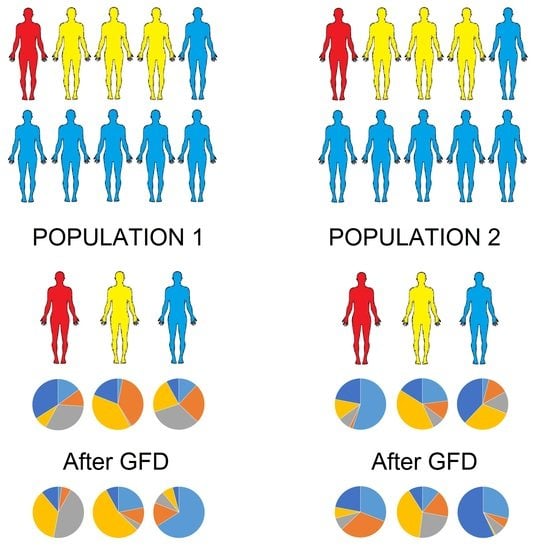
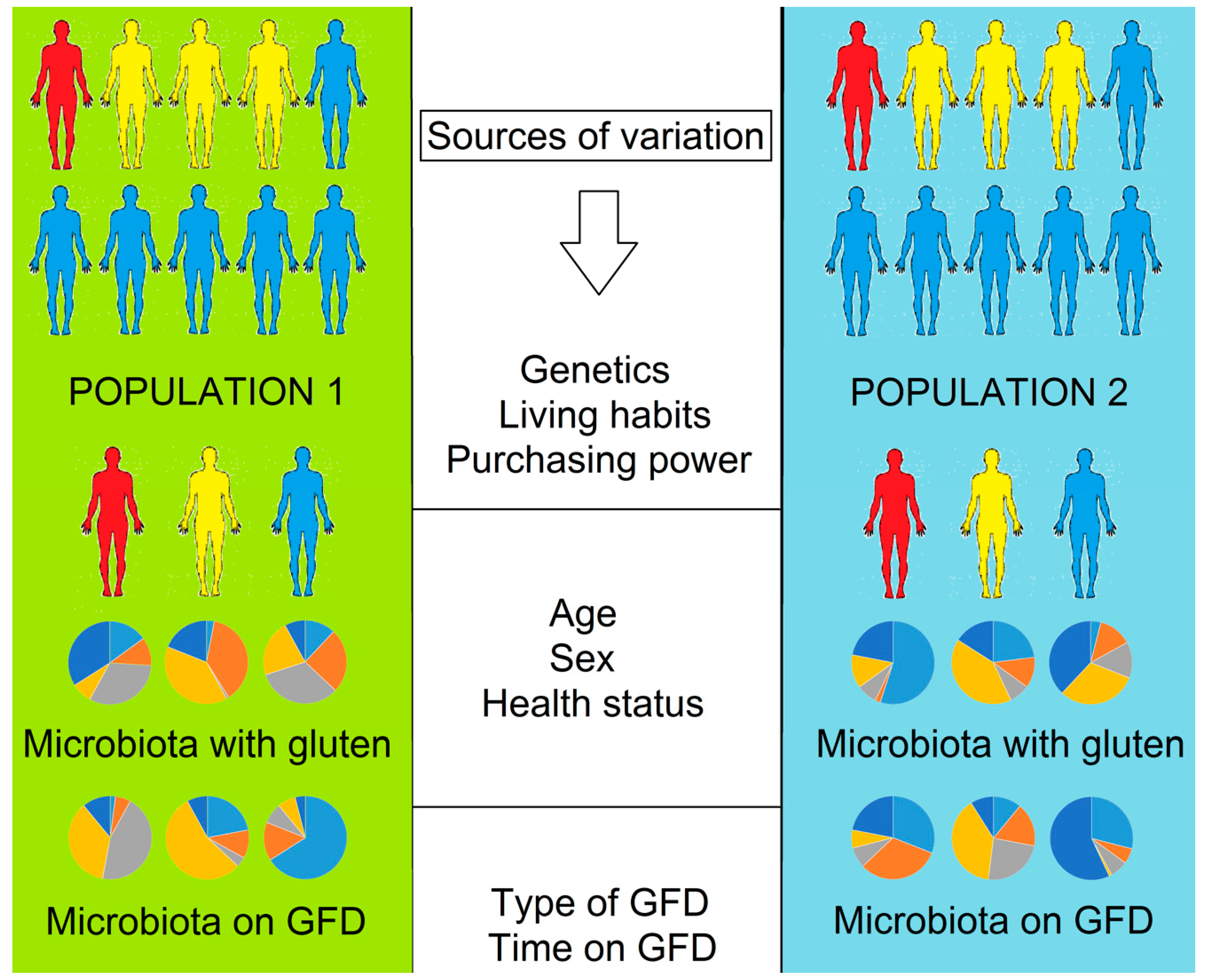
The Effect of Gluten-Free Diet on Health and the Gut Microbiota Cannot Be Extrapolated from One Population to Others
Abstract
:1. Introduction
2. Gluten-Related Disorders and Celiac Disease
3. GRD and the Gut Microbiota
4. Effect of GFD on the Gut Microbiome
5. The Issue of Extrapolation
5.1. Individuality and Over Time Variability of the Gut Microbiota
5.2. Dietary Differences in Gluten-Containing and GFD
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Douglas, A.E.; Werren, J.H. Holes in the hologenome: Why host-microbe symbioses are not holobionts. mBio 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Maslowski, K.M.; Mackay, C.R. Diet, gut microbiota and immune responses. Nat. Immunol. 2011, 12, 5–9. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The effect of diet on the human gut microbiome: A metagenomic analysis in humanized gnotobiotic mice. Sci. Transl. Med. 2009, 1. [Google Scholar] [CrossRef] [PubMed]
- Faith, J.J.; McNulty, N.P.; Rey, F.E.; Gordon, J.I. Predicting a human gut microbiota’s response to diet in gnotobiotic mice. Science 2011, 333, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Conlon, M.A.; Bird, A.R. The impact of diet and lifestyle on gut microbiota and human health. Nutrients 2015, 7, 17–44. [Google Scholar] [CrossRef] [PubMed]
- Le Doare, K.; Holder, B.; Bassett, A.; Pannaraj, P.S. Mother’s milk: A purposeful contribution to the development of the infant microbiota and immunity. Front. Immunol. 2018, 9, 361. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Clemente, J.C.; Ursell, L.K.; Parfrey, L.W.; Knight, R. The impact of the gut microbiota on human health: An integrative view. Cell. 2012, 148, 1258–1270. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Bäckhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell. Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef] [PubMed]
- Forrest, E.A.; Wong, M.; Nama, S.; Sharma, S. Celiac crisis, a rare and profound presentation of celiac disease: A case report. BMC Gastroenterol. 2018, 18, 59. [Google Scholar] [CrossRef] [PubMed]
- De Re, V.; Magris, R.; Cannizzaro, R. New insights into the pathogenesis of celiac disease. Front. Med. 2017, 4, 137. [Google Scholar] [CrossRef] [PubMed]
- Cukrowska, B.; Sowińska, A.; Bierła, J.B.; Czarnowska, E.; Rybak, A.; Grzybowska-Chlebowczyk, U. Intestinal epithelium, intraepithelial lymphocytes and the gut microbiota—Key players in the pathogenesis of celiac disease. World J. Gastroenterol. 2017, 23, 7505–7518. [Google Scholar] [CrossRef] [PubMed]
- Rampertab, S.D.; Pooran, N.; Brar, P.; Singh, P.; Green, P.H. Trends in the presentation of celiac disease. Am. J. Med. 2006, 119, 355.e9–355.e14. [Google Scholar] [CrossRef] [PubMed]
- Gujral, N.; Freeman, H.J.; Thomson, A.B.R. Celiac disease: Prevalence, diagnosis, pathogenesis and treatment. World J. Gastroenterol. 2012, 18, 6036–6059. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.B.; Uhde, M.; Green, P.H.; Alaedini, A. Autoantibodies in the extraintestinal manifestations of celiac disease. Nutrients 2018, 10, 1123. [Google Scholar] [CrossRef] [PubMed]
- Ramírez-Cervantes, K.L.; Remes-Troche, J.M.; Milke-García, M.P.; Romero, V.; Uscanga, L.F. Characteristics and factors related to quality of life in Mexican Mestizo patients with celiac disease. BMC Gastroenterol. 2015, 15, 4. [Google Scholar] [CrossRef] [PubMed]
- Verdu, E.F.; Armstrong, D.; Murray, J.A. Between celiac disease and irritable bowel syndrome: The “no man’s land” of gluten sensitivity. Am. J. Gastroenterol. 2009, 104, 1587–1594. [Google Scholar] [CrossRef] [PubMed]
- Lammers, K.M.; Herrera, M.G.; Dodero, V.I. Translational chemistry meets gluten-related disorders. Chem. Open 2018, 7, 217–232. [Google Scholar] [CrossRef] [PubMed]
- Reunala, T.; Salmi, T.T.; Hervonen, K.; Kaukinen, K.; Collin, P. Dermatitis herpetiformis: A common extraintestinal manifestation of coeliac disease. Nutrients 2018, 10, 602. [Google Scholar] [CrossRef] [PubMed]
