Personalized Nutrition—Genes, Diet, and Related Interactive Parameters as Predictors of Cancer in Multiethnic Colorectal Cancer Families
Abstract
:1. Introduction
2. Materials and Method
2.1. Study Population and Setting
2.2. Demographic and Genetic Measurements
2.3. Dietary Indexes
2.4. Data Analysis
3. Results
3.1. Characteristics of Study Participants
3.2. Dietary Parameters
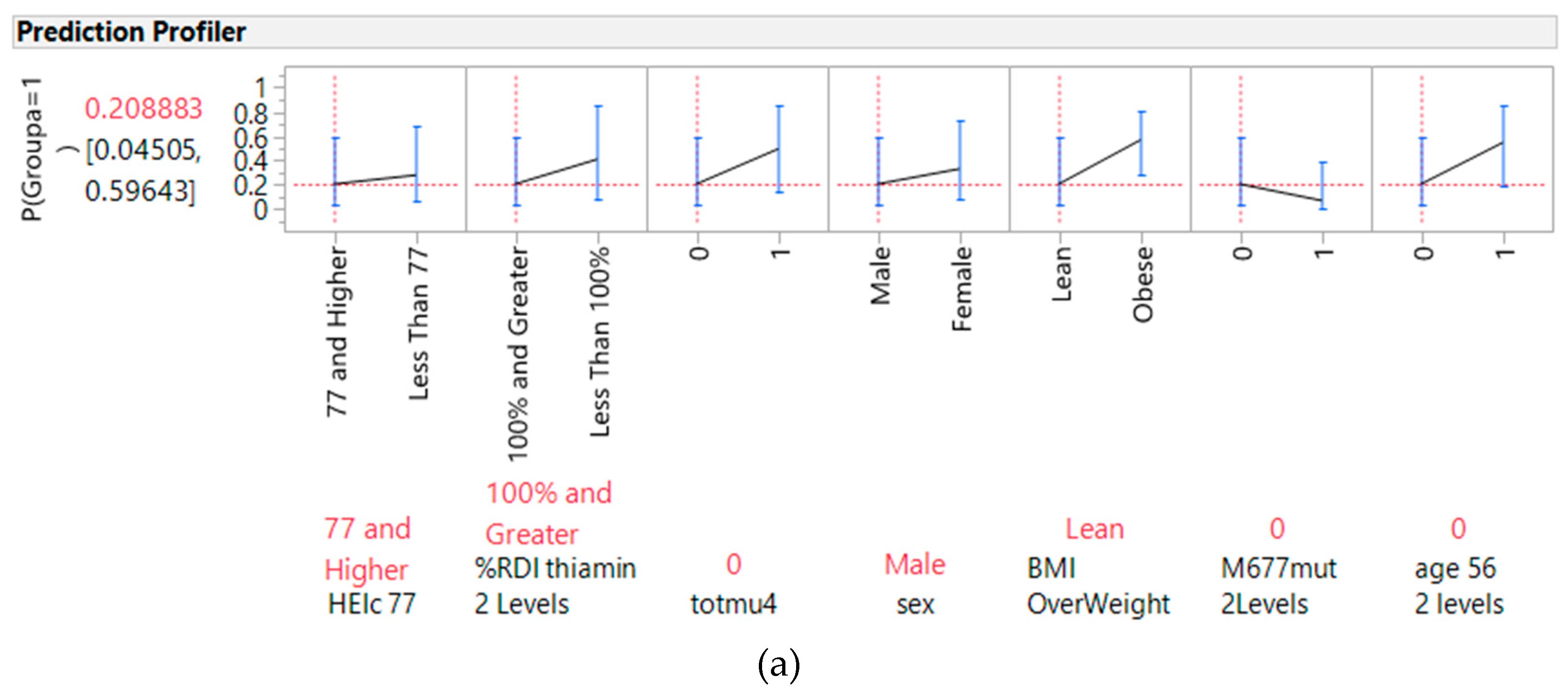
3.3. Most Influential Predictors of Variables of Importance
3.4. Predictors of Cancer from Genes, Diet, and Interactive Parameters
3.5. Predictive Model
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Magalhães, B.; Peleteiro, B.; Lunet, N. Dietary patterns and colorectal cancer: Systematic review and meta-analysis. Eur. J. Cancer Prev. 2012, 21, 15–23. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Jin, X.; Man, C.; Gao, Z.; Wang, X. Meta-analysis of the association between the inflammatory potential of diet and colorectal cancer risk. Oncotarget 2017, 8, 59592–59600. [Google Scholar] [CrossRef] [PubMed]
- Tárraga López, P.J.; Albero, J.S.; Rodríguez-Montes, J.A. Primary and secondary prevention of colorectal cancer. Clin. Med. Insights Gastroenterol. 2014, 7, 33–46. [Google Scholar] [CrossRef]
- Cavicchia, P.P.; Steck, S.E.; Hurley, T.G.; Hussey, J.R.; Ma, Y.; Ockene, I.S.; Hebert, J.R. A new dietary inflammatory index predicts interval changes in serum high-sensitivity C.-reactive protein. J. Nutr. 2009, 139, 2365–2372. [Google Scholar] [CrossRef] [PubMed]
- Shivappa, N.; Steck, S.E.; Hurley, T.G.; Hussey, J.R.; Hebert, J.R. Designing and developing a literature-derived, population-based dietary inflammatory index. Public. Health Nutr. 2014, 17, 1689–1696. [Google Scholar] [CrossRef] [PubMed]
- Johnson, C.M.; Wei, C.; Ensor, J.E.; Smolenski, D.J.; Amos, C.I.; Levin, B.; Berry, D.A. Meta-analyses of colorectal cancer risk factors. Cancer Causes Control 2013, 24, 1207–1222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WCRF-AICR Continuous Update Project. Diet, Nutrition, Physical Activity and Colorectal Cancer. 2017. World Cancer Research Fund International/American Institute for Cancer Research. Available online: http://www.aicr.org/continuous-update-project/reports/colorectal-cancer-2017-report.pdf (accessed on 31 March 2018).
- Holden, D.J.; Harris, R.; Porterfield, D.S.; Jones, D.E.; Morgan, L.C.; Reuland, D.; Gilehrist, M.; Viswanathan, M.; Lohr, K.N.; Lynda-MdDonald, B. Enhancing the use of quality of colorectal cancer screening. Evid. Rep. Technol. Assess. 2010, 190, 1–195. [Google Scholar]
