallholder Farming Systems. Remote Sens. 2022, 14, 518. [Google Scholar] [CrossRef]
- Roßberg, D.; Jörg, E.; Falke, K. SIMONTO: Ein neues Ontogenesemodell für Wintergetreide und Winterraps. Nachrichtenblatt Des Dtsch. Pflanzenschutzdienstes 2005, 57, 74–80. [Google Scholar]
- Kumudini, S.; Andrade, F.H.; Boote, K.; Brown, G.; Dzotsi, K.; Edmeades, G.; Gocken, T.; Goodwin, M.; Halter, A.; Hammer, G.; et al. Predicting maize phenology: Intercomparison of functions for developmental response to temperature. Agron. J. 2014, 106, 2087–2097. [Google Scholar] [CrossRef]


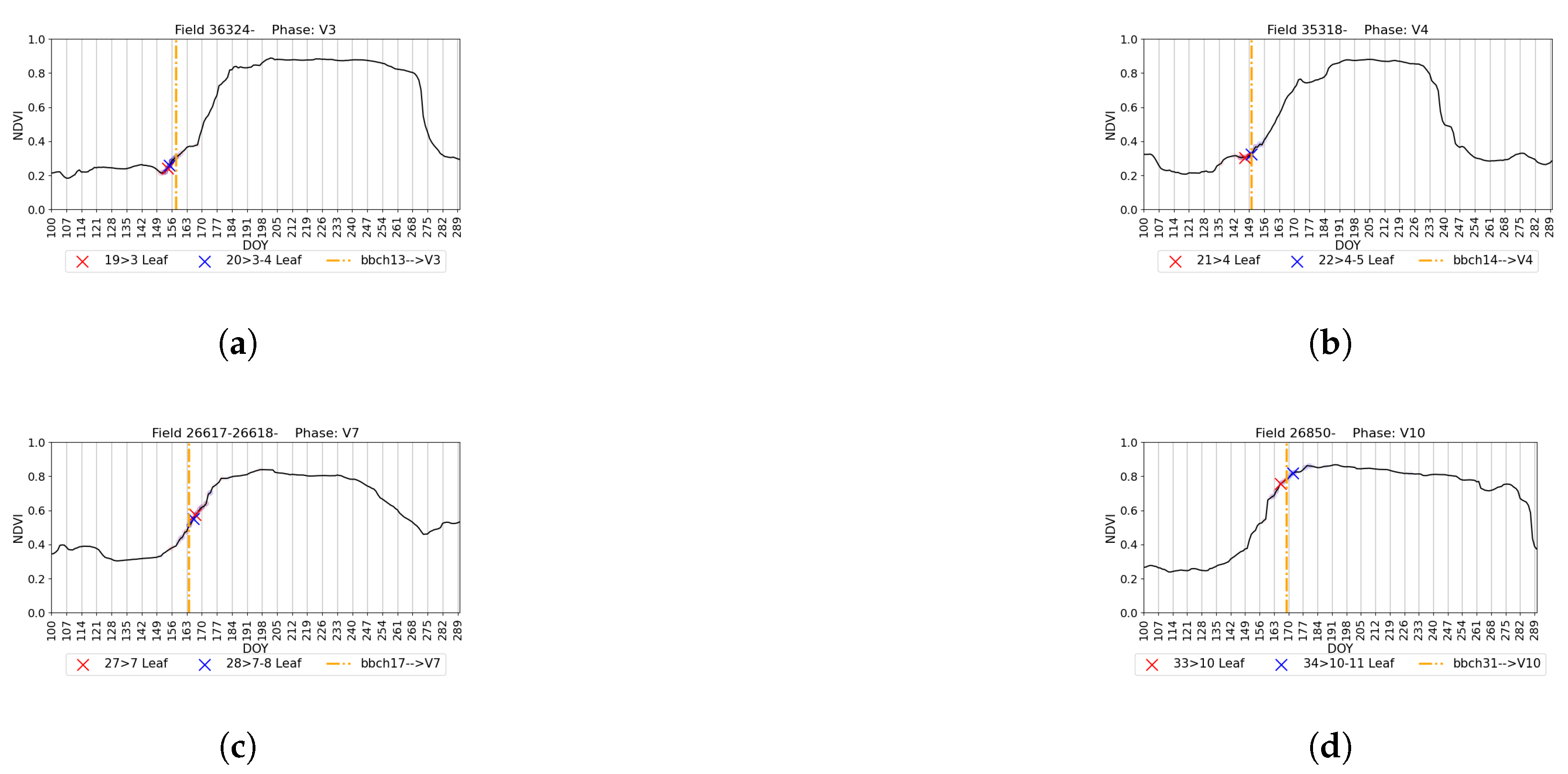
| Macro-Stage | Corresponding BBCH [39] | Starting Micro-Stage | Ending Micro-Stage | Number of Micro-Stage |
|---|---|---|---|---|
| VE | BBCH10 | Emerging | Seedling—1 Leaf | 4 |
| V1 | BBCH11 | 1-leaf | 1–2-leaf | 2 |
| V2 | BBCH12 | 2-leaf | 2–3-leaf | 2 |
| V3 | BBCH13 | 3-leaf | 3–4-leaf | 2 |
| V4 | BBCH14 | 4 leaf | 4–5-leaf | 2 |
| V5 | BBCH15 | 5-leaf | 5–6-leaf | 2 |
| V6 | BBCH16 | 6-leaf | 6–7-leaf | 2 |
| V7 | BBCH17 | 7-leaf | 7–8-leaf | 2 |
| V8 | BBCH18 | 8-leaf | 8–9-leaf | 2 |
| V9 | BBCH19 | 9-leaf | 9–10-leaf | 2 |
| V10 | BBCH31 | 10-leaf | 10–11-leaf | 2 |
| V11 | N/A | 11-leaf | 11–12-leaf | 2 |
| V12 | N/A | 12-leaf | 12–13-leaf | 2 |
| V13 | N/A | 13-leaf | 13–14-leaf | 2 |
| V14 | N/A | 14-leaf | 14–15-leaf | 2 |
| V15 | N/A | 15-leaf | 15–16-leaf | 2 |
| V16 | N/A | 16-leaf | 16-leaf—Tassel | 3 |
| VT | BBCH59 | Tassel | Silk—Brown Silk | 3 |
| R1 | BBCH63 | Silking | Silking—Blister | 3 |
| R2 | BBCH71 | Blister | Blister—Milk | 2 |
| R3 | BBCH75 | Milk | Milk—Dough | 2 |
| R4 | BBCH85 | Dough | Dough—Early Dent | 4 |
| R5 | BBCH86 | Early Dent | Black Layer | 19 |
| R6 | BBCH87 | Maturity | Maturity | 1 |
| Index | Formula | Vegetation Property |
|---|---|---|
| NDVI | (NIR − Red)/(NIR + Red) | Productivity |
| EVI [41] | 2.5 × (NIR − Red)/((NIR + 6 × Red − 7.5 × Blue) + 1) | Greenness |
| EVI2 [42] | 2.5 × (NIR − Red)/(NIR + 2.4 × Red + 1) | Greenness |
| kNDVI [43] | tanh(((NIR − Red)/(NIR + Red))2) | Productivity |
| MCARI [44] | (1.2 × (2.5 × (NIR − Red) − 1.3 × (NIR − Green))) | Leaf chlorophyll concentration |
| CVI [45] | NIR × (Red/(Green2)) | Leaf chlorophyll content |
| NDWI [46] | (Green − NIR)/(Green + NIR) | Water content |
| Max Error | ||||
|---|---|---|---|---|
| Algorithm | 1 Day | 5 Day | 10 Day | 15 Day |
| Method 1 | 486 (15%) | 1628 (50%) | 2550 (79%) | 3029 (94%) |
| Method 2 | 652 (20%) | 1832 (57%) | 2645 (82%) | 2994 (93%) |
| Method 3 (proposed) | 636 (20%) | 2023 (63%) | 2900 (90%) | 3136 (97%) |
| Phenology Stages | RMSE (Days) | MedAE (Days) | MAE (Days) | ||||
|---|---|---|---|---|---|---|---|
| Macro | Micro | NDVI | MCARI | NDVI | MCARI | NDVI | MCARI |
| V1 | 1-leaf | 7.6 | 6.7 | 6.0 | 4.0 | 6.2 | 5.6 |
| V4 | 4-leaf | 4.6 | 4.3 | 3.0 | 3.0 | 5.5 | 4.3 |
| V4 | 4–5-leaf | 4.6 | 4.6 | 3.0 | 2.5 | 3.3 | 3.3 |
| V6 | 6-leaf | 4.8 | 4.8 | 2.0 | 2.0 | 3.2 | 3.2 |
| VT | Silk—Brown Silk | 6.8 | 3.9 | 5.5 | 3.0 | 5.6 | 3.1 |
| VT | Tassel | 7.2 | 5.1 | 6.0 | 4.0 | 6.5 | 4.3 |
| VT | Tassel—Silk | 7.2 | 3.9 | 5.0 | 3.0 | 5.7 | 3.3 |
| R1 | Pollen Shed | 6.9 | 5.8 | 5.0 | 3.0 | 5.5 | 4.3 |
| R1 | Silking—Blister | 5.6 | 5.1 | 5.0 | 2.0 | 4.6 | 3.8 |
| R5 | 2/3 Milk Line | 10.7 | 9.4 | 6.0 | 4.0 | 7.8 | 6.9 |
| R5 | Early–Mid Dent | 6.3 | 7.8 | 4.5 | 6.0 | 4.8 | 6.4 |
| R5 | Early Dent | 5.6 | 7.0 | 4.0 | 6.0 | 4.6 | 6.0 |
| R5 | Full Dent | 7.3 | 8.7 | 5.0 | 6.0 | 5.8 | 6.6 |
| R5 | Mid–Full Dent | 7.4 | 8.5 | 4.0 | 6.0 | 5.4 | 6.9 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Senaras, C.; Grady, M.; Rana, A.S.; Nieto, L.; Ciampitti, I.; Holden, P.; Davis, T.; Wania, A. Detection of Maize Crop Phenology Using Planet Fusion. Remote Sens. 2024, 16, 2730. https://doi.org/10.3390/rs16152730
Senaras C, Grady M, Rana AS, Nieto L, Ciampitti I, Holden P, Davis T, Wania A. Detection of Maize Crop Phenology Using Planet Fusion. Remote Sensing. 2024; 16(15):2730. https://doi.org/10.3390/rs16152730
Chicago/Turabian StyleSenaras, Caglar, Maddie Grady, Akhil Singh Rana, Luciana Nieto, Ignacio Ciampitti, Piers Holden, Timothy Davis, and Annett Wania. 2024. "Detection of Maize Crop Phenology Using Planet Fusion" Remote Sensing 16, no. 15: 2730. https://doi.org/10.3390/rs16152730
APA StyleSenaras, C., Grady, M., Rana, A. S., Nieto, L., Ciampitti, I., Holden, P., Davis, T., & Wania, A. (2024). Detection of Maize Crop Phenology Using Planet Fusion. Remote Sensing, 16(15), 2730. https://doi.org/10.3390/rs16152730