- Rodrigo, L.; Beteta-Gorriti, V.; Alvarez, N.; Gómez de Castro, C.; de Dios, A.; Palacios, L.; Santos-Juanes, J. Cutaneous and mucosal manifestations associated with celiac disease. Nutrients 2018, 10, 800. [Google Scholar] [CrossRef] [PubMed]
- Ciccocioppo, R.; Panelli, S.; Conti Bellocchi, M.C.; Cangemi, G.C.; Frulloni, L.; Capelli, E.; Corazza, G.R. The transcriptomic analysis of circulating immune cells in a celiac family unveils further insights into disease pathogenesis. Front. Med. 2018, 5, 182. [Google Scholar] [CrossRef] [PubMed]
- Singh, P.; Arora, A.; Strand, T.A.; Leffler, D.A.; Catassi, C.; Green, P.H.; Kelly, C.P.; Ahuja, V.; Makharia, G.K. Global prevalence of celiac disease: Systematic review and meta-analysis. Clin. Gastroenterol. Hepatol. 2018, 16, 823–836. [Google Scholar] [CrossRef] [PubMed]
- Teresi, S.; Crapisi, M.; Vallejo, M.D.; Castellaneta, S.P.; Francavilla, R.; Iacono, G.; Ravelli, A.; Menegazzi, O.; Louali, M.; Catassi, C. Celiac disease seropositivity in Saharawi children: A follow-up and family study. J. Pediatr. Gastroenterol. Nutr. 2010, 50, 506–509. [Google Scholar] [CrossRef] [PubMed]
- Haridy, J.; Lewis, D.; Newnham, E.D. Investigational drug therapies for coeliac disease—Where to from here? Expert Opin. Investig. Drugs 2018, 27, 225–233. [Google Scholar] [CrossRef] [PubMed]
- Ontiveros, N.; Rodriguez-Bellegarrigue, C.I.; Galicia-Rodriguez, G.; Vergara-Jimenez, M.J.; Zepeda-Gomez, E.M.; Aramburo-Galvez, J.G.; Gracia-Valenzuela, M.H.; Cabrera-Chavez, F. Prevalence of self-reported gluten-related disorders and adherence to a gluten-free diet in Salvadoran adult population. Int. J. Environ. Res. Public Health 2018, 15, 786. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Calabuig, M.; Sanz, Y. Differences between the fecal microbiota of coeliac infants and healthy controls. Curr. Issues Intest. Microbiol. 2007, 8, 9–14. [Google Scholar] [PubMed]
- Sanz, Y.; Sánchez, E.; Marzotto, M.; Calabuig, M.; Torriani, S.; Dellaglio, F. Differences in faecal bacterial communities in coeliac and healthy children as detected by PCR and denaturing gradient gel electrophoresis. FEMS Immunol. Med. Microbiol. 2007, 51, 562–568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Medina, M.; De Palma, G.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Bifidobacterium strains suppress in vitro the pro-inflammatory milieu triggered by the large intestinal microbiota of coeliac patients. J. Inflamm. 2008, 5, 19. [Google Scholar] [CrossRef] [PubMed]
- Sánchez, E.; Nadal, I.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Reduced diversity and increased virulence-gene carriage in intestinal enterobacteria of coeliac children. BMC Gastroenterol. 2008, 8, 50. [Google Scholar] [CrossRef] [PubMed]
- Ou, G.; Hedberg, M.; Hörstedt, P.; Baranov, V.; Forsberg, G.; Drobni, M.; Sandström, O.; Wai, S.N.; Johansson, I.; Hammarström, M.L.; et al. Proximal small intestinal microbiota and identification of rod-shaped bacteria associated with childhood celiac disease. Am. J. Gastroenterol. 2009, 104, 3058–3067. [Google Scholar] [CrossRef] [PubMed]
- Sanz, Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult humans. Gut Microbes 2010, 1, 135–137. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cinova, J.; De Palma, G.; Stepankova, R.; Kofronova, I.; Kverka, M.; Sanz, Y.; Tuckova, L. Role of intestinal bacteria in gliadin-induced changes in intestinal mucosa: Study in germ-free rats. PLoS ONE 2011, 6, e16169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sjöberg, V.; Sandström, O.; Hedberg, M.; Hammarström, S.; Hernell, O.; Hammarström, M.-L. Intestinal T-cell responses in celiac disease —Impact of celiac disease associated bacteria. PLoS ONE 2013, 8, e53414. [Google Scholar] [CrossRef] [PubMed]
- Wacklin, P.; Kaukinen, K.; Tuovinen, E.; Collin, P.; Lindfors, K.; Partanen, J.; Mäki, M.; Mättö, J. The duodenal microbiota composition of adult celiac disease patients is associated with the clinical manifestation of the disease. Inflamm. Bowel Dis. 2013, 19, 934–941. [Google Scholar] [PubMed]
- Olivares, M.; Benítez-Páez, A.; de Palma, G.; Capilla, A.; Nova, E.; Castillejo, G.; Varea, V.; Marcos, A.; Garrote, J.A.; Polanco, I.; et al. Increased prevalence of pathogenic bacteria in the gut microbiota of infants at risk of developing celiac disease: the PROFICEL study. Gut Microbes 2018. [Google Scholar] [CrossRef] [PubMed]
- Ashorn, S.; Raukola, H.; Välineva, T.; Ashorn, M.; Wei, B.; Braun, J.; Rantala, I.; Kaukinen, K.; Luukkaala, T.; Collin, P.; et al. Elevated serum anti-Saccharomyces cerevisiae, anti-I2 and anti-OmpW antibody levels in patients with suspicion of celiac disease. J. Clin. Immunol. 2008, 28, 486–494. [Google Scholar] [PubMed]
- Harnett, J.; Myers, S.P.; Rolfe, M. Significantly higher faecal counts of the yeasts candida and saccharomyces identified in people with coealic disease. Gut Pathog. 2017, 9, 26. [Google Scholar] [CrossRef] [PubMed]
- Sovran, B.; Planchais, J.; Jegou, S.; Straube, M.; Lamas, B.; Natividad, J.M.; Agus, A.; Dupraz, L.; Glodt, J.; Da Costa, G.; et al. Enterobacteriaceae are essential for the modulation of colitis severity by fungi. Microbiome 2018, 6, 152. [Google Scholar] [CrossRef] [PubMed]
- D’Arienzo, R.; Stefanile, R.; Maurano, F.; Mazzarella, G.; Ricca, E.; Troncone, R.; Auricchio, S.; Rossi, M. Immunomodulatory effects of Lactobacillus casei administration in a mouse model of gliadin-sensitive enteropathy. Scand. J. Immunol. 2011, 74, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Primec, M.; Klemenak, M.; Di Gioia, D.; Aloisio, I.; BozziCionci, N.; Quagliariello, A.; Gorenjak, M.; Mičetić-Turk, D.; Langerholc, T. Clinical intervention using Bifidobacterium strains in celiac disease children reveals novel microbial modulators of TNF-α and short-chain fatty acids. Clin. Nutr. 2018. [Google Scholar] [CrossRef] [PubMed]
- Smecuol, E.; Hwang, H.J.; Sugai, E.; Corso, L.; Cherñavsky, A.C.; Bellavite, F.P.; González, A.; Vodánovich, F.; Moreno, M.L.; Vázquez, H.; et al. Exploratory, randomized, double-blind, placebo-controlled study on the effects of Bifidobacteriuminfantis natren life start strain super strain in active celiac disease. J. Clin. Gastroenterol. 2013, 47, 139–147. [Google Scholar] [CrossRef] [PubMed]
- Pinto-Sánchez, M.I.; Smecuol, E.C.; Temprano, M.P.; Sugai, E.; González, A.; Moreno, M.L.; Huang, M.L.; Bercik, P.; Cabanne, A.; Vázquez, H.; et al. Bifidobacteriuminfantis NLS super strain reduces the expression of α-defensin-5, a marker of Innate Immunity, in the mucosa of active celiac disease patients. J. Clin. Gastroenterol. 2017, 51, 814–817. [Google Scholar] [PubMed]
- Martinello, F.; Roman, C.F.; Souza, P.A. Effects of probiotic intake on intestinal bifidobacteria of celiac patients. Arq. Gastroenterol. 2017, 54, 85–90. [Google Scholar] [CrossRef] [PubMed]
- Harnett, J.; Myers, S.P.; Rolfe, M. Probiotics and the microbiome in celiac disease: A randomised controlled trial. Evid. Based Complement. Alternat. Med. 2016, 2016, 9048574. [Google Scholar] [CrossRef] [PubMed]
- Lagier, J.C.; Dubourg, G.; Million, M.; Cadoret, F.; Bilen, M.; Fenollar, F.; Levasseur, A.; Rolain, J.M.; Fournier, P.E.; Raoult, D. Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 2018, 16, 540–550. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Mazcorro, J.F.; Rivera-Gutierrez, X.; Cobos-Quevedo, O.J.; Grube-Pagola, P.; Meixueiro-Daza, A.; Hernandez-Flores, K.; Cabrera-Jorge, F.J.; Vivanco-Cid, H.; Dowd, S.E.; Remes-Troche, J.M. Consumption of gluten-free diet for four weeks influences the gut microbiota in patients with celiac disease and non-celiac gluten sensitivity. Unpublished work. 2018.
- Bonder, M.J.; Tigchelaar, E.F.; Cai, X.; Trynka, G.; Cenit, M.C.; Hrdlickova, B.; Zhong, H.; Vatanen, T.; Gevers, D.; Wijmenga, C.; et al. The influence of a short-term gluten-free diet on the human gut microbiome. Genome Med. 2016, 8, 45. [Google Scholar] [CrossRef] [PubMed]
- Tjellström, B.; Högberg, L.; Stenhammar, L.; Fälth-Magnusson, K.; Magnusson, K.-E.; Norin, E.; Sundqvist, T.; Midtvedt, T. Faecal short-chain fatty acid pattern in childhood coeliac disease is normalised after more than one year’s gluten-free diet. Microb. Ecol. Health Dis. 2013, 24, 20905. [Google Scholar] [CrossRef] [PubMed]
- Nistal, E.; Caminero, A.; Vivas, S.; Ruiz de Morales, J.M.; Sáenz de Miera, L.E.; Rodríguez-Aparicio, L.B.; Casqueiro, J. Differences in faecal bacteria populations and faecal bacteria metabolism in healthy adults and celiac disease patients. Biochimie 2012, 94, 1724–1729. [Google Scholar] [CrossRef] [PubMed]
- Di Cagno, R.; De Angelis, M.; De Pasquale, I.; Ndagijimana, M.; Vernocchi, P.; Ricciuti, P.; Gagliaradi, F.; Laghi, L.; Crecchio, C.; Guerzoni, M.E.; et al. Duodenal and faecalmicrobiota of celiac children: Molecular, phenotype and metabolome characterization. BMC Microbiol. 2011, 11, 219. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Nadal, I.; Medina, M.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Intestinal dysbiosis and reduced immunoglobulin-coated bacteria associated with coeliac disease in children. BMC Microbiol. 2010, 10, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schippa, S.; Iebba, V.; Barbato, M.; Di Nardo, G.; Totino, V.; Checchi, M.P.; Longhi, C.; Maiella, G.; Cucchiara, S.; Conte, M.P. A distinctive ‘microbial signature’ in celiac pediatric patients. BMC Microbiol. 2010, 10, 175. [Google Scholar] [CrossRef] [PubMed]
- De Palma, G.; Nadal, I.; Collado, M.C.; Sanz, Y. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects. Br. J. Nutr. 2009, 102, 1154–1160. [Google Scholar] [CrossRef] [PubMed]
- Bertini, I.; Calabrò, A.; De Carli, V.; Luchinat, C.; Nepi, S.; Porfirio, B.; Renzi, D.; Saccenti, E.; Tenori, L. The metabonomic signature of celiac disease. J. Proteome Res. 2009, 8, 170–177. [Google Scholar] [CrossRef] [PubMed]
- Collado, M.C.; Donat, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Specific duodenal and faecal bacterial groups associated with paediatric coeliac disease. J. Clin. Pathol. 2009, 62, 264–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Di Cagno, R.; Rizzello, C.G.; Gagliardi, F.; Ricciuti, P.; Ndagijimana, M.; Francavilla, R.; Guerzoni, M.E.; Crecchio, C.; Gobbetti, M.; De Angelis, M. Different fecal microbiotas and volatile organic compounds in treated and untreated children with celiac disease. Appl. Environ. Microbiol. 2009, 75, 3963–3971. [Google Scholar] [CrossRef] [PubMed]
- Nadal, I.; Donant, E.; Ribes-Koninckx, C.; Calabuig, M.; Sanz, Y. Imbalance in the composition of the duodenal microbiota of children with coeliac disease. J. Med. Microbiol. 2007, 56, 1669–1674. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjellström, B.; Stenhammar, L.; Högberg, L.; Fälth-Magnusson, K.; Magnusson, K.E.; Midtvedt, T.; Sundqvist, T.; Norin, E. Gut microflora associated characteristics in children with celiac disease. Am. J. Gastroenterol. 2005, 100, 2784–2788. [Google Scholar] [CrossRef] [PubMed]
- Lukjancenko, O.; Wassenaar, T.M.; Ussery, D.W. Comparison of 61 sequenced Escherichia coli genomes. Microb. Ecol. 2010, 60, 708–720. [Google Scholar] [CrossRef] [PubMed]
- Koeppel, A.F.; Wu, M. Surprisingly extensive mixed phylogenetic and ecological signals among bacterial Operational Taxonomic Units. Nucleic Acids Res. 2013, 41, 5175–5188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Callahan, B.J.; McMurdie, P.J.; Holmes, S.P. Exact sequence variants should replace operational taxonomic units in marker-gene data analysis. ISME J. 2017, 11, 2639–2643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, L.; Chen, X.; Skogerbø, G.; Zhang, P.; Chen, R.; He, S.; Huang, D.-W. The human microbiome: A hot spot of microbial horizontal gene transfer. Genomics 2012, 100, 265–270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smillie, C.S.; Smith, M.B.; Friedman, J.; Cordero, O.X.; David, L.A.; Alm, E.J. Ecology drives a global network of gene exchange connecting the human microbiome. Nature 2011, 480, 241–244. [Google Scholar] [CrossRef] [PubMed]
- Sipos, R.; Székely, A.J.; Palatinszky, M.; Révész, S.; Márialigeti, K.; Nikolausz, M. Effect of primer mismatch, annealing temperature and PCR cycle number on 16S rRNA gene-targeting bacterial community analysis. FEMS Microbiol. Ecol. 2007, 60, 341–350. [Google Scholar] [CrossRef] [PubMed]
- Swidsinski, A.; Loening-Baucke, V.; Vaneechoutte, M.; Doerffel, Y. Active Crohn’s disease and ulcerative colitis can be specifically diagnosed and monitored based on the biostructure of the fecal flora. Inflamm. Bowel Dis. 2008, 14, 147–161. [Google Scholar] [CrossRef] [PubMed]
- Donaldson, G.P.; Lee, S.M.; Mazmanian, S.K. Gut biogeography of the bacterial microbiota. Nat. Rev. Microbiol. 2016, 14, 20–32. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Takahashi, R.; Nishi, T.; Sakamoto, M.; Benno, Y. Molecular analysis of jejunal, ileal, caecal and recto-sigmodial human colonic microbiota using 16S rRNA gene libraries and terminal restriction fragment length polymorphism. J. Med. Microbiol. 2005, 54, 1093–1101. [Google Scholar] [CrossRef] [PubMed]
- Shortt, C.; Hasselwander, O.; Meynier, A.; Nauta, A.; Fernández, E.N.; Putz, P.; Rowland, I.; Swann, J.; Türk, J.; Vermeiren, J.; et al. Systematic review of the effects of the intestinal microbiota on selected nutrients and non-nutrients. Eur. J. Nutr. 2018, 57, 25–49. [Google Scholar] [CrossRef] [PubMed]
- Wald, C.; Wu, C. Of mice and women: the bias in animal models. Science 2010, 327, 1571–1572. [Google Scholar] [CrossRef] [PubMed]
- Camacho-Ortiz, A.; Gutiérrez-Delgado, E.M.; Garcia-Mazcorro, J.F.; Mendoza-Olazarán, S.; Martínez-Meléndez, A.; Palau-Davila, L.; Baines, S.D.; Maldonado-Garza, H.; Garza-González, E. Randomized clinical trial to evaluate the effect of fecal microbiota transplant for initial Clostridium difficile infection in intestinal microbiome. PLoS ONE 2017, 12, e0189768. [Google Scholar] [CrossRef] [PubMed]
- Chong, C.Y.L.; Bloomfield, F.H.; O’Sullivan, J.M. Factors affecting gastrointestinal microbiome development in neonates. Nutrients 2018, 10, 74. [Google Scholar] [CrossRef] [PubMed]
- Dethlefsen, L.; Relman, D.A. Incomplete recovery and individualized responses of the human distal gut microbiota to repeated antibiotic perturbation. Proc. Natl. Acad. Sci. USA 2011, 108, 4554–4561. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.; Gonzalez, A.; Ackermann, G.; Wendel, D.; Vázquez-Baeza, Y.; Jansson, J.K.; Gordon, J.I.; Knight, R. Meta-analyses of studies of the human microbiota. Genome Res. 2013, 23, 1704–1714. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McCartney, A.L.; Wenzhi, W.; Tannock, G.W. Molecular analysis of the composition of the bifidobacterial and Lactobacillus microflora of humans. Appl. Environ. Microbiol. 1996, 62, 4608–4613. [Google Scholar] [PubMed]
- The Human Microbiome Project Consortium. Structure, function and diversity of the healthy human microbiome. Nature 2012, 486, 207–214. [Google Scholar] [Green Version]
- Ursell, L.K.; Clemente, J.C.; Rideout, J.R.; Gevers, D.; Caporaso, J.G.; Knight, R. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J. Allergy Clin. Immunol. 2012, 129, 1204–1208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Markle, J.G.M.; Frank, D.N.; Mortin-Toth, S.; Robertson, C.E.; Feazel, L.M.; Rolle-Kampczyk, U.; von Bergen, M.; McCoy, K.D.; Macpherson, A.J.; Danska, J.S. Sex differences in the gut microbiome drive hormone-dependent regulation of autoimmunity. Science 2013, 339, 1084–1088. [Google Scholar] [PubMed]
- Ciacci, C.; Cirillo, M.; Sollazzo, R.; Savino, G.; Sabbatini, F.; Mazzacca, G. Gender and clinical presentation in adult celiac disease. Scand. J. Gastroenterol. 1995, 30, 1077–1081. [Google Scholar] [CrossRef] [PubMed]
- Lahti, L.; Salojärvi, J.; Salonen, A.; Scheffer, M.; de Vos, W.M. Tipping elements in the human intestinal ecosystem. Nat. Commun. 2014, 5, 4344. [Google Scholar] [CrossRef] [PubMed]
- Moreno-Estrada, A.; Gignoux, C.R.; Fernandez-Lopez, J.C.; Zakharia, F.; Sikora, M.; Contreras, A.V.; Acuña-Alonzo, V.; Sandoval, K.; Eng, C.; Romero-Hidalgo, S.; et al. The genetics of Mexico recapitulates native American substructure and affects biomedical traits. Science 2014, 344, 1280–1285. [Google Scholar] [CrossRef] [PubMed]
- Rehman, A.; Rausch, P.; Wang, J.; Skieceviciene, J.; Kiudelis, G.; Bhagalia, K.; Amarapurkar, D.; Kupcinskas, L.; Schreiber, S.; Rosenstiel, P.; et al. Geographical patterns of the standing and active human gut microbiome in health and IBD. Gut 2015, 65, 238–248. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Lauber, C.L.; Costello, E.K.; Berg-Lyons, D.; Gonzalez, A.; Stombaugh, J.; Knights, D.; Gajer, P.; Ravel, J.; Fierer, N.; et al. Moving pictures of the human microbiome. Genome Biol. 2011, 12, R50. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Winter, S.E.; Bäumler, A.J. Why related bacterial species bloom simultaneously in the gut: Principles underlying the ‘like will to like’ concept. Cell. Microbiol. 2014, 16, 179–184. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, R.F.; McCririck, M.Y. Gluten-free diets. Br. Med. J. 1958, 2, 299–301. [Google Scholar] [CrossRef] [PubMed]
- Jackson, F.W. Effects of a gluten-free diet on gut microbiota and immune function in healthy adult human subjects—comment by Jackson. Br. J. Nutr. 2010, 104, 773. [Google Scholar] [CrossRef] [PubMed]
- Bathrellou, E.; Kontogianni, M.D.; Panagiotakos, D.B. Celiac disease and non-celiac gluten or wheat sensitivity and health in later life: A review. Maturitas 2018, 112, 29–33. [Google Scholar] [CrossRef] [PubMed]
- Morreale, F.; Angelino, D.; Pellegrini, N. Designing a score-based method for the evaluation of the nutritional quality of the gluten-free bakery products and their gluten-containing counterparts. Plant Foods Hum. Nutr. 2018, 73, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Larretxi, I.; Simon, E.; Benjumea, L.; Miranda, J.; Bustamante, M.A.; Lasa, A.; Eizaguirre, F.J.; Churruca, I. Gluten-free-rendered products contribute to imbalanced diets in children and adolescents with celiac disease. Eur. J. Nutr. 2018. [Google Scholar] [CrossRef] [PubMed]
- Newberry, C.; McKnight, L.; Sarav, M.; Pickett-Blakely, O. Going gluten free: The history and nutritional implications of today’s most popular diet. Curr. Gastroenterol. Rep. 2017, 19, 54. [Google Scholar] [CrossRef] [PubMed]
- Thompson, T. Thiamin, riboflavin, and niacin contents of the gluten-free diet: Is there cause for concern? J. Am. Diet. Assoc. 1999, 99, 858–862. [Google Scholar] [CrossRef]
- Mariani, P.; Viti, M.G.; Montuori, M.; La Vecchia, A.; Cipolletta, E.; Calvani, L.; Bonamico, M. The gluten-free diet: A nutritional risk factor for adolescents with celiac disease? J. Pediatr. Gastroenterol. Nutr. 1998, 27, 519–523. [Google Scholar] [CrossRef] [PubMed]
- Tortora, R.; Capone, P.; De Stefano, G.; Imperatore, N.; Gerbino, N.; Donetto, S.; Monaco, V.; Caporaso, N.; Rispo, A. Metabolic syndrome in patients with coeliac disease on a gluten-free diet. Aliment. Pharmacol. Ther. 2015, 41, 352–359. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Potter, M.D.E.; Brienesse, S.C.; Walker, M.M.; Boyle, A.; Talley, N.J. Effect of the gluten-free diet on cardiovascular risk factors in patients with coeliac disease: A systematic review. J. Gastroenterol. Hepatol. 2018, 33, 781–791. [Google Scholar] [CrossRef] [PubMed]
- Ercolini, D.; Francavilla, R.; Vannini, L.; De Filippis, F.; Capriati, T.; Di Cagno, R.; Iacono, G.; De Angelis, M.; Gobbetti, M. From an imbalance to a new imbalance: Italian-style gluten-free diet alters the salivary microbiota and metabolome of African celiac children. Sci. Rep. 2015, 5, 18571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Verma, A.K.; Gatti, S.; Galeazzi, T.; Monachesi, C.; Padella, L.; Del Baldo, G.; Annibali, R.; Lionetti, E.; Catassi, C. Gluten contamination in naturally or labeled gluten-free products marketed in Italy. Nutrients 2017, 9, 115. [Google Scholar] [CrossRef] [PubMed]
- Cabrera-Chaves, F.; Iametti, S.; Miriani, M.; de la Barca, A.M.; Mamone, G.; Bonomi, F. Maize prolamins resistant to peptic-tryptic digestion maintain immune-recognition by IgA from some celiac disease patients. Plant Foods Hum. Nutr. 2012, 67, 24. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M. Long-Term Prospects for the Global Rice Economy, in Rice in Global Markets and Sustainable Production Systems; Food and Agriculture Organization of the United Nations: Rome, Italy, 2004. [Google Scholar]
- Catassi, C.; Gatti, S.; Fasano, A. The new epidemiology of celiac disease. J. Pediatr. Gastroenterol. Nutr. 2014, 59, S7–S9. [Google Scholar] [CrossRef] [PubMed]
- Walker, A.J.; Curtis, H.J.; Bacon, S.; Croker, R.; Goldacre, B. Trends, geographical variation and factors associated with prescribing of gluten-free foods in English primary care: A cross-sectional study. BMJ Open 2018, 8, e021312. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Zhang, H.; Luoto, S.; Ren, X. Gluten-free living in China: The characteristics, food choices and difficulties in following a gluten-free diet—An online survey. Appetite 2018, 127, 242–248. [Google Scholar] [CrossRef] [PubMed]
- Halmos, E.P.; Deng, M.; Knowles, S.R.; Sainsbury, K.; Mullan, B.; Tye-Din, J.A. Food knowledge and psychological state predict adherence to a gluten-free diet in a survey of 5310 Australians and New Zealanders with coeliac disease. Aliment. Pharmacol. Ther. 2018. [Google Scholar] [CrossRef] [PubMed]
- Norsa, L.; Branchi, F.; Bravo, M.; Ferretti, F.; Roncoroni, L.; Somalvico, F.; Conte, D.; Bardella, M.T.; Fabiano, S.; Barigelletti, G.; et al. Celiac disease 30 years after diagnosis: Struggling with gluten-free adherence or gaining gluten tolerance? J. Pediatr. Gastroenterol. Nutr. 2018. [Google Scholar] [CrossRef] [PubMed]
| Patients Characteristics | Sample Collection and Analysis | Methods | Main Finding(s) | Reference (Year of Publication) |
|---|---|---|---|---|
| 30 subjects (21–73 years old) with CD (n = 6), NCGS (n = 12) and controls (n = 12) from Veracruz, México | Samples were obtained at baseline and after 4 weeks on a GFD | 16S rDNA sequencing using the Illumina MiSeq platform | Pseudomonas was higher in duodenum of CD patients after 4 weeks on GFD | [49] (Unpublished) |
| 21 healthy adults (16–61 years old) from Groningen, The Netherlands | Nine samples were obtained from each participant at baseline, during and after 4 weeks on GFD | 16S rDNA sequencing using 454 pyrosequencing | Veillonellaceae (class Clostridia, Firmicutes) was reduced on GFD; 21 predicted pathway activity scores showed significant association to the change in diet | [50] (2016) |
| 53 young subjects (0.5–18 years old) with CD at presentation; 74 young subjects (1–18 years old) with CD on GFD for less than 1 year; 25 subjects (3–33 years old) with CD on GFD for more than 1 year from Norrköping, Sweden | One fecal sample was obtained once from each subject | Gas liquid chromatography for SCFA measurement | Fecal SCFA levels were higher in CD patients on GFD for < 1 year compared to those on GFD > 1 year | [51] (2013) |
| 10 untreated CD patients, 11 treated CD patients and 11 healthy adults from Leon, Spain | Samples were obtained in normal gluten diet and in GFD | DGGE and gas-liquid chromatography of SCFAs | Microbial communities of treated CD clustered together with those of healthy adults | [52] (2012) |
| 19 CD children (6–12 years old) on GFD for at least 2 years and 15 non-celiac children from Bari, Apulia, Italy | Duodenal biopsies and fecal samples were obtained once from each subject | DGGE and culture-based methods | 2 years of GFD does not fully restore the microbiota and metabolome of CD children | [53] (2010) |
| 24 untreated CD patients (2–12 years old) on a normal-gluten containing diet; 18 treated CD patients (1–12 years old) on GFD for at least 2 years; 20 healthy children (2–11 years old) without known gluten intolerance from Valencia, Spain | One fecal sample was obtained once from each subject | FISH, flow cytometry and immunoglobulin-coated bacterial analysis | CD patients have lower levels of IgA-coated bacteria thus providing new insights into the relationship between the gut microbiota and host immune defenses | [54] (2010) |
| 20 children with CD (1.2–16.1years old) before and after at least 9 months on GFD, and 10 controls (7.8–20.8 years old) from Rome, Italy | Biopsies from the second part of the duodenum from CD children before and after at least 9 months on GFD; duodenal biopsies from the controls undergoing upper GI endoscopy for functional dyspepsia | TGGE | Number of bands was higher in active and inactive states compared to controls, implying higher biodiversity | [55] (2010) |
| 10 healthy adults (23–40 years old) from Valencia, Spain | One fecal sample was obtained once from each subject at baseline and after 1 month on GFD | FISH and qPCR | Reduction of “beneficial” bacteria and the ability of fecal samples to stimulate the host’s immunity | [56] (2009) |
| 34 CD patients at diagnosis and after 12 months on GFD, and 34 healthy controls from Fiorentino, Italy | Serum and urine samples were obtained once from each subject | Nuclear Magnetic Resonance (NMR) of urine and serum samples | After 12 months of GFD, all but one patient was classified as healthy | [57] (2009) |
| Group 1 (30 untreated CD patients on a normal gluten-containing diet, 56–61 months old); group 2 (18 treated CD patients with a GFD for at least 2 years, 64–58 months old); group 3 (30 control children without gluten intolerance, 45–49 months old) from Valencia, Spain | 30 fecal and 25 duodenal biopsies from Group 1; 18 fecal and 8 biopsy samples from Group 2; 30 fecal and 8 biopsy samples from Group 3 | qPCR for a small group of selected microbes | Duodenal and fecal microbiota is partially restored after long-term (>2 years) GFD | [58] (2009) |
| Seven symptom-free CD patients on GFD for at least 2 years; seven CD patients on gluten-containing diet; seven children with no known food intolerance (6–12 years old) from Bari, Apulia, Italy | Each child provided 3 fecal samples over an unknown period of time. The samples were mixed | DGGE and culture-based techniques; gas chromatography-mass spectrometry for VOCs | CD is associated with differences in fecal microbiota and biochemistry | [59] (2009) |
| 20 CD patients (1.6–12 years old) and 10 symptom-free CD patients who had been on GFD for 1–2 years (2–8 years old) and 8 control children (2–7.8 years old) from Valencia, Spain | An unknown number of biopsy specimens was obtained once from each subject | FISH and flow cytometry for a few selected bacterial groups in duodenum | Ratio of Lactobacillus-Bifidobacterium to Bacteroides-E. coli was reduced in CD patients with either active or inactive disease compared to controls | [60] (2007) |
| 36 children with CD at presentation, 47 patients on GFD for at least 3 months, and 42 healthy controls from Stockholm, Sweden | One fecal sample was obtained once for each subject | Gas-liquid chromatography of SCFAs in fecal samples | Difference between children on GFD and controls regarding acetic, i-butyric, i-valeric acid, and total SCFAs | [61] (2005) |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Garcia-Mazcorro, J.F.; Noratto, G.; Remes-Troche, J.M. The Effect of Gluten-Free Diet on Health and the Gut Microbiota Cannot Be Extrapolated from One Population to Others. Nutrients 2018, 10, 1421. https://doi.org/10.3390/nu10101421
Garcia-Mazcorro JF, Noratto G, Remes-Troche JM. The Effect of Gluten-Free Diet on Health and the Gut Microbiota Cannot Be Extrapolated from One Population to Others. Nutrients. 2018; 10(10):1421. https://doi.org/10.3390/nu10101421
Chicago/Turabian StyleGarcia-Mazcorro, Jose F., Giuliana Noratto, and Jose M. Remes-Troche. 2018. "The Effect of Gluten-Free Diet on Health and the Gut Microbiota Cannot Be Extrapolated from One Population to Others" Nutrients 10, no. 10: 1421. https://doi.org/10.3390/nu10101421
APA StyleGarcia-Mazcorro, J. F., Noratto, G., & Remes-Troche, J. M. (2018). The Effect of Gluten-Free Diet on Health and the Gut Microbiota Cannot Be Extrapolated from One Population to Others. Nutrients, 10(10), 1421. https://doi.org/10.3390/nu10101421