- Visser, A.; Vrieling, A.; Murugesu, L.; Hoogerbrugge, N.; Kampman, E.; Hoedjes, M. Determinants of adherence to recommendations for cancer prevention among Lynch Syndrome mutation carriers: A qualitative exploration. PLoS ONE 2017, 12, e0178205. [Google Scholar] [CrossRef] [PubMed]
- Campbell, P.T.; Curtin, K.; Ulrich, C.M.; Samowitz, W.S.; Bigler, J.; Velicer, C.M.; Caan, B.; Potter, J.D.; Slattery, M.L. Mismatch repair polymorphisms and risk of colon cancer, tumour microsatellite instability and interactions with lifestyle factors. Gut 2009, 58. [Google Scholar] [CrossRef] [PubMed]
- Bishehsari, F.; Mahdavinia, M.; Vacca, M.; Malekzadeh, R.; Mariani-Costantini, R. Epidemiological transition of colorectal cancer in developing countries: Environmental factors, molecular pathways, and opportunities for prevention. World J. Gastroenterol. 2014, 20, 6055–6072. [Google Scholar] [CrossRef] [PubMed]
- Shiao, S.P.; Yu, C.H. Meta-Prediction of MTHFR Gene polymorphism mutations and associated risk for colorectal cancer. Biol. Res. Nurs. 2016, 18, 357–369. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.-M.; Chen, Z.-F.; Young, L.; Shiao, S.P.K. Meta-prediction of the effects of Methylenetetrahydrofolate reductase gene polymorphisms and air pollution on risk of Alzheimer’s disease. Int. J. Environ. Res. Public Health 2017, 14, 63. [Google Scholar] [CrossRef] [PubMed]
- Lien, S.-Y.A.; Young, L.; Gau, B.-S.; Shiao, S.P.K. Meta-prediction of MTHFR gene polymorphism-mutations, air pollution, and risks of leukemia among world populations. Oncotarget 2017, 8, 4387–4398. [Google Scholar] [CrossRef] [PubMed]
- Gonzales, M.C.; Yu, P.-J.; Shiao, S.P.K. Meta-prediction of MTHFR gene polymorphism-mutations and air pollution as risk factors for breast cancer. Nurs. Res. 2017, 66, 152–163. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.L.; Yang, H.L.; Shiao, S.P.K. Meta-prediction of MTHFR gene polymorphisms and air pollution on the risk of hypertensive disorders in pregnancy worldwide. Int. J. Environ. Res. Public Health 2018, 15, 326. [Google Scholar] [CrossRef] [PubMed]
