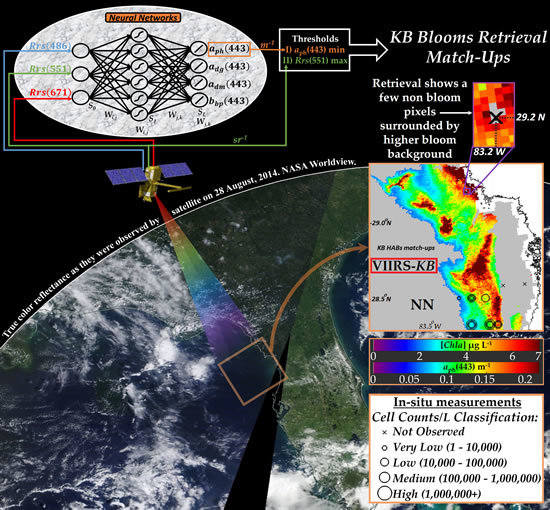
Satellite Retrievals of Karenia brevis Harmful Algal Blooms in the West Florida Shelf Using Neural Networks and Comparisons with Other Techniques
Abstract
:1. Introduction
2. Materials and Methods
Neural Network Algorithm Background
Synthetic Dataset
Algorithm Training
3. Results and Discussion
3.1. VIIRS Retrievals of Rrs551 and aph443 and Determination of Limiting Values of Rrs551max and aph443min in a KB Bloom Environment
3.2. Comparisons of NN KB HABs with Other Retrieval Techniques
3.2.1. Comparison of NN KB HABs Retrieval Techniques with Those Using the Normalized Fluorescence Height (nFLH) and Red Band Difference (RBD) Techniques
3.2.2. Comparisons of MODIS-A NN Retrievals with Red Band Difference (RBD) Index Retrievals
3.2.3. Comparisons of VIIRS NN Retrievals with VIIRS Ocean Color Chlorophyll-a (OCI/OC3) Retrievals
3.2.4. Comparisons of VIIRS NN Retrievals with Red/Green Chlorophyll-a Index (RGCI) Retrievals
3.3. Comparisons of VIIRS NN, OCI/OC3 and RGCI Retrievals with in Situ Cell Count Measurements
3.3.1. Evaluation of VIIRS KB HABs Retrievals against in Situ Measurement for Match-Ups Occurring over 2012–2015 Period
3.3.2. Evaluation of NN KB HABs Retrievals for Specific Bloom Events
4. Summary and Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| KB | Karenia brevis |
| HABs | Harmful algal blooms |
| WFS | West Florida Shelf |
| VIIRS | Visible Infrared Imaging Radiometer Suite |
| MODIS-A | Moderate Resolution Imaging Spectroradiometer Aqua |
| MERIS | MEdium Resolution Imaging Spectrometer |
| aph | Absorption coefficient due to phytoplankton particulates (m−1) |
| adg | Absorption coefficient due to non-phytoplankton particulates and dissolved substances, adm + ag (m−1) |
| adm | Absorption coefficient due to non-phytoplankton particulates (m−1) |
| aw: | Absorption coefficient due to water (m−1) |
| at | Total absorption coefficient, aph + adm + ag + aw (m−1) |
| bbp | Backscattering coefficient due to particulates (m−1) |
| bbw | Backscattering coefficient due to water (m−1) |
| bb | Total backscattering coefficient, bbp + bbw (m−1) |
| [Chla] | Chlorophyll-a concentration (µg·L−1) |
| CDOM | Color dissolved organic matter (ppm) |
| NAP | Non-phytoplankton particulate concentration (g·m−3) |
| AOP | Apparent optical properties |
| IOP | Inherent Optical properties |
| RT | Radiative transfer |
| Rrs | Above-surface remote-sensing reflectance (sr−1) |
| nLw | Normalized water leaving radiance (W·m−2·µm·sr−1) |
| MLPNN | Multi Layer perceptron neural network |
| NN | Neural network |
| NN [Chla] | NN deriving [Chla] from Rrs as inputs |
| NOMAD | NASA bio-Optical Marine Algorithm Data set [21]. |
| nFLH | normalized fluorescence height Algorithm [54]. |
| OC | Ocean Color |
| OC3 | Chlorophyll-a concentration (µg·L−1) derived using VIIRS and MODIS algorithm [39,40,41]. |
| OCI | Chlorophyll-a concentration (µg·L−1) derived using VIIRS and MODIS algorithm [39]. |
| RGCI | Red Green chlorophyll-a Index |
| ɛ | Estimate of the standard deviation of the error |
| N | Number of points |
| µ | Mean value |
| σ | standard deviation |
Appendix A
A1. NN Algorithm Background and Directions for Implementation for aph443 Retrieval
A.1.1. Background
