Chlorogenic Acid Targeting of the AKT PH Domain Activates AKT/GSK3?/FOXO1 Signaling and Improves Glucose Metabolism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
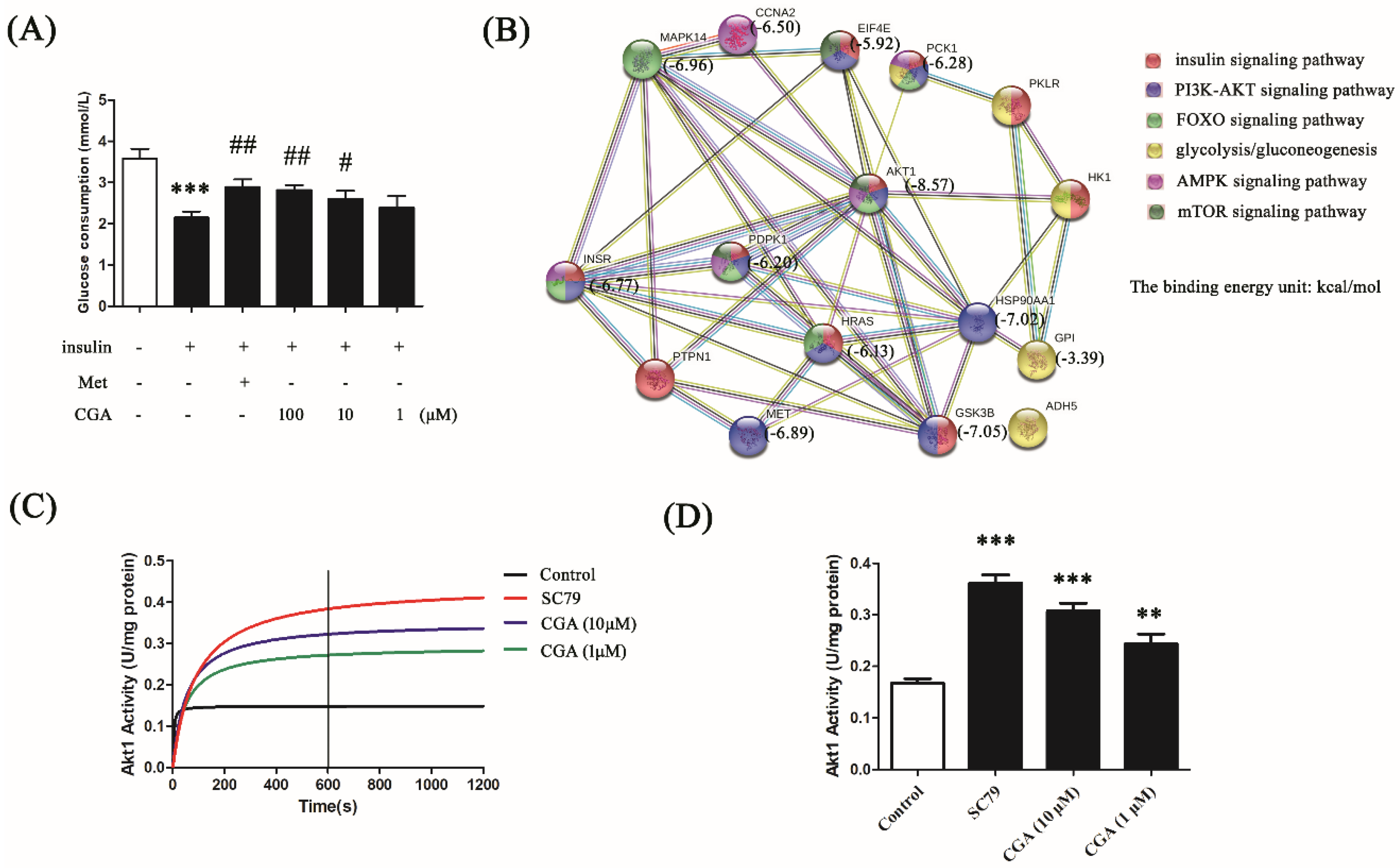
2.3. Glucose Consumption
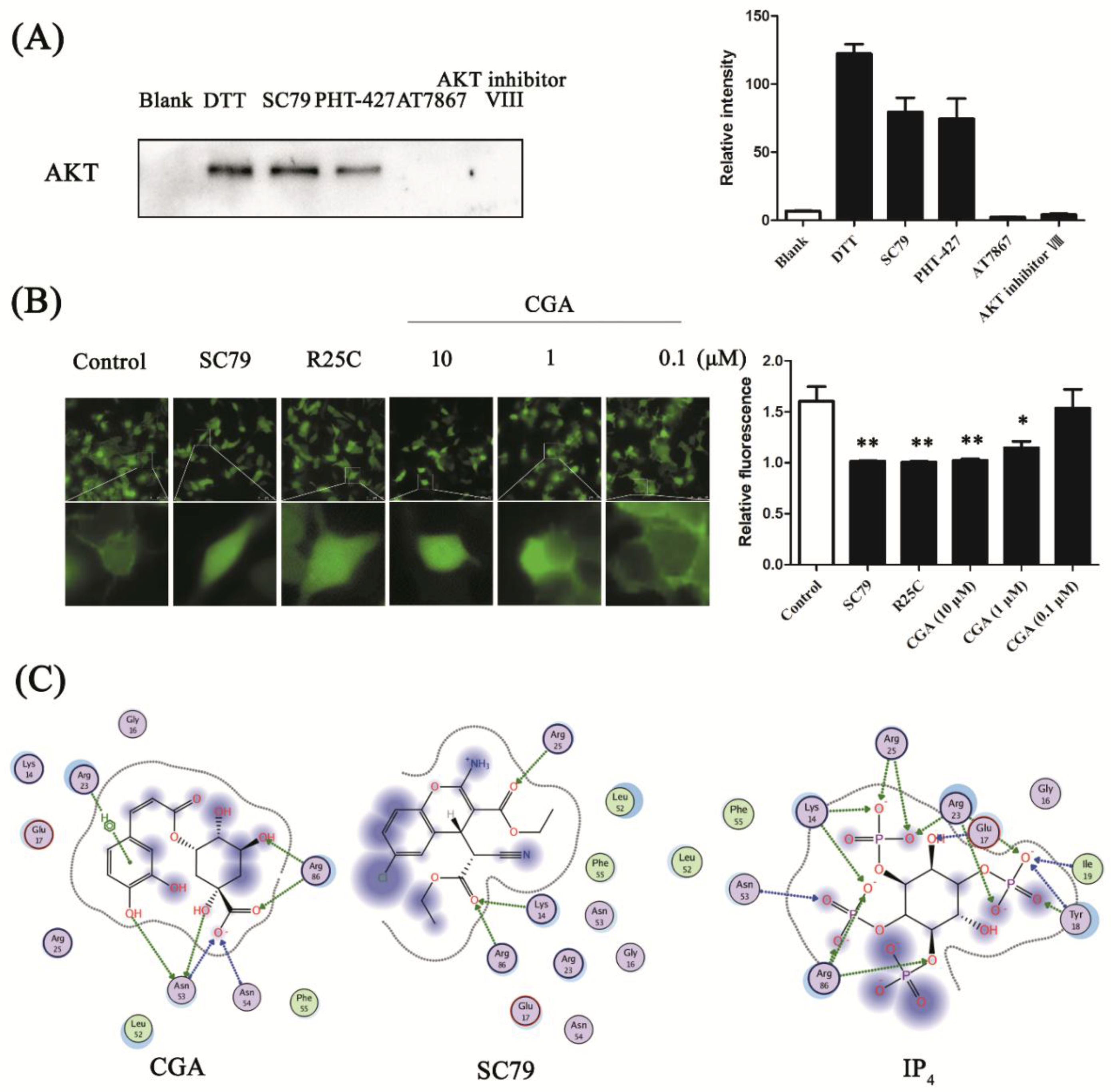
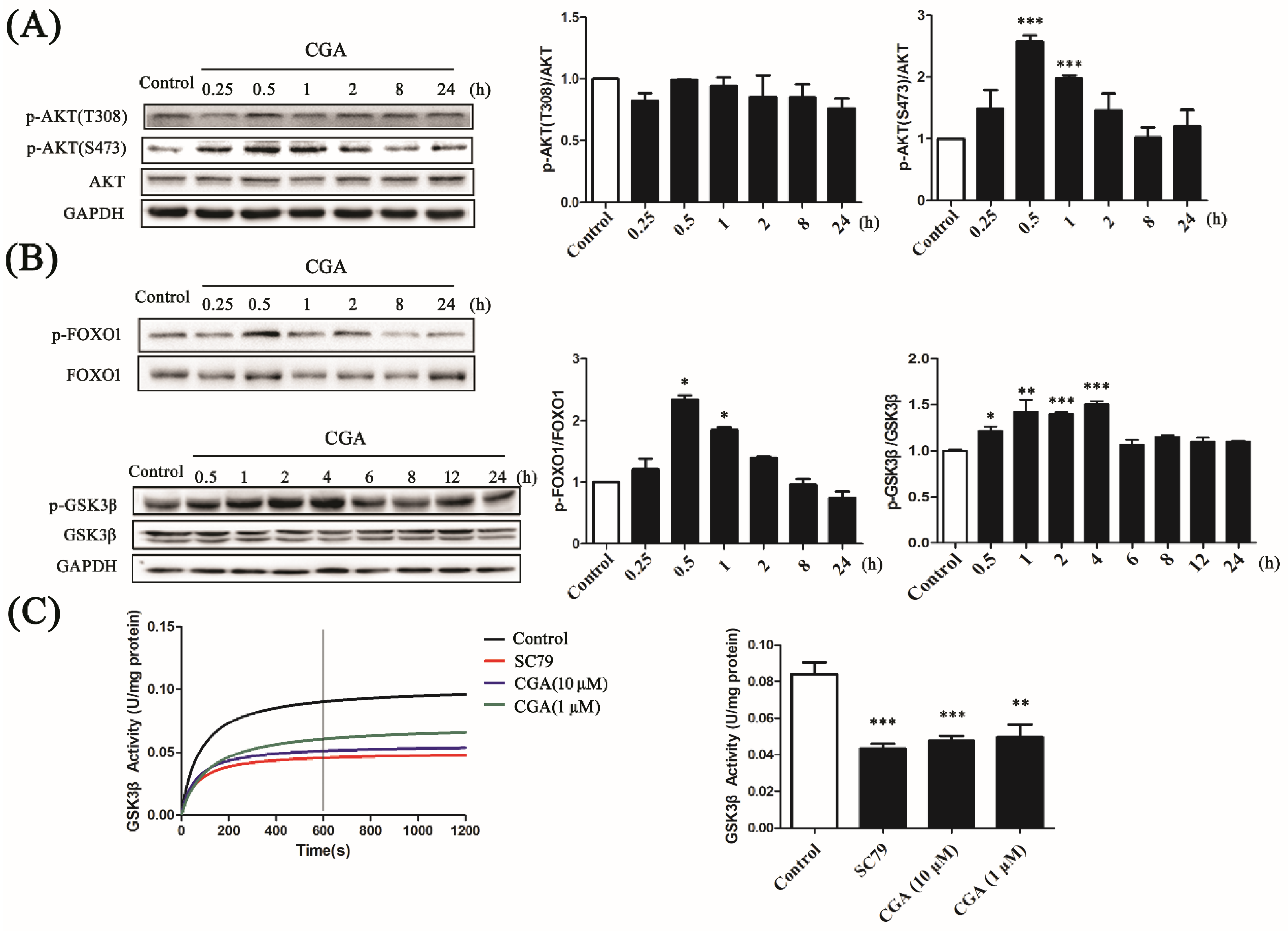
2.4. Enzymatic Activity Determination
2.5. Target Prediction and Molecular Docking
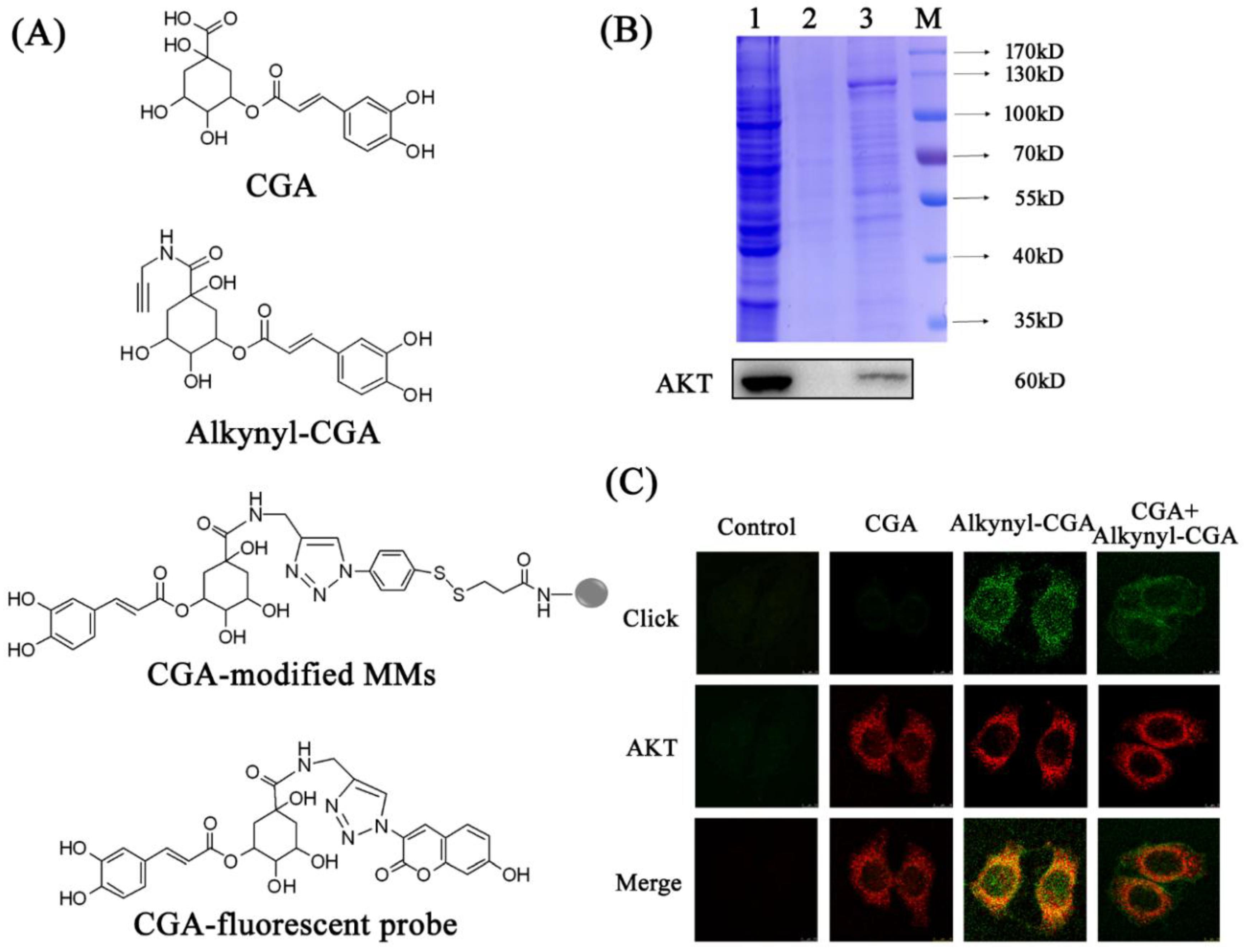
2.6. Target Fishing
2.7. Co-Localization of CGA with Target Protein
2.8. Competitive Test against AKT Using CGA-Modified MMs
2.9. Membrane Translocation of the PH-Domain
2.10. SDS-PAGE and Western Blot Analysis
2.11. Statistical Analysis
3. Results
3.1. CGA Regulates Glucose Metabolism by Increasing AKT Activity
3.2. Identification of AKT as a Target of CGA
3.3. CGA Binds to the PH Domain of AKT
3.4. CGA Phosphorylates AKT and Regulates Its Downstream Factors
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Nardini, M.; Cirillo, E.; Natella, F.; Scaccini, C. Absorption of phenolic acids in humans after coffee consumption. J. Agric. Food Chem. 2002, 50, 5735–5741. [Google Scholar] [CrossRef] [PubMed]
- Loader, T.B.; Taylor, C.G.; Zahradka, P.; Jones, P.J. Chlorogenic acid from coffee beans: Evaluating the evidence for a blood pressure-regulating health claim. Nutr. Rev. 2017, 75, 114–133. [Google Scholar] [CrossRef] [PubMed]
- Lee, A.H.; Tan, L.; Hiramatsu, N.; Ishisaka, A.; Alfonso, H.; Tanaka, A.; Uemura, N.; Fujiwara, Y.; Takechi, R. Plasma concentrations of coffee polyphenols and plasma biomarkers of diabetes risk in healthy Japanese women. Nutr. Diabetes 2016, 6, e212. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Liang, M.; Li, W.; Li, K.; Li, P.; Hu, Y.; Yang, Z. Protective effects of Eucommia ulmoides Oliv. bark and leaf on amyloid beta-induced cytotoxicity. Environ. Toxicol. Pharmacol. 2009, 28, 342–349. [Google Scholar] [CrossRef] [PubMed]