- Cadet, J.; Douki, T.; Ravanat, J.L. Oxidatively generated base damage to cellular DNA. Free Radic. Biol. Med. 2010, 49, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Hair, J.M.; Terzoudi, G.I.; Hatzi, V.I.; Lehockey, K.A.; Srivastava, D.; Wang, W.; Pantelias, G.E.; Georgakilas, A.G. BRCA1 role in the mitigation of radiotoxicity and chromosomal instability through repair of clustered DNA lesions. Chem. Biol. Interact. 2010, 188, 350–358. [Google Scholar] [CrossRef] [PubMed]
- Baccarelli, A.; Cassano, P.A.; Litonjua, A.; Park, S.K.; Suh, H.; Sparrow, D.; Vokonas, P.; Schwartz, J. Cardiac autonomic dysfunction: Effects from particulate air pollution and protection by dietary methyl nutrients and metabolic polymorphisms. Circulation 2008, 117, 1802–1809. [Google Scholar] [CrossRef] [PubMed]
- Taioli, E.; Garza, M.A.; Ahn, Y.O.; Bishop, D.T.; Bost, J.; Budai, B.; Chen, K.; Gemignani, F.; Keku, T.; Lima, C.S.; et al. Meta- and pooled analyses of the methylenetetrahydrofolate reductase (MTHFR) C677T polymorphism and colorectal cancer: A HuGE-GSEC review. Am. J. Epidemiol. 2009, 170, 1207–1221. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, D.A.; Stern, S.J.; Matok, I.; Moretti, M.E.; Sarkar, M.; Adams-Webber, T.; Koren, G. Folate intake, MTHFR polymorphisms, and the risk of colorectal cancer: A systematic review and meta-analysis. J. Cancer Epidemiol. 2012, 2012, 952508. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wen, X.; Wu, W.; Guo, Y.; Cui, W. Elevated Homocysteine Level and Folate Deficiency Associated with Increased Overall Risk of Carcinogenesis: Meta-Analysis of 83 Case-Control Studies Involving 35,758 Individuals. PLoS ONE 2015, 10, e0123423. [Google Scholar] [CrossRef] [PubMed]
- Anderson, O.S.; Sant, K.E.; Dolinoy, D.C. Nutrition and epigenetics: An interplay of dietary methyl donors, one-carbon metabolism and DNA methylation. J. Nutr. Biochem. 2012, 23, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Ryan-Harshman, M.; Aldoori, W. Diet and colorectal cancer: Review of the evidence. Can. Fam. Phys. 2007, 53, 1913–1920. [Google Scholar]
- Turati, F.; Bravi, F.; Di Maso, M.; Bosetti, C.; Polesel, J.; Serraino, D.; Dalmartello, M.; Giacosa, A.; Montella, M.; Tavani, A.; et al. Adherence to the World Cancer Research Fund/American Institute for Cancer Research recommendations and colorectal cancer risk. Eur. J. Cancer 2017, 85, 86–94. [Google Scholar] [CrossRef] [PubMed]
- Tabung, F.K.; Brown, L.S.; Fung, T.T. Dietary Patterns and Colorectal Cancer Risk: A Review of 17 Years of Evidence (2000–2016). Curr. Colorectal Cancer Rep. 2017, 13, 440–454. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.R.; Abar, L.; Chan, D.S.M.; Vingeliene, S.; Polemiti, E.; Stevens, C.; Greenwood, D.; Norat, T. Foods and beverages and colorectal cancer risk: A systematic review and meta-analysis of cohort studies, an update of the evidence of the WCRF-AICR Continuous Update Project. Ann. Oncol. 2017, 28, 1788–1802. [Google Scholar] [CrossRef] [PubMed]
- Klai, S.; Fekih-Mrissa, N.; El Housaini, S.; Kaabechi, N.; Nsiri, B.; Rachdi, R.; Gritli, N. Association of MTHFR A1298C polymorphism (but not of MTHFR C677T) with elevated homocysteine levels and placental vasculopathies. Blood Coagul. Fibrinolysis 2011, 22, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Yang, B.; Liu, Y.; Li, Y.; Fan, S.; Zhi, X.; Lu, X.; Wang, D.; Zheng, Q.; Wang, Y.; Wang, Y. Geographical distribution of MTHFR C677T, A1298C and MTRR A66G gene polymorphisms in China: Findings from 15357 adults of Han nationality. PLoS ONE 2013, 8, e57917. [Google Scholar] [CrossRef] [PubMed]
- Frosst, P.; Blom, H.J.; Milos, R.; Goyette, P.; Sheppard, C.A.; Matthews, R.G.; Boers, G.J.; den Heijer, M.; Kluijtmans, L.A.; van den Heuvel, L.P.; et al. A candidate genetic risk factor for vascular disease: A common mutation in methylenetetrahydrofolate reductase. Nat. Genet. 1995, 10, 111–113. [Google Scholar] [CrossRef] [PubMed]
- Yaliwal, L.V.; Desai, R.M. Methylenetetrahydrofolate reductase mutations, a genetic cause for familial recurrent neural tube defects. Indian J. Hum. Genet. 2012, 18, 122–124. [Google Scholar] [CrossRef] [PubMed]
- Ravegnini, G.; Zolezzi Moraga, J.M.; Maffei, F.; Musti, M.; Zenesini, C.; Simeon, V.; Sammarini, G.; Festi, D.; Hrelia, P.; Angelini, S. Simultaneous analysis of SEPT9 promoter methylation status, micronuclei frequency, and folate-related gene polymorphisms: The potential for a novel blood-based colorectal cancer biomarker. Int. J. Mol. Sci. 2015, 16, 28486–28497. [Google Scholar] [CrossRef] [PubMed]
- Selhub, J.; Rosenberg, I.H. Excessive folic acid intake and relation to adverse health outcome. Biochimie 2016, 126, 71–78. [Google Scholar] [CrossRef] [PubMed]
- Cheng, T.Y.; Makar, K.W.; Neuhouser, M.L.; Miller, J.W.; Song, X.; Brown, E.C.; Beresford, S.A.; Zheng, Y.; Poole, E.M.; Galbraith, R.L.; et al. Folate-mediated one-carbon metabolism genes and interactions with nutritional factors on colorectal cancer risk: Women’s Health Initiative Observational Study. Cancer 2015, 121, 3684–3691. [Google Scholar] [CrossRