Isolation and Characterization of Potentially Probiotic Bacterial Strains from Mice: Proof of Concept for Personalized Probiotics
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
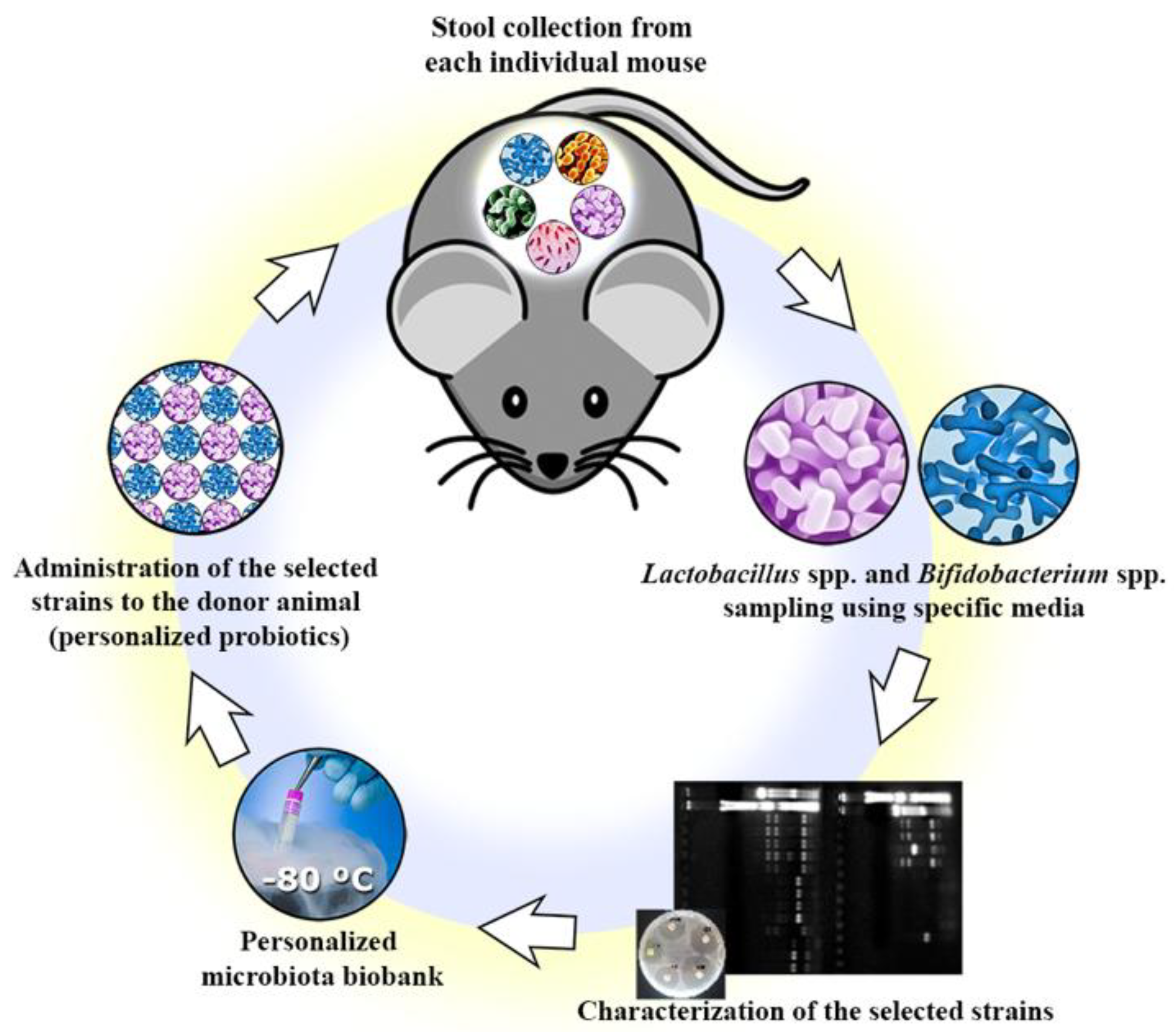
2.2. Isolation of Commensal Bacteria Strains
2.3. Preliminary Identification
2.4. Genera Confirmation
2.5. Evaluation of Survival in Simulated Gastrointestinal Conditions
2.6. Antibiotic Susceptibility Test
2.7. Dextran Sodium Sulfate (DSS)-Induced Colitis Experiment
2.8. Tissue Collection
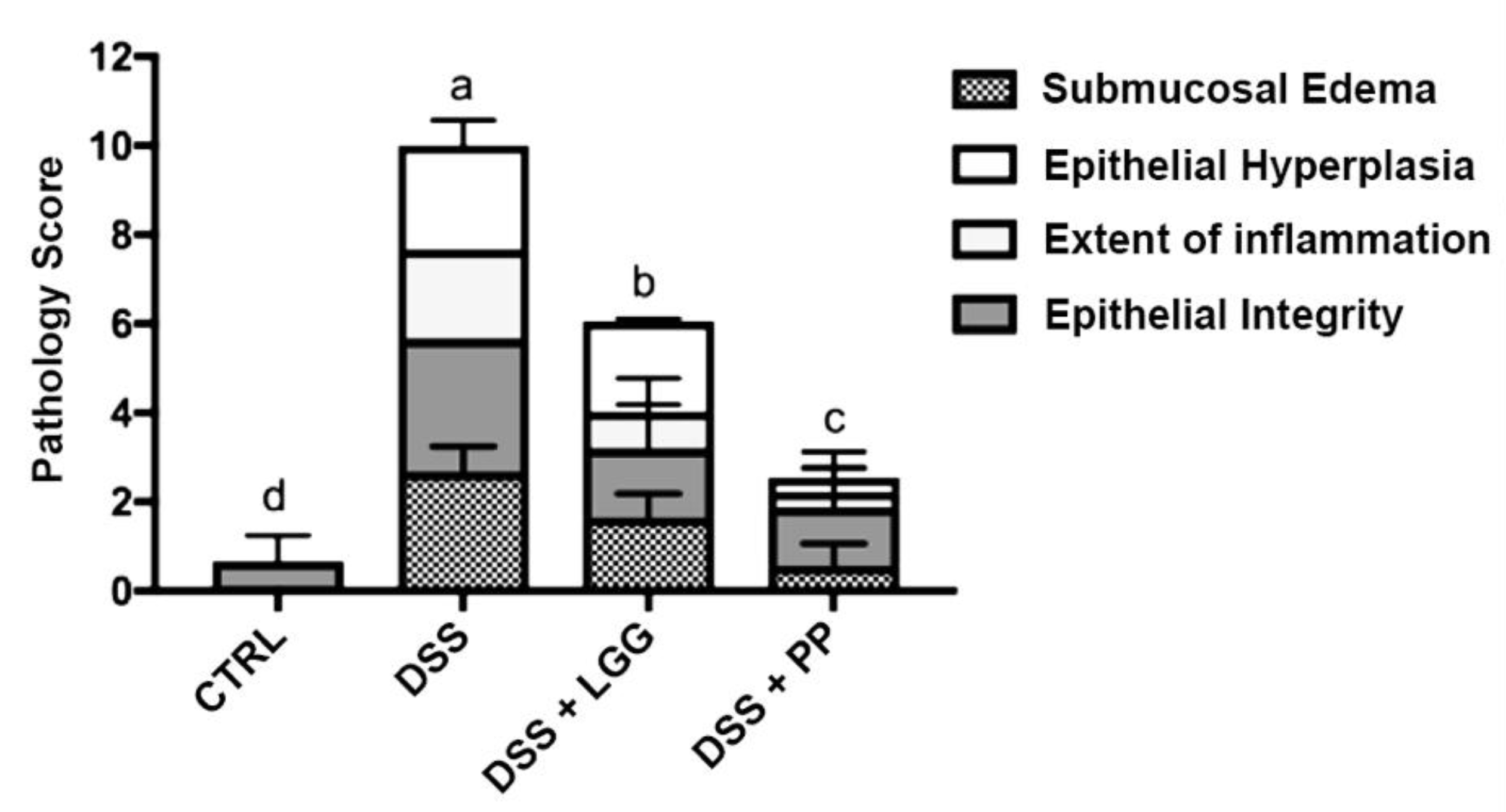
2.9. Histopathological Scoring
2.10. RNA Extraction and Quantitative Real-Time PCR
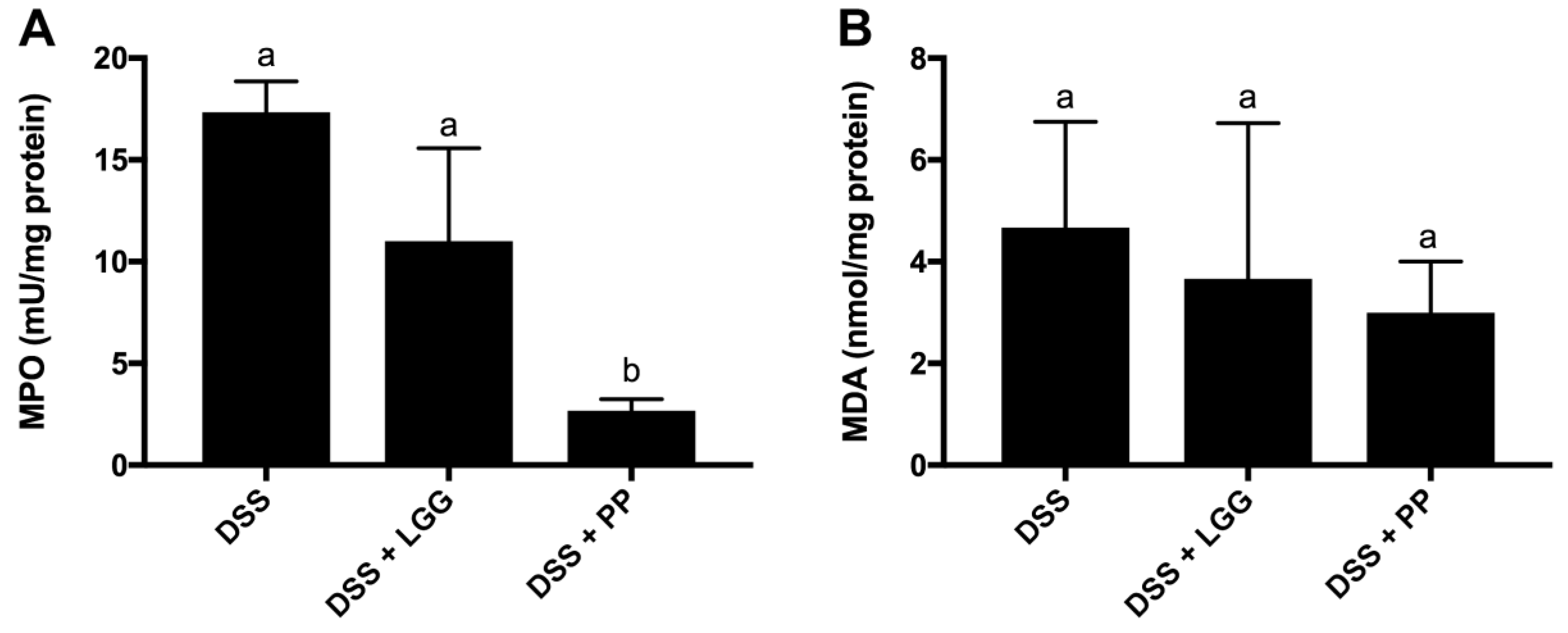
2.11. Myeloperoxidase (MPO) and Malondialdehyde (MDA) Activity
2.12. Statistical Analysis
3. Results
3.1. Isolation and Genera Confirmation
3.2. In Vitro Tests Demonstrate a Potential Probiotic Effect of the Isolated Strains
3.3. Personalized Commensal Strains Protect Mice against Acute Dextran Sodium Sulfate-Induced Colitis
3.4. Personalized Probiotic Therapy Positively Modulates the Host Immune Response during DSS-Colitis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References and Notes
- Cénit, M.C.; Matzaraki, V.; Tigchelaar, E.F.; Zhernakova, A. Rapidly expanding knowledge on the role of the gut microbiome in health and disease. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 1981–1992. [Google Scholar] [CrossRef] [PubMed]
- Lozupone, C.A.; Stombaugh, J.I.; Gordon, J.I.; Jansson, J.K.; Knight, R. Diversity, stability and resilience of the human gut microbiota. Nature 2012, 489, 220–230. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dicksved, J.; Halfvarson, J.; Rosenquist, M.; Järnerot, G.; Tysk, C.; Apajalahti, J.; Engstrand, L.; Jansson, J.K. Molecular analysis of the gut microbiota of identical twins with Crohn’s disease. ISME J. 2008, 2, 716–727. [Google Scholar] [CrossRef] [PubMed]
- Frank, D.N.; St Amand, A.L.; Feldman, R.A.; Boedeker, E.C.; Harpaz, N.; Pace, N.R. Molecular-phylogenetic characterization of microbial community imbalances in human inflammatory bowel diseases. Proc. Natl. Acad. Sci. USA 2007, 104, 13780–13785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rodiño-Janeiro, B.K.; Vicario, M.; Alonso-Cotoner, C.; Pascua-García, R.; Santos, J. A Review of Microbiota and Irritable Bowel Syndrome: Future in Therapies. Adv. Ther. 2018, 35, 289–310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef] [PubMed]
- Wu, X.; Ma, C.; Han, L.; Nawaz, M.; Gao, F.; Zhang, X.; Yu, P.; Zhao, C.; Li, L.; Zhou, A.; et al. Molecular Characterisation of the Faecal Microbiota in Patients with Type II Diabetes. Curr. Microbiol. 2010, 61, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.-C.; Arévalo, A.; Stiemsma, L.; Dimitriu, P.; Chico, M.E.; Loor, S.; Vaca, M.; Boutin, R.C.T.; Morien, E.; Jin, M.; et al. Associations between infant fungal and bacterial dysbiosis and childhood atopic wheeze in a nonindustrialized setting. J. Allergy Clin. Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Ohigashi, S.; Sudo, K.; Kobayashi, D.; Takahashi, O.; Takahashi, T.; Asahara, T.; Nomoto, K.; Onodera, H. Changes of the intestinal microbiota, short chain fatty acids, and fecal pH in patients with colorectal cancer. Dig. Dis. Sci. 2013, 58, 1717–1726. [Google Scholar] [CrossRef] [PubMed]
- Lupton, J.R. Microbial Degradation Products Influence Colon Cancer Risk: The Butyrate Controversy. J. Nutr. 2004, 134, 479–482. [Google Scholar] [CrossRef] [PubMed]
- Lau, L.H.S.; Wong, S.H. Microbiota, Obesity and NAFLD. Adv. Exp. Med. Biol. 2018, 1061, 111–125. [Google Scholar] [PubMed]
- Jia, W.; Rajani, C. The Influence of Gut Microbial Metabolism on the Development and Progression of Non-alcoholic Fatty Liver Disease. Adv. Exp. Med. Biol. 2018, 1061, 95–110. [Google Scholar] [PubMed]
- Gonzalez, A.; Stombaugh, J.; Lozupone, C.; Turnbaugh, P.J.; Gordon, J.I.; Knight, R. The mind-body-microbial continuum. Dialogues Clin. Neurosci. 2011, 13, 55–62. [Google Scholar] [PubMed]
- Pulikkan, J.; Maji, A.; Dhakan, D.B.; Saxena, R.; Mohan, B.; Anto, M.M.; Agarwal, N.; Grace, T.; Sharma, V.K. Gut Microbial Dysbiosis in Indian Children with Autism Spectrum Disorders. Microb. Ecol. 2018. [Google Scholar] [CrossRef] [PubMed]
- Levy, M.; Kolodziejczyk, A.A.; Thaiss, C.A.; Elinav, E. Dysbiosis and the immune system. Nat. Rev. Immunol. 2017, 17, 219–232. [Google Scholar] [CrossRef] [PubMed]
- Petersen, C.; Round, J.L. Defining dysbiosis and its influence on host immunity and disease. Cell. Microbiol. 2014, 16, 1024–1033. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooks, K.B.; O’Malley, M.A. Dysbiosis and Its Discontents. MBio 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Vangay, P.; Ward, T.; Gerber, J.S.; Knights, D. Antibiotics, pediatric dysbiosis, and disease. Cell Host Microbe 2015, 17, 553–564. [Google Scholar] [CrossRef] [PubMed]
- Celiberto, L.S.; Graef, F.A.; Healey, G.R.; Bosman, E.S.; Jacobson, K.; Sly, L.M.; Vallance, B.A. Inflammatory Bowel Disease and Immunonutrition: Novel Therapeutic Approaches Through Modulation of Diet and the Gut Microbiome. Immunology 2018. [Google Scholar] [CrossRef] [PubMed]
- Martín, R.; Chain, F.; Miquel, S.; Motta, J.-P.; Vergnolle, N.; Sokol, H.; Langella, P. Using murine colitis models to analyze probiotics–host interactions. FEMS Microbiol. Rev. 2017, 41, S49–S70. [Google Scholar] [CrossRef] [PubMed]
- Borchers, A.T.; Selmi, C.; Meyers, F.J.; Keen, C.L.; Gershwin, M.E. Probiotics and immunity. J. Gastroenterol. 2009, 44, 26–46. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Khalesi, S.; Bellissimo, N.; Vandelanotte, C.; Williams, S.; Stanley, D.; Irwin, C. A review of probiotic supplementation in healthy adults: Helpful or hype? Eur. J. Clin. Nutr. 2018. [Google Scholar] [CrossRef] [PubMed]
- Fedorak, R.N.; Madsen, K.L. Probiotics and the management of inflammatory bowel disease. Inflamm. Bowel Dis. 2004, 10, 286–299. [Google Scholar] [CrossRef] [PubMed]
- Boirivant, M.; Strober, W. The mechanism of action of probiotics. Curr. Opin. Gastroenterol. 2007, 23, 679–692. [Google Scholar] [CrossRef] [PubMed]
- Marco, M.L.; Pavan, S.; Kleerebezem, M. Towards understanding molecular modes of probiotic action. Curr. Opin. Biotechnol. 2006, 17, 204–210. [Google Scholar] [CrossRef] [PubMed]
- Celiberto, L.S.; Bedani, R.; Rossi, E.A.; Cavallini, D.C.U. Probiotics: The Scientific Evidence in the Context of Inflammatory Bowel Disease. Crit. Rev. Food Sci. Nutr. 2015, 1759–1768. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Guarner, F.; Reid, G.; Gibson, G.R.; Merenstein, D.J.; Pot, B.; Morelli, L.; Canani, R.B.; Flint, H.J.; Salminen, S.; et al. The International Scientific Association for Probiotics and Prebiotics consensus statement on the scope and appropriate use of the term probiotic. Nat. Rev. Gastroenterol. Hepatol. 2014, 11, 506–514. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reid, G. Probiotics: Definition, scope and mechanisms of action. Best Pract. Res. Clin. Gastroenterol. 2016, 30, 17–25. [Google Scholar] [CrossRef] [PubMed]
- Parker, E.A.; Roy, T.; D’Adamo, C.R.; Wieland, L.S. Probiotics and gastrointestinal conditions: An overview of evidence from the Cochrane Collaboration. Nutrition 2018, 45, 125–134.e11. [Google Scholar] [CrossRef] [PubMed]
- Sommer, F.; Anderson, J.M.; Bharti, R.; Raes, J.; Rosenstiel, P. The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 2017, 15, 630–638. [Google Scholar] [CrossRef] [PubMed]
- Martin, R.; Nauta, A.J.; Ben Amor, K.; Knippels, L.M.J.; Knol, J.; Garssen, J. Early life: Gut microbiota and immune development in infancy. Benef. Microbes 2010, 1, 367–382. [Google Scholar] [CrossRef] [PubMed]
- Dominguez-bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McNaughton, S.J. Diversity and Stability of Ecological Communities: A Comment on the Role of Empiricism in Ecology. Am. Nat. 1977, 111, 515–525. [Google Scholar] [CrossRef]
- Naeem, S.; Li, S. Biodiversity enhances ecosystem reliability. Nature 1997, 390, 507–509. [Google Scholar] [CrossRef]
- Jameson, J.L.; Longo, D.L. Precision Medicine—Personalized, Problematic, and Promising. N. Engl. J. Med. 2015, 372, 2229–2234. [Google Scholar] [CrossRef] [PubMed]
- Kort, R. Personalized therapy with probiotics from the host by TripleA. Trends Biotechnol. 2014, 32, 291–293. [Google Scholar] [CrossRef] [PubMed]
- Pelczar, M.J. Microbiology; Tata McGraw-Hill: New York, NY, USA, 2003; ISBN 0074623206. [Google Scholar]
- Heilig, H.G.H.J.; Zoetendal, E.G.; Vaughan, E.E.; Marteau, P.; Akkermans, A.D.L.; de Vos, W.M. Molecular Diversity of Lactobacillus spp. and Other Lactic Acid Bacteria in the Human Intestine as Determined by Specific Amplification of 16S Ribosomal DNA Molecular Diversity of Lactobacillus spp. and Other Lactic Acid Bacteria in the Human Intestine. Appl. Environ. Microbiol. 2002, 68, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Vincent, D.; Roy, D.; Mondou, F.; Déry, C. Characterization of bifidobacteria by random DNA amplification. Int. J. Food Microbiol. 1998, 43, 185–193. [Google Scholar] [CrossRef]
- Liserre, A.M.; Ré, M.I.; Franco, B.D.G.M. Microencapsulation of Bifidobacterium animalis subsp. lactis in Modified Alginate-chitosan Beads and Evaluation of Survival in Simulated Gastrointestinal Conditions. Food Biotechnol. 2007, 21, 1–16. [Google Scholar] [CrossRef]
- Buriti, F.C.A.; Castro, I.A.; Saad, S.M.I. Viability of Lactobacillus acidophilus in synbiotic guava mousses and its survival under in vitro simulated gastrointestinal conditions. Int. J. Food Microbiol. 2010, 137, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Clinical and Laboratory Standards Institute.
- Performance Standards for Antimicrobial Susceptibility Testing An informational supplement for global application developed through the Clinical and Laboratory Standards Institute.
- Segers, M.E.; Lebeer, S. Towards a better understanding of Lactobacillus rhamnosus GG—Host interactions. Microb. Cell Fact. 2014, 13, S7. [Google Scholar] [CrossRef] [PubMed]
- Szajewska, H.; Kołodziej, M. Systematic review with meta-analysis: Lactobacillus rhamnosus GG in the prevention of antibiotic-associated diarrhoea in children and adults. Aliment. Pharmacol. Ther. 2015, 42, 1149–1157. [Google Scholar] [CrossRef] [PubMed]
- Cooper, H.S.; Murthy, S.N.; Shah, R.S.; Sedergran, D.J. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab. Investig. 1993, 69, 238–249. [Google Scholar] [PubMed]
- Ryz, N.R.; Lochner, A.; Bhullar, K.; Ma, C.; Huang, T.; Bhinder, G.; Bosman, E.; Wu, X.; Innis, S.M.; Jacobson, K.; et al. Dietary vitamin D3 deficiency alters intestinal mucosal defense and increases susceptibility to Citrobacter rodentium-induced colitis. Am. J. Physiol. Gastrointest. Liver Physiol. 2015, 309, G730–G742. [Google Scholar] [CrossRef] [PubMed]
- Zarepour, M.; Bhullar, K.; Montero, M.; Ma, C.; Huang, T.; Velcich, A.; Xia, L.; Vallance, B.A. The mucin Muc2 limits pathogen burdens and epithelial barrier dysfunction during Salmonella enterica serovar Typhimurium colitis. Infect. Immun. 2013, 81, 3672–3683. [Google Scholar] [CrossRef] [PubMed]
- Eissa, N.; Hussein, H.; Wang, H.; Rabbi, M.F.; Bernstein, C.N.; Ghia, J.-E. Stability of Reference Genes for Messenger RNA Quantification by Real-Time PCR in Mouse Dextran Sodium Sulfate Experimental Colitis. PLoS ONE 2016, 11, e0156289. [Google Scholar] [CrossRef] [PubMed]
- Schmittgen, T.D.; Livak, K.J. Analyzing real-time PCR data by the comparative C(T) method. Nat. Protoc. 2008, 3, 1101–1108. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, E.J.C.; Tyrrell, K.L.; Citron, D.M. Lactobacillus Species: Taxonomic Complexity and Controversial Susceptibilities. Clin. Infect. Dis. 2015, 60, S98–S107. [Google Scholar] [CrossRef] [PubMed]
- Muñoa, F.J.; Pares, R. Selective medium for isolation and enumeration of Bifidobacterium spp. Appl. Environ. Microbiol. 1988, 54, 1715–1718. [Google Scholar] [PubMed]
- Xiao, L.; Feng, Q.; Liang, S.; Sonne, S.B.; Xia, Z.; Qiu, X.; Li, X.; Long, H.; Zhang, J.; Zhang, D.; et al. A catalog of the mouse gut metagenome. Nat. Biotechnol. 2015, 33, 1103–1108. [Google Scholar] [CrossRef] [PubMed]
- Perše, M.; Cerar, A. Dextran sodium sulphate colitis mouse model: Traps and tricks. J. Biomed. Biotechnol. 2012, 2012, 718617. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Chen, L.; Chen, L.; Ren, X.; Ge, H.; Li, B.; Ma, G.; Ke, X.; Zhu, J.; Li, L.; et al. Potential probiotic characterization of Lactobacillus reuteri from traditional Chinese highland barley wine and application for room-temperature-storage drinkable yogurt. J. Dairy Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Zmora, N.; Zilberman-Schapira, G.; Suez, J.; Mor, U.; Dori-Bachash, M.; Bashiardes, S.; Kotler, E.; Zur, M.; Regev-Lehavi, D.; Brik, R.B.-Z.; et al. Personalized Gut Mucosal Colonization Resistance to Empiric Probiotics Is Associated with Unique Host and Microbiome Features. Cell 2018, 174, 1388–1405.e21. [Google Scholar] [CrossRef] [PubMed]
- De Vuyst, L.; Neysens, P. The sourdough microflora: Biodiversity and metabolic interactions. Trends Food Sci. Technol. 2005, 16, 43–56. [Google Scholar] [CrossRef]
- Ehrmann, M.A.; Müller, M.R.A.; Vogel, R.F. Molecular analysis of sourdough reveals Lactobacillus mindensis sp. nov. Int. J. Syst. Evol. Microbiol. 2003, 53, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Hayford, A.E.; Petersen, A.; Vogensen, F.K.; Jakobsen, M. Use of conserved randomly amplified polymorphic DNA (RAPD) fragments and RAPD pattern for characterization of Lactobacillus fermentum in Ghanaian fermented maize dough. Appl. Environ. Microbiol. 1999, 65, 3213–3221. [Google Scholar] [PubMed]
- Venturi, M.; Guerrini, S.; Granchi, L.; Vincenzini, M. Typing of Lactobacillus sanfranciscensis isolates from traditional sourdoughs by combining conventional and multiplex RAPD–PCR profiles. Int. J. Food Microbiol. 2012, 156, 122–126. [Google Scholar] [CrossRef] [PubMed]
- FAO/WHO. Joint FAO/WHO Working Group on Drafting Guidelines for the Evaluation of Probiotics in Food; FAO/WHO: London, ON, Canada, 2002. [Google Scholar]
- Gueimonde, M.; Sánchez, B.; de los Reyes-Gavilán, C.G.; Margolles, A. Antibiotic resistance in probiotic bacteria. Front. Microbiol. 2013, 4, 202. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.S.; Pillidge, C.J.; Gopal, P.K.; Gill, H.S. Antibiotic susceptibility profiles of new probiotic Lactobacillus and Bifidobacterium strains. Int. J. Food Microbiol. 2005, 98, 211–217. [Google Scholar] [CrossRef] [PubMed]
- Mayrhofer, S.; Mair, C.; Kneifel, W.; Domig, K.J. Susceptibility of Bifidobacteria of Animal Origin to Selected Antimicrobial Agents. Chemother. Res. Pract. 2011, 2011, 1–6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eichele, D.D.; Kharbanda, K.K. Dextran sodium sulfate colitis murine model: An indispensable tool for advancing our understanding of inflammatory bowel diseases pathogenesis. World J. Gastroenterol. 2017, 23, 6016–6029. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- König, J.; Wells, J.; Cani, P.D.; García-Ródenas, C.L.; MacDonald, T.; Mercenier, A.; Whyte, J.; Troost, F.; Brummer, R.-J. Human Intestinal Barrier Function in Health and Disease. Clin. Transl. Gastroenterol. 2016, 7, e196. [Google Scholar] [CrossRef] [PubMed]
- Michielan, A.; D’Incà, R. Intestinal Permeability in Inflammatory Bowel Disease: Pathogenesis, Clinical Evaluation, and Therapy of Leaky Gut. Mediators Inflamm. 2015, 2015, 628157. [Google Scholar] [CrossRef] [PubMed]
- Chang, J.; Leong, R.W.; Wasinger, V.C.; Ip, M.; Yang, M.; Phan, T.G. Impaired Intestinal Permeability Contributes to Ongoing Bowel Symptoms in Patients With Inflammatory Bowel Disease and Mucosal Healing. Gastroenterology 2017, 153, 723–731.e1. [Google Scholar] [CrossRef] [PubMed]
- Fukui, H. Increased Intestinal Permeability and Decreased Barrier Function: Does It Really Influence the Risk of Inflammation? Inflamm. Intest. Dis. 2016, 1, 135–145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Al Mijan, M.; Lim, B.O. Diets, functional foods, and nutraceuticals as alternative therapies for inflammatory bowel disease: Present status and future trends. World J. Gastroenterol. 2018, 24, 2673–2685. [Google Scholar] [CrossRef] [PubMed]
- Celiberto, L.S.; Bedani, R.; Dejani, N.N.; de Medeiros, A.I.; Sampaio Zuanon, J.A.; Spolidorio, L.C.; Tallarico Adorno, M.A.; Amâncio Varesche, M.B.; Carrilho Galvão, F.; Valentini, S.R.; et al. Effect of a probiotic beverage consumption (Enterococcus faecium CRL 183 and Bifidobacterium longum ATCC 15707) in rats with chemically induced colitis. PLoS ONE 2017, 12, e0175935. [Google Scholar] [CrossRef] [PubMed]
- Seo, S.; Shin, J.S.; Lee, W.S.; Rhee, Y.K.; Cho, C.W.; Hong, H.D.; Lee, K.T. Anti-colitis effect of Lactobacillus sakei K040706 via suppression of inflammatory responses in the dextran sulfate sodium-induced colitis mice model. J. Funct. Foods 2017, 29, 256–268. [Google Scholar] [CrossRef]
- Shigemori, S.; Watanabe, T.; Kudoh, K.; Ihara, M.; Nigar, S.; Yamamoto, Y.; Suda, Y.; Sato, T.; Kitazawa, H.; Shimosato, T. Oral delivery of Lactococcus lactis that secretes bioactive heme oxygenase-1 alleviates development of acute colitis in mice. Microb. Cell Fact. 2015, 14, 189. [Google Scholar] [CrossRef] [PubMed]
- Park, J.-S.; Joe, I.; Rhee, P.D.; Jeong, C.-S.; Jeong, G. A lactic acid bacterium isolated from kimchi ameliorates intestinal inflammation in DSS-induced colitis. J. Microbiol. 2017, 55, 304–310. [Google Scholar] [CrossRef] [PubMed]
- Camuesco, D.; Peran, L.; Comalada, M.; Nieto, A.; Di Stasi, L.C.; Rodriguez-Cabezas, M.E.; Concha, A.; Zarzuelo, A.; Galvez, J. Preventative effects of lactulose in the trinitrobenzenesulphonic acid model of rat colitis. Inflamm. Bowel Dis. 2005, 11, 265–271. [Google Scholar] [CrossRef] [PubMed]