- Gokcen, B.B.; Sanlier, N. Coffee consumption and disease correlations. Crit. Rev. Food Sci. Nutr. 2017, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Hayat, K.; Iqbal, H.; Malik, U.; Bilal, U.; Mushtaq, S. Tea and its consumption: Benefits and risks. Crit. Rev. Food Sci. Nutr. 2015, 55, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Ong, K.W.; Hsu, A.; Tan, B.K. Chlorogenic acid stimulates glucose transport in skeletal muscle via AMPK activation: A contributor to the beneficial effects of coffee on diabetes. PLoS ONE 2012, 7, e32718. [Google Scholar] [CrossRef] [PubMed]
- Tajik, N.; Tajik, M.; Mack, I.; Enck, P. The potential effects of chlorogenic acid, the main phenolic components in coffee, on health: A comprehensive review of the literature. Eur. J. Nutr. 2017, 56, 2215–2244. [Google Scholar] [CrossRef] [PubMed]
- Farah, A.; Monteiro, M.; Donangelo, C.M.; Lafay, S. Chlorogenic acids from green coffee extract are highly bioavailable in humans. J. Nutr. 2008, 138, 2309–2315. [Google Scholar] [CrossRef] [PubMed]
- Monente, C.; Ludwig, I.A.; Stalmach, A.; de Pena, M.P.; Cid, C.; Crozier, A. In vitro studies on the stability in the proximal gastrointestinal tract and bioaccessibility in Caco-2 cells of chlorogenic acids from spent coffee grounds. Int. J. Food Sci. Nutr. 2015, 66, 657–664. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuniga, L.Y.; Aceves-de la Mora, M.C.A.; Gonzalez-Ortiz, M.; Ramos-Nunez, J.L.; Martinez-Abundis, E. Effect of Chlorogenic Acid Administration on Glycemic Control, Insulin Secretion, and Insulin Sensitivity in Patients with Impaired Glucose Tolerance. J. Med. Food 2018, 21, 469–473. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.J.; Zhu, Q.; Zhong, Y.L.; Xu, S.H.; Wang, Z. Chlorogenic Acid Maintains Glucose Homeostasis through Modulating the Expression of SGLT-1, GLUT-2, and PLG in Different Intestinal Segments of Sprague-Dawley Rats Fed a High-Fat Diet. Biomed. Environ. Sci. 2015, 28, 894–903. [Google Scholar] [CrossRef] [PubMed]
- Sanchez, M.B.; Miranda-Perez, E.; Verjan, J.C.G.; de Los Angeles Fortis Barrera, M.; Perez-Ramos, J.; Alarcon-Aguilar, F.J. Potential of the chlorogenic acid as multitarget agent: Insulin-secretagogue and PPAR alpha/gamma dual agonist. Biomed. Pharmacother. 2017, 94, 169–175. [Google Scholar] [CrossRef] [PubMed]
- Song, J.J.; Wang, Q.; Du, M.; Li, T.G.; Chen, B.; Mao, X.Y. Casein glycomacropeptide-derived peptide IPPKKNQDKTE ameliorates high glucose-induced insulin resistance in HepG2 cells via activation of AMPK signaling. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef]
- Wang, K.; Cao, P.; Wang, H.; Tang, Z.; Wang, N.; Wang, J.; Zhang, Y. Chronic administration of Angelica sinensis polysaccharide effectively improves fatty liver and glucose homeostasis in high-fat diet-fed mice. Sci. Rep. 2016, 6, 26229. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saltiel, A.R.; Kahn, C.R. Insulin signalling and the regulation of glucose and lipid metabolism. Nature 2001, 414, 799–806. [Google Scholar] [CrossRef] [PubMed]
- Kamat, P.K.; Kalani, A.; Rai, S.; Tota, S.K.; Kumar, A.; Ahmad, A.S. Streptozotocin Intracerebroventricular-Induced Neurotoxicity and Brain Insulin Resistance: A Therapeutic Intervention for Treatment of Sporadic Alzheimer’s Disease (sAD)-Like Pathology. Mol. Neurobiol. 2016, 53, 4548–4562. [Google Scholar] [CrossRef] [PubMed]
- Montori-Grau, M.; Pedreira-Casahuga, R.; Boyer-Diaz, Z.; Lassot, I.; García-Martínez, C.; Orozco, A.; Cebrià, J.; Osorio-Conles, O.; Chacón, M.R.; Vendrell, J.; et al. GNIP1 E3 ubiquitin ligase is a novel player in regulating glycogen metabolism in skeletal muscle. Metabolism 2018, 83, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Hirota, K.; Sakamaki, J.-I.; Ishida, J.; Shimamoto, Y.; Nishihara, S.; Kodama, N.; Ohta, K.; Yamamoto, M.; Tanimoto, k.; Fukamizu, A. A Combination of HNF-4 and Foxo1 Is Required for Reciprocal Transcriptional Regulation of Glucokinase and Glucose-6-phosphatase Genes in Response to Fasting and Feeding. J. Biol. Chem. 2008, 283, 32432–32441. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yarushkin, A.A.; Kachaylo, E.M.; Pustylnyak, V.O. The constitutive androstane receptor activator 4-[(4R,6R)-4,6-diphenyl-1,3-dioxan-2-yl]-N,N-dimethylaniline inhibits the gluconeogenic genes PEPCK and G6Pase through the suppression of HNF4alpha and FOXO1 transcriptional activity. Br. J. Pharmacol. 2013, 168, 1923–1932. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.N.; Iyer, S.; Ring, R.; Almeida, M. The Role of FoxOs in Bone Health and Disease. Curr. Top. Dev. Biol. 2018, 127, 149–163. [Google Scholar] [CrossRef] [PubMed]
- Chu, Y.; Gomez Rosso, L.; Huang, P.; Wang, Z.; Xu, Y.; Yao, X.; Bao, M.; Yan, J.; Song, H.; Wang, G. Liver Med23 ablation improves glucose and lipid metabolism through modulating FOXO1 activity. Cell Res. 2014, 24, 1250–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Muller, Y.L.; Hanson, R.L.; Wiessner, G.; Nieboer, L.; Kobes, S.; Piaggi, P.; Abdussamad, M.; Okani, C.; Knowler, W.C.; Bogardus, C.; et al. Assessing FOXO1A as a potential susceptibility locus for type 2 diabetes and obesity in American Indians. Obesity 2015, 23, 1960–1965. [Google Scholar] [CrossRef] [PubMed]
- Yin, J.; Hu, R.M.; Chen, M.D.; Tang, J.; Li, F.; Yang, Y.; Chen, J. Effects of berberine on glucose metabolism in vitro. Metabolism 2002, 51, 1439–1443. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhang, Y.; Wang, Z.; Shen, Y.; Zhang, M.; Nie, Q.; Hou, Y.; Bai, G. Ursolic Acid, a Natural Nutraceutical Agent, Targets Caspase3 and Alleviates Inflammation-Associated Downstream Signal Transduction. Mol. Nutr. Food Res. 2017, 61. [Google Scholar] [CrossRef] [PubMed]
- Jo, H.; Mondal, S.; Tan, D.; Nagata, E.; Takizawa, S.; Sharma, A.K.; Hou, Q.; Shanmugasundaram, K.; Prasad, A.; Tung, J.K.; et al. Small molecule-induced cytosolic activation of protein kinase Akt rescues ischemia-elicited neuronal death. Proc. Natl. Acad. Sci. USA 2012, 109, 10581–10586. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meuillet, E.J.; Zuohe, S.; Lemos, R.; Ihle, N.; Kingston, J.; Watkins, R.; Moses, S.A.; Zhang, S.; Du-Cuny, L.; Herbst, R.; et al. Molecular pharmacology and antitumor activity of PHT-427, a novel Akt/phosphatidylinositide-dependent protein kinase 1 pleckstrin homology domain inhibitor. Mol. Cancer Ther. 2010, 9, 706–717. [Google Scholar] [CrossRef] [PubMed]
- Grimshaw, K.M.; Hunter, L.J.; Yap, T.A.; Heaton, S.P.; Walton, M.I.; Woodhead, S.J.; Fazal, L.; Reule, M.; Davies, T.G.; Seavers, L.C.; et al. AT7867 is a potent and oral inhibitor of AKT and p70 S6 kinase that induces pharmacodynamic changes and inhibits human tumor xenograft growth. Mol. Cancer Ther. 2010, 9, 1100–1110. [Google Scholar] [CrossRef] [PubMed]
- Lindsley, C.W.; Zhao, Z.; Leister, W.H.; Robinson, R.G.; Barnett, S.F.; Defeo-Jones, D.; Jones, R.E.; Hartman, G.D.; Huff, J.R.; Huber, H.E.; et al. Allosteric Akt (PKB) inhibitors: Discovery and SAR of isozyme selective inhibitors. Bioorg. Med. Chem. Lett. 2005, 15, 761–764. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Rajala, A.; Wiechmann, A.F.; Anderson, R.E.; Rajala, R.V. Activation and membrane binding of retinal protein kinase Balpha/Akt1 is regulated through light-dependent generation of phosphoinositides. J. Neurochem. 2008, 107, 1382–1397. [Google Scholar] [CrossRef] [PubMed]
- Thomas, C.C.; Deak, M.; Alessi, D.R.; van Aalten, D.M. High-resolution structure of the pleckstrin homology domain of protein kinase b/akt bound to phosphatidylinositol (3,4,5)-trisphosphate. Curr. Biol. 2002, 12, 1256–1262. [Google Scholar] [CrossRef]
- Han, D.; Chen, W.; Gu, X.; Shan, R.; Zou, J.; Liu, G.; Shahid, M.; Gao, J.; Han, B. Cytoprotective effect of chlorogenic acid against hydrogen peroxide-induced oxidative stress in MC3T3-E1 cells through PI3K/Akt-mediated Nrf2/HO-1 signaling pathway. Oncotarget 2017, 8, 14680–14692. [Google Scholar] [CrossRef] [PubMed]
- Pearce, L.R.; Komander, D.; Alessi, D.R. The nuts and bolts of AGC protein kinases. Nat. Rev. Mol. Cell Biol. 2010, 11, 9–22. [Google Scholar] [CrossRef] [PubMed]
- He, W.; Zhao, A.; Zou, J.; Luo, X.; Lin, X.; Wang, L.; Lin, C. Synthesis, In Vitro Coagulation Activities and Molecular Docking Studies on Three L-Histidine Amide Derivatives. Chem. Res. Chin. Univ. 2018, 34, 90–94. [Google Scholar] [CrossRef]
- Najafov, A.; Shpiro, N.; Alessi, D.R. Akt is efficiently activated by PIF-pocket- and PtdIns(3,4,5)P3-dependent mechanisms leading to resistance to PDK1 inhibitors. Biochem. J. 2012, 448, 285–295. [Google Scholar] [CrossRef] [PubMed]
- Case, N.; Thomas, J.; Sen, B.; Styner, M.; Xie, Z.; Galior, K.; Rubin, J. Mechanical regulation of glycogen synthase kinase 3beta (GSK3beta) in mesenchymal stem cells is dependent on Akt protein serine 473 phosphorylation via mTORC2 protein. J. Biol. Chem. 2011, 286, 39450–39456. [Google Scholar] [CrossRef] [PubMed]
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gao, J.; He, X.; Ma, Y.; Zhao, X.; Hou, X.; Hao, E.; Deng, J.; Bai, G. Chlorogenic Acid Targeting of the AKT PH Domain Activates AKT/GSK3?/FOXO1 Signaling and Improves Glucose Metabolism. Nutrients 2018, 10, 1366. https://doi.org/10.3390/nu10101366
Gao J, He X, Ma Y, Zhao X, Hou X, Hao E, Deng J, Bai G. Chlorogenic Acid Targeting of the AKT PH Domain Activates AKT/GSK3?/FOXO1 Signaling and Improves Glucose Metabolism. Nutrients. 2018; 10(10):1366. https://doi.org/10.3390/nu10101366
Chicago/Turabian StyleGao, Jie, Xin He, Yuejiao Ma, Xuezhi Zhao, Xiaotao Hou, Erwei Hao, Jiagang Deng, and Gang Bai. 2018. "Chlorogenic Acid Targeting of the AKT PH Domain Activates AKT/GSK3?/FOXO1 Signaling and Improves Glucose Metabolism" Nutrients 10, no. 10: 1366. https://doi.org/10.3390/nu10101366
APA StyleGao, J., He, X., Ma, Y., Zhao, X., Hou, X., Hao, E., Deng, J., & Bai, G. (2018). Chlorogenic Acid Targeting of the AKT PH Domain Activates AKT/GSK3?/FOXO1 Signaling and Improves Glucose Metabolism. Nutrients, 10(10), 1366. https://doi.org/10.3390/nu10101366