A.1.2. Synthetic Dataset
A.1.3. NN Training
A.1.4. Testing the NN

A2. Retrieval of aph443 from Rrs486, 551 and 671 nm Using NN
- Retrieval of NN aph443 in (m−1) units, is obtained by denormalizing the NN output to obtain Equation (a1). below:where âph443 is the normalized phytoplankton absorption at 443 nm obtained by NN and carries out the final weighted sum activity and biases associated with the NN output aph443 m−1 and inputs Rrs486, Rrs551 and Rrs671 nm shown below in (Equation (a3)). The µo (λ) and σo (λ) are the mean and standard deviation of the NN âph443 outputs respectively, obtained from the simulated datasets, and shown in Table A1.
- Neural Network parameters needed for the above calculations:The first step shown in (Equation (a2)) below, requires input reflectance’s Rrs (λ), expressed as base 10 logarithm, standardized by removing, from each Rrs (λ), mean (µi(λ)) of input values, of the simulated dataset, and then scaling the difference by the standard deviation (σi(λ)) of input values, of the simulated dataset, as shown in Equation (a2). Table A1 shows the relevant mean and standard deviation input (for each corresponding Rrs wavelength) and output values. (This procedure is also applied as the NN is trained using normalized Rrs (λ) values, so that it is equally sensitive to all inputs, avoiding conditioning problems).
| log10(Rrs486) | log10(Rrs551) | log10(Rrs671) | log10(aph443) | ||
|---|---|---|---|---|---|
| µi | µo | ||||
| σi | σo |
References
- NOAA’s National Ocean Service. Harmful Algal Blooms. Available online: http://oceanservice.noaa.gov/hazards/hab (accessed on 9 March 2015).
- Tomlinson, M.C.; Wynne, T.T.; Stumpf, R.P. An evaluation of remote sensing techniques for enhanced detection of the toxic dinoflagellate Karenia brevis. Remote Sens. Environ. 2009, 113, 598–609. [Google Scholar] [CrossRef]
- Soto, I.M.; Cannizzaro, J.; Muller-Karger, F.E.; Hu, C.; Wolny, J.; Goldgof, D. Evaluation and optimization of remote sensing techniques for detection of Karenia brevis blooms on the West Florida Shelf. Remote Sens. Environ. 2015, 170. [Google Scholar] [CrossRef]
- Soto-Ramos, I.M. Harmful Algal Blooms of the West Florida Shelf and Campeche Bank: Visualization and Quantification Using Remote Sensing Methods. Ph.D. Thesis, University of South Florida, Tampa, FL, USA, January 2013. [Google Scholar]
- Amin, R.; Gilerson, A.; Gross, B.; Moshary, F.; Ahmed, S. MODIS and MERIS detection of dinoflagellate blooms using the RBD technique. Proc. SPIE 2009, 7473, 747304. [Google Scholar]
- Amin, R.; Zhou, J.; Gilerson, A.; Gross, B.; Moshary, F.; Ahmed, S. Novel optical techniques for detecting and classifying toxic dinoflagellate Karenia brevis blooms using satellite imagery. Opt. Express 2009, 17, 9126–9144. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Muller-Karger, F.E.; Taylor, C.; Carder, K.L.; Kelble, C.; Johns, E.; Heil, C.A. Red tide detection and tracing using MODIS fluorescence data: A regional example in SW Florida coastal waters. Remote Sens. Environ. 2005, 97, 311–321. [Google Scholar] [CrossRef]
- Hu, C.; Barnes, B.B.; Qi, L.; Corcoran, A.A. A harmful algal bloom of Karenia brevis in the Northeastern Gulf of Mexico as revealed by MODIS and VIIRS: A comparison. Sensors 2015, 15, 2873–2887. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.A.; Minnett, P.J.; Banzon, V.F.; Baringer, W.; Heil, C.A. Long-term evaluation of three satellite ocean color algorithms for identifying harmful algal blooms (Karenia brevis) along the west coast of Florida: A matchup assessment. Remote Sens. Environ. 2011, 115, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Carvalho, G.A.; Minnett, P.J.; Fleming, L.E.; Banzon, V.F.; Baringer, W. Satellite remote sensing of harmful algal blooms: A new multi-algorithm method for detecting the Florida Red Tide (Karenia brevis). Harmful Algae 2010, 9, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Blondeau-Patissier, D.; Gower, J.F.; Dekker, A.G.; Phinn, S.R.; Brando, V.E. A review of ocean color remote sensing methods and statistical techniques for the detection, mapping and analysis of phytoplankton blooms in coastal and open oceans. Progress Oceanogr. 2014, 123, 123–144. [Google Scholar] [CrossRef] [Green Version]
- Ioannou, I.; Gilerson, A.; Gross, B.; Moshary, F.; Ahmed, S. Neural network approach to retrieve the inherent optical properties of the ocean from observations of MODIS. Appl. Opt. 2011, 50, 3168–3186. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, I.; Gilerson, A.; Gross, B.; Moshary, F.; Ahmed, S. Deriving ocean color products using neural networks. Remote Sens. Environ. 2013, 134, 78–91. [Google Scholar] [CrossRef]
- Ioannou, I.; Gilerson, A.; Ondrusek, M.; Hlaing, S.; Foster, R.; El-Habashi, A.; Bastani, K.; Ahmed, S. Remote estimation of in water constituents in coastal waters using neural networks. Proc. SPIE 2014, 9240. [Google Scholar] [CrossRef]
- El-habashi, A.; Ahmed, S. Neural network algorithms for retrieval of harmful algal blooms in the west Florida shelf from VIIRS satellite observations and comparisons with other techniques, without the need for a fluorescence channel. Proc. SPIE 2015. [Google Scholar] [CrossRef]
- Le, C.; Hu, C. A hybrid approach to estimate chromophoric dissolved organic matter in turbid estuaries from satellite measurements: A case study for Tampa Bay. Opt. Express 2013, 21, 18849–18871. [Google Scholar] [CrossRef] [PubMed]
- Cannizzaro, J.P.; Carder, K.L.; Chen, F.R.; Heil, C.A.; Vargo, G.A. A novel technique for detection of the toxic dinoflagellate, Karenia brevis, in the Gulf of Mexico from remotely sensed ocean color data. Cont. Shelf Res. 2008, 28, 137–158. [Google Scholar] [CrossRef]
- Cannizzaro, J.P.; Hu, C.; English, D.C.; Carder, K.L.; Heil, C.A.; Müller-Karger, F.E. Detection of Karenia brevis blooms on the west Florida shelf using in situ backscattering and fluorescence data. Harmful Algae 2009, 8, 898–909. [Google Scholar] [CrossRef]
- Aires, F.; Prigent, C.; Rossow, W.B.; Rothstein, M. A neural network approach including first guess for retrieval of atmospheric water vapor, cloud liquid water path, surface temperature, and emissivities over land from satellite microwave observations. J. Geophys. Res. 2001, 106, 14887–14907. [Google Scholar] [CrossRef]
- Aires, F.; Prigent, C.; Rossow, W.B. Neural network uncertainty assessment using Bayesian statistics: A remote sensing application. Neural Comput. 2004, 16, 2415–2458. [Google Scholar] [CrossRef] [PubMed]
- Werdell, P.J.; Bailey, S.W. An improved in situ bio-optical data set for ocean color algorithm development and satellite data product validation. Remote Sens. Environ. 2005, 98, 122–140. [Google Scholar] [CrossRef]
- IOCCG. Remote Sensing of Inherent Optical Properties: Fundamentals, Tests of Algorithms, and Applications; Report 5; Lee, Z.-P., Ed.; IOCCG: Bay Saint Louis, MS, USA, 2006; pp. 57–62. [Google Scholar]
- Lee, Z.P.; Carder, K.L.; Arnone, R.A. Deriving inherent optical properties from water color: A multiband quasi-analytical algorithm for optically deep waters. Appl. Opt. 2002, 41, 5755–5772. [Google Scholar] [CrossRef] [PubMed]
- Morel, A. Optical modeling of the upper ocean in relation to its biogenous matter content (case I waters). J. Geophys. Res. 1988, 93, 10749–10768. [Google Scholar] [CrossRef]
- Gordon, H.R.; Brown, O.B.; Evans, R.H.; Brown, J.W.; Smith, R.C.; Baker, K.S.; Clark, D.K. A semi-analytic radiance model of ocean color. J. Geophys. Res. 1988, 93, 10909–10924. [Google Scholar] [CrossRef]
- Morel, A.; Maritorena, S. Bio-optica l properties of oceanic waters: A reappraisal. J. Geophys. Res. 2001, 106, 7163–7180. [Google Scholar] [CrossRef]
- Lee, Z.P. International Ocean-Colour Coordinating Group. Available online: http://www.ioccg.org/groups/lee_data.pdf (accessed on 9 March 2015).
- Bricaud, A.; Babin, M.; Morel, A.; Claustre, H. Variability in the chlorophyll-specific absorption coefficients of natural phytoplankton: Analysis and parameterization. J. Geophys. Res. 1995, 100, 13321–13332. [Google Scholar] [CrossRef]
- Bukata, R.P.; Jerome, J.H.; Kondratyev, K.Y.; Pozdnyakov, D.V. Optical Properties and Remote Sensing of Inland and Coastal Waters; CRC Press: Boca Raton, FL, USA, 1995. [Google Scholar]
- Stramski, D.; Bricaud, A.; Morel, A. Modeling the inherent optical properties of the ocean based on the detailed composition of the planktonic community. Appl. Opt. 2001, 40, 2929–2945. [Google Scholar] [CrossRef] [PubMed]
- Pope, R.; Fry, E. Absorption spectrum 380–700 nm of pure waters: II. Integrating cavity measurements. Appl. Opt. 1997, 36, 8710–8723. [Google Scholar] [CrossRef] [PubMed]
- Morel, A. Optical properties of pure water and pure sea water. In Optical Aspects of Oceanography; Jerlov, N.G., Nielsen, E.S., Eds.; Academic Press: New York, NY, USA, 1974; pp. 1–24. [Google Scholar]
- Ciotti, A.M.; Lewis, M.R.; Cullen, J.J. Assessment of the relationships between dominant cell size in natural phytoplankton communities and the spectral shape of the absorption coefficient. Limnol. Oceanogr. 2002, 47, 404–417. [Google Scholar] [CrossRef]
- Mobley, C.D. Light and Water: Radiative Transfer in Natural Waters; Academic Press: New York, NY, USA, 1994. [Google Scholar]
- Babin, M.; Morel, A.; Fournier-Sicre, V.; Fell, F.; Stramski, D. Light scattering properties of marine particles in coastal and oceanic waters as related to the particle mass concentration. Limnol. Oceanogr. 2003, 48, 843–859. [Google Scholar] [CrossRef]
- Mobley, C.D.; Sundman, L.K. HYDROLIGHT 4.2; Sequoia Scientific, Inc.: Bellevue, WA, USA, 2001. [Google Scholar]
- Matthew Dornback. Harmful Algal Blooms Observing System. Available online: http://habsos.noaa.gov/ (accessed on 9 March 2015).
- NASA Ocean Biology (OB). Suomi-NPP Visible Infrared Imaging/Radiometer Suite (VIIRS) and Moderate Resolution Imaging Spectroradiometer (MODIS-Aqua) Ocean Color Data; NASA OB.DAAC: Greenbelt, MD, USA, 2014. [Google Scholar]
- Hu, C.; Lee, Z.; Franz, B. Chlorophyll a algorithms for oligotrophic oceans: A novel approach based on three-band reflectance difference. J. Geophys. Res. 2012, 117. [Google Scholar] [CrossRef]
- O’Reilley, J.E.; Maritorena, S.; O’Brien, M.C.; Siegel, D.A.; Toole, D.; Menzies, D.; Smith, R.C.; Mueller, J.L.; Mitchell, B.G.; Kahru, M.; et al. SeaWiFS Postlaunch Calibration and Validation Analyses; Part 3; Hooker, S.B., Firestone, E.R., Eds.; NASA Goddard Space Flight Center: Hampton, VA, USA, 2000; p. 49. [Google Scholar]
- O’Reilly, J.E.; Maritorena, S.; Mitchell, B.G.; Siegel, D.A.; Carder, K.L.; Garver, S.A.; Kahru, M.; McClain, C.R. Ocean color chlorophyll algorithms for SeaWiFS. J. Geophys. Res. 1998, 103. [Google Scholar] [CrossRef]
- Qi, L.; Hu, C.; Cannizzaro, J.; Corcoran, A.A.; English, D.; Le, C. VIIRS observations of a Karenia brevis bloom in the northeastern gulf of Mexico in the absence of a fluorescence band. IEEE Geosci. Remote Sens. Lett. 2015, 12, 2213–2217. [Google Scholar] [CrossRef]
- Stumpf, R.P.; Tomlinson, M.C.; Calkins, J.A.; Kirkpatrick, B.; Fisher, K.; Nierenberg, K.; Currier, R.; Wynne, T.T. Skill assessment for an operational algal bloom forecast system. J. Mar. Syst. 2009, 76, 151–161. [Google Scholar] [CrossRef] [PubMed]
- Tester, P.A.; Shea, D.; Kibler, S.R.; Varnam, S.M.; Black, M.D.; Litaker, R.W. Relationships among water column toxins, cell abundance and chlorophyll-a concentrations during Karenia brevis blooms. Cont. Shelf Res. 2008, 28, 59–72. [Google Scholar] [CrossRef]
- Craig, S.E.; Lohrenz, S.E.; Lee, Z.; Mahoney, K.L.; Kirkpatrick, G.J.; Schofield, O.M.; Steward, R.G. Use of hyperspectral remote sensing reflectance for detection and assessment of the harmful alga, Karenia brevis. Appl. Opt. 2006, 45, 5414–5425. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, R.P.; Tomlinson, M.C. Remote sensing of harmful algal blooms. In Remote Sensing of Coastal Aquatic Environments; Miller, R.L., Del Castillo, C.E., McKee, B.A., Eds.; Springer: Dordrecht, The Netherlands, 2005; pp. 277–296. [Google Scholar]
- Wynne, T.T.; Stumpf, R.P.; Tomlinson, M.C.; Ransibrahmanakul, V.; Villareal, T.A. Detecting Karenia brevis blooms and algal resuspension in the western Gulf of Mexico with satellite ocean color imagery. Harmful Algae 2005, 4, 992–1003. [Google Scholar] [CrossRef]
- Tomlinson, M.C.; Stumpf, R.P.; Ransibrahmanakul, V.; Truby, E.W.; Kirkpatrick, G.J.; Pederson, B.A.; Vargo, G.A.; Heil, C.A. Evaluation of the use of SeaWiFS imagery for detecting Karenia brevis harmful algal blooms in the eastern Gulf of Mexico. Remote Sens. Environ. 2004, 91, 293–303. [Google Scholar] [CrossRef]
- Stumpf, R.P.; Culver, M.E.; Tester, P.A.; Tomlinson, M.; Kirkpatrick, G.J.; Pederson, B.A.; Truby, E.; Ransibrahmanakul, V.; Soracco, M. Monitoring Karenia brevis blooms in the Gulf of Mexico using satellite ocean color imagery and other data. Harmful Algae 2003, 2, 147–160. [Google Scholar] [CrossRef]
- Mahoney, K.L. backscattering of Light by Karenia Brevis and Implications for Optical Detection and Monitoring. Available online: http://aquila.usm.edu/theses_dissertations/3545/ (accessed on 9 March 2015).
- Stumpf, R.P. Applications of satellite ocean color sensors for monitoring and predicting harmful algal blooms. Hum. Ecol. Risk Assess. Int. J. 2001, 7, 1363–1368. [Google Scholar] [CrossRef]
- Carder, K.; Steward, R.G. A remote-sensing reflectance model of a red-tide dinoflagellate off west Florida. Limnol. Oceanogr. 1985, 30, 286–298. [Google Scholar] [CrossRef]
- NASA’s OceanColor Web by the Ocean Biology Processing Group (OBPG) at NASA’s Goddard Space Flight Center. Available online: http://oceancolor.gsfc.nasa.gov/cms/atbd/chlor_a/ (accessed on 9 March 2015).
- Behrenfeld, M.J.; Westberry, T.K.; Boss, E.S.; O'Malley, R.T.; Siegel, D.A.; Wiggert, J.D.; Franz, B.A.; McClain, C.R.; Feldman, G.C.; Doney, S.C.; et al. Satellite-detected fluorescence reveals global physiology of ocean phytoplankton. Biogeosciences 2009, 6, 779–795. [Google Scholar] [CrossRef]
- Letelier, R.M.; Abbott, M.R. An analysis of chlorophyll fluorescence algorithms for the Moderate Resolution Imaging Spectrometer (MODIS). Remote Sens. Environ. 1996, 58, 215–223. [Google Scholar] [CrossRef]
- Gilerson, A.; Zhou, J.; Hlaing, S.; Ioannou, I.; Schalles, J.; Gross, B.; Moshary, F.; Ahmed, S. Fluorescence component in the reflectance spectra from coastal waters. Dependence on water composition. Opt. Express 2007, 15, 15702–15721. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Fell, F.; Liu, Z.S.; Preusker, R.; Fischer, J.; He, M.X. Evaluating the performance of artificial neural network techniques for pigment retrieval from ocean color in Case I waters. J. Geophys. Res. Oceans 2003, 108. [Google Scholar] [CrossRef]
- Gross, L.; Thiria, S.; Frouin, R.; Mitchell, B.G. Artificial neural networks for modeling the transfer function between marine reflectance and phytoplankton pigment concentration. J. Geophys. Res. 2000, 105, 3483–3495. [Google Scholar] [CrossRef]
- Matthews, M.W.; Bernard, S.; Winter, K. Remote sensing of cyanobacteria-dominant algal blooms and water quality parameters in Zeekoevlei, a small hypertrophic lake, using MERIS. Remote Sens. Environ. 2010, 114, 2070–2087. [Google Scholar] [CrossRef]
- Schiller, H.; Doerffer, R. Neural network for emulation of an inverse model operational derivation of Case II water properties from MERIS data. Int. J. Remote Sens. 1999, 20, 1735–1746. [Google Scholar] [CrossRef]
- Doerffer, R.; Schiller, H.; Krasemann, H. MERIS Case 2 Water Validation Early Results North Sea/Helgoland/German Bight; ESA Special Publication: Paris, France, 2003; p. 99. [Google Scholar]
- Schiller, H.; Doerffer, R. Improved determination of coastal water constituent concentrations from MERIS data. IEEE Trans. Geosci. Remote Sens. 2005, 43, 1585–1591. [Google Scholar] [CrossRef]
- Park, Y.J.; Ruddick, K. Detecting Algae Blooms in EUROPEAN Waters. In ENVISAT Symposium; European Space Agency Special Publication SP-636: Paris, France, 2007; pp. 23–27. [Google Scholar]
- Bishop, C.M. Neural Networks for Pattern Recognition; Oxford University Press: Oxford, UK, 1995. [Google Scholar]
- Levenberg, K. A method for the solution of certain non-linear problems in least squares. Q. Appl. Math. 1944, 2, 164–168. [Google Scholar]
- Foresee, F.D.; Hagan, M.T. Gauss-Newton approximation to Bayesian regularization. IEEE International Joint Conference on Neural Networks: Houston, TX, USA, 1997. [Google Scholar]
- Mackay, D.J.C. Bayesian interpolation. Neural Comput. 1992, 4, 415–447. [Google Scholar] [CrossRef]
- Marquardt, D.W. An algorithm for the least-squares estimation of nonlinear parameters. SIAM J. Appl. Math. 1963, 11, 431–441. [Google Scholar] [CrossRef]
- Lawrence, J. Data preparation for a neural network. AI Expert 1991, 6, 34–41. [Google Scholar]
- Zaneveld, J.R.V.; Moore, C.; Barnard, A.H.; Walsh, I. Correction and analysis of spectral absorption data taken with the WET Labs ac-s. In Proceedings of the Ocean Optics XVII, Perth, Australia, 25–29 October 2004; pp. 25–29.

















| y-axis | x-axis | R2 | Slope & Intercept | ɛ | N | |
|---|---|---|---|---|---|---|
| OCI/OC3 [Chla] (µg·L−1) | VIIRS | NN-[Chla] (µg·L−1) | 0.90 | 0.960.22 | 0.33 | 5755 |
| RGCI [Chla] (µg·L−1) | 0.72 | 1.500.22 | 0.95 | 5755 | ||
| nFLH [Chla]-KB (W·m−2·µm−1·sr−1) | MODIS | 0.71 | 0.800.36 | 0.43 | 2274 | |
| OCI/OC3 [Chla] (µg·L−1) | 0.50 | 0.480.70 | 0.57 | 2274 | ||
| RGCI [Chla] (µg·L−1) | 0.50 | 0.550.98 | 0.57 | 2274 | ||
| NN [Chla] (µg·L−1) | In situ |  | 0.32 | 0.26 | 94 | |
| OCI/OC3 [Chla] (µg·L−1) | 0.28 | 0.24 | 94 | |||
| RGCI [Chla] (µg·L−1) | 0.17 | 0.45 | 94 | |||
| NN [Chla] (µg·L−1) |  | 0.69 | 0.18 | 21 | ||
| OCI/OC3 [Chla] (µg·L−1) | 0.33 | 0.18 | 21 | |||
| RGCI [Chla] (µg·L−1) | 0.62 | 0.37 | 21 | |||
| NN [Chla] (µg·L−1) |  | 0.82 | 0.16 | 12 | ||
| OCI/OC3 [Chla] (µg·L−1) | 0.44 | 0.17 | 12 | |||
| RGCI [Chla] (µg·L−1) | 0.70 | 0.38 | 12 |
| Rrs551 | NN aph443 | Equivalent NN [Chla] Value |
|---|---|---|
| ≤0.006 sr−1 | ≥0.061 m−1 | ≥1.27 µg L−1 |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
El-habashi, A.; Ioannou, I.; Tomlinson, M.C.; Stumpf, R.P.; Ahmed, S. Satellite Retrievals of Karenia brevis Harmful Algal Blooms in the West Florida Shelf Using Neural Networks and Comparisons with Other Techniques. Remote Sens. 2016, 8, 377. https://doi.org/10.3390/rs8050377
El-habashi A, Ioannou I, Tomlinson MC, Stumpf RP, Ahmed S. Satellite Retrievals of Karenia brevis Harmful Algal Blooms in the West Florida Shelf Using Neural Networks and Comparisons with Other Techniques. Remote Sensing. 2016; 8(5):377. https://doi.org/10.3390/rs8050377
Chicago/Turabian StyleEl-habashi, Ahmed, Ioannis Ioannou, Michelle C. Tomlinson, Richard P. Stumpf, and Sam Ahmed. 2016. "Satellite Retrievals of Karenia brevis Harmful Algal Blooms in the West Florida Shelf Using Neural Networks and Comparisons with Other Techniques" Remote Sensing 8, no. 5: 377. https://doi.org/10.3390/rs8050377
APA StyleEl-habashi, A., Ioannou, I., Tomlinson, M. C., Stumpf, R. P., & Ahmed, S. (2016). Satellite Retrievals of Karenia brevis Harmful Algal Blooms in the West Florida Shelf Using Neural Networks and Comparisons with Other Techniques. Remote Sensing, 8(5), 377. https://doi.org/10.3390/rs8050377