- Osman, N.; Adawi, D.; Molin, G.; Ahrne, S.; Berggren, A.; Jeppsson, B. Bifidobacterium infantis strains with and without a combination of oligofructose and inulin attenuate inflammation in DSS-induced colitis in rats. BMC Gastroenterol. 2006, 6, 31. [Google Scholar] [CrossRef] [PubMed]
- Krieglstein, C.F.; Anthoni, C.; Cerwinka, W.H.; Stokes, K.Y.; Russell, J.; Grisham, M.B.; Granger, D.N. Role of Blood- and Tissue-Associated Inducible Nitric-Oxide Synthase in Colonic Inflammation. Am. J. Pathol. 2007, 170, 490–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Alex, P.; Zachos, N.C.; Nguyen, T.; Gonzales, L.; Chen, T.-E.; Conklin, L.S.; Centola, M.; Li, X. Distinct Cytokine Patterns Identified from Multiplex Profiles of Murine DSS and TNBS-Induced Colitis. Inflamm. Bowel Dis. 2009, 15, 341–352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramasamy, S.; Nguyen, D.D.; Eston, M.A.; Alam, S.N.; Moss, A.K.; Ebrahimi, F.; Biswas, B.; Mostafa, G.; Chen, K.T.; Kaliannan, K.; et al. Intestinal alkaline phosphatase has beneficial effects in mouse models of chronic colitis. Inflamm. Bowel Dis. 2011, 17, 532–542. [Google Scholar] [CrossRef] [PubMed] [Green Version]






| Animal | Strain | log10CFU/mL | ||||
|---|---|---|---|---|---|---|
| 0 h | 2 h | 6 h | Gram | Catalase | ||
| 1 | Bifidobacterium spp. | 8.85 ± 0.02 | 8.76 ± 0.04 | 8.10 ± 0.10 | + | − |
| Bifidobacterium spp. | 8.97 ± 0.01 | 8.44 ± 0.02 | 8.12 ± 0.11 | + | − | |
| Lactobacillus spp. | 9.96 ± 0.01 | 9.35 ± 0.04 | 9.19 ± 0.08 | + | − | |
| 2 | Bifidobacterium spp. | 9.18 ± 0.08 | 8.95 ± 0.03 | 8.18 ± 0.15 | + | − |
| Bifidobacterium spp. | 9.99 ± 0.01 | 8.64 ± 0.12 | 8.60 ± 0.05 | + | − | |
| Lactobacillus spp. | 10.17 ± 0.08 | 9.62 ± 0.02 | 9.35 ± 0.04 | + | − | |
| 3 | Bifidobacterium spp. | 9.35 ± 0.04 | 9.11 ± 0.02 | 8.55 ± 0.06 | + | − |
| Bifidobacterium spp. | 8.96 ± 0.03 | 8.55 ± 0.06 | 8.12 ± 0.11 | + | − | |
| Lactobacillus spp. | 9.62 ± 0.02 | 9.40 ± 0.06 | 8.91 ± 0.04 | + | − | |
| 4 | Bifidobacterium spp. | 8.96 ± 0.03 | 8.72 ± 0.02 | 8.38 ± 0.09 | + | − |
| Bifidobacterium spp. | 9.65 ± 0.03 | 9.34 ± 0.09 | 8.85 ± 0.02 | + | − | |
| Lactobacillus spp. | 10.43 ± 0.06 | 9.97 ± 0.01 | 9.85 ± 0.05 | + | − | |
| 5 | Bifidobacterium spp. | 8.89 ± 0.03 | 8.10 ± 0.10 | 8.07 ± 0.03 | + | − |
| Bifidobacterium spp. | 9.96 ± 0.02 | 9.12 ± 0.01 | 8.95 ± 0.01 | + | − | |
| Lactobacillus spp. | 9.48 ± 0.06 | 8.89 ± 0.03 | 8.06 ± 0.07 | + | − | |
| 6 | Bifidobacterium spp. | 9.17 ± 0.09 | 8.95 ± 0.01 | 8.55 ± 0.06 | + | − |
| Bifidobacterium spp. | 9.84 ± 0.04 | 9.40 ± 0.06 | 9.34 ± 0.09 | + | − | |
| Lactobacillus spp. | 9.34 ± 0.09 | 8.91 ± 0.04 | 8.55 ± 0.06 | + | − | |
| 7 | Bifidobacterium spp. | 8.76 ± 0.07 | 8.38 ± 0.09 | 8.07 ± 0.03 | + | − |
| Bifidobacterium spp. | 9.66 ± 0.01 | 9.36 ± 0.08 | 8.85 ± 0.02 | + | − | |
| Lactobacillus spp. | 9.96 ± 0.02 | 9.68 ± 0.06 | 8.77 ± 0.05 | + | − | |
| 8 | Bifidobacterium spp. | 8.85 ± 0.02 | 8.44 ± 0.02 | 8.24 ± 0.08 | + | − |
| Bifidobacterium spp. | 9.41 ± 0.04 | 9.11 ± 0.01 | 8.89 ± 0.03 | + | − | |
| Lactobacillus spp. | 9.12 ± 0.01 | 8.60 ± 0.05 | 8.10 ± 0.10 | + | − | |
| 9 | Bifidobacterium spp. | 8.97 ± 0.01 | 8.64 ± 0.12 | 8.31 ± 0.07 | + | − |
| Bifidobacterium spp. | 8.34 ± 0.04 | 8.21 ± 0.06 | 8.06 ± 0.02 | + | − | |
| Lactobacillus spp. | 8.89 ± 0.03 | 8.55 ± 0.06 | 8.06 ± 0.07 | + | − | |
| 10 | Bifidobacterium spp. | 9.41 ± 0.04 | 8.98 ± 0.01 | 8.74 ± 0.03 | + | − |
| Bifidobacterium spp. | 8.91 ± 0.04 | 8.72 ± 0.02 | 8.60 ± 0.05 | + | − | |
| Lactobacillus spp. | 9.65 ± 0.02 | 9.61 ± 0.08 | 8.63 ± 0.02 | + | − | |
| Animal | Strains | CRO | IPM | ATM | ERI | VAN | CLO | TET | NIT | NOR | CIP |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Bifidobacterium spp. | 29.0 | 27.0 | 9.0 | 32.0 | 23.0 | 32.0 | 37.0 | 25.0 | 23.0 | 26.0 |
| Bifidobacterium spp. | 20.0 | 29.0 | 0.0 | 35.0 | 25.0 | 34.0 | 39.0 | 25.0 | 26.0 | 27.0 | |
| Lactobacillus spp. | 25.0 | 25.0 | 0.0 | 33.0 | 27.0 | 32.0 | 34.0 | 29.0 | 24.0 | 25.0 | |
| 2 | Bifidobacterium spp. | 27.0 | 31.0 | 0.0 | 35.0 | 25.0 | 30.0 | 35.0 | 26.0 | 29.0 | 30.0 |
| Bifidobacterium spp. | 26.0 | 30.0 | 0.0 | 31.0 | 23.0 | 32.0 | 32.0 | 20.0 | 21.0 | 23.0 | |
| Lactobacillus spp. | 25.0 | 34.0 | 0.0 | 32.0 | 29.0 | 30.0 | 32.0 | 28.0 | 25.0 | 28.0 | |
| 3 | Bifidobacterium spp. | 29.0 | 32.0 | 0.0 | 34.0 | 22.0 | 32.0 | 35.0 | 25.0 | 23.0 | 24.0 |
| Bifidobacterium spp. | 28.0 | 33.0 | 0.0 | 33.0 | 26.0 | 32.0 | 40.0 | 22.0 | 24.0 | 26.0 | |
| Lactobacillus spp. | 27.0 | 29.0 | 0.0 | 30.0 | 24.0 | 30.0 | 35.0 | 27.0 | 29.0 | 21.0 | |
| 4 | Bifidobacterium spp. | 25.0 | 29.0 | 11.0 | 35.0 | 25.0 | 31.0 | 27.0 | 24.0 | 27.0 | 27.0 |
| Bifidobacterium spp. | 18.0 | 28.0 | 0.0 | 36.0 | 27.0 | 38.0 | 40.0 | 24.0 | 26.0 | 29.0 | |
| Lactobacillus spp. | 23.0 | 25.0 | 0.0 | 31.0 | 26.0 | 31.0 | 31.0 | 25.0 | 28.0 | 23.0 | |
| 5 | Bifidobacterium spp. | 28.0 | 28.0 | 2.0 | 33.0 | 25.0 | 35.0 | 39.0 | 25.0 | 25.0 | 28.0 |
| Bifidobacterium spp. | 28.0 | 32.0 | 0.0 | 36.0 | 25.0 | 32.0 | 35.0 | 25.0 | 27.0 | 30.0 | |
| Lactobacillus spp. | 30.0 | 30.0 | 0.0 | 34.0 | 24.0 | 32.0 | 32.0 | 25.0 | 26.0 | 24.0 | |
| 6 | Bifidobacterium spp. | 26.0 | 27.0 | 0.0 | 39.0 | 26.0 | 31.0 | 35.0 | 25.0 | 25.0 | 29.0 |
| Bifidobacterium spp. | 28.0 | 32.0 | 0.0 | 35.0 | 25.0 | 32.0 | 36.0 | 24.0 | 27.0 | 30.0 | |
| Lactobacillus spp. | 31.0 | 26.0 | 0.0 | 36.0 | 24.0 | 30.0 | 34.0 | 24.0 | 24.0 | 32.0 | |
| 7 | Bifidobacterium spp. | 20.0 | 30.0 | 0.0 | 36.0 | 26.0 | 37.0 | 40.0 | 26.0 | 26.0 | 30.0 |
| Bifidobacterium spp. | 26.0 | 30.0 | 1.0 | 37.0 | 26.0 | 32.0 | 36.0 | 23.0 | 27.0 | 30.0 | |
| Lactobacillus spp. | 25.0 | 25.0 | 0.0 | 37.0 | 23.0 | 30.0 | 35.0 | 23.0 | 29.0 | 30.0 | |
| 8 | Bifidobacterium spp. | 29.0 | 30.0 | 0.0 | 34.0 | 27.0 | 39.0 | 36.0 | 23.0 | 26.0 | 29.0 |
| Bifidobacterium spp. | 26.0 | 31.0 | 1.0 | 37.0 | 27.0 | 31.0 | 37.0 | 28.0 | 26.0 | 29.0 | |
| Lactobacillus spp. | 27.0 | 32.0 | 0.0 | 37.0 | 25.0 | 30.0 | 39.0 | 27.0 | 27.0 | 32.0 | |
| 9 | Bifidobacterium spp. | 30.0 | 36.0 | 0.0 | 37.0 | 26.0 | 31.0 | 36.0 | 24.0 | 26.0 | 30.0 |
| Bifidobacterium spp. | 16.0 | 30.0 | 0.0 | 39.0 | 27.0 | 35.0 | 41.0 | 21.0 | 26.0 | 29.0 | |
| Lactobacillus spp. | 29.0 | 26.0 | 0.0 | 36.0 | 25.0 | 33.0 | 39.0 | 23.0 | 23.0 | 31.0 | |
| 10 | Bifidobacterium spp. | 25.0 | 28.0 | 8.0 | 36.0 | 25.0 | 31.0 | 37.0 | 25.0 | 27.0 | 30.0 |
| Bifidobacterium spp. | 25.0 | 29.0 | 9.0 | 37.0 | 27.0 | 30.0 | 34.0 | 25.0 | 26.0 | 29.0 | |
| Lactobacillus spp. | 27.0 | 25.0 | 0.0 | 35.0 | 26.0 | 34.0 | 33.0 | 28.0 | 24.0 | 31.0 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Celiberto, L.S.; Pinto, R.A.; Rossi, E.A.; Vallance, B.A.; Cavallini, D.C.U. Isolation and Characterization of Potentially Probiotic Bacterial Strains from Mice: Proof of Concept for Personalized Probiotics. Nutrients 2018, 10, 1684. https://doi.org/10.3390/nu10111684
Celiberto LS, Pinto RA, Rossi EA, Vallance BA, Cavallini DCU. Isolation and Characterization of Potentially Probiotic Bacterial Strains from Mice: Proof of Concept for Personalized Probiotics. Nutrients. 2018; 10(11):1684. https://doi.org/10.3390/nu10111684
Chicago/Turabian StyleCeliberto, Larissa S., Roseli Aparecida Pinto, Elizeu Antonio Rossi, Bruce A. Vallance, and Daniela C. U. Cavallini. 2018. "Isolation and Characterization of Potentially Probiotic Bacterial Strains from Mice: Proof of Concept for Personalized Probiotics" Nutrients 10, no. 11: 1684. https://doi.org/10.3390/nu10111684
APA StyleCeliberto, L. S., Pinto, R. A., Rossi, E. A., Vallance, B. A., & Cavallini, D. C. U. (2018). Isolation and Characterization of Potentially Probiotic Bacterial Strains from Mice: Proof of Concept for Personalized Probiotics. Nutrients, 10(11), 1684. https://doi.org/10.3390/nu10111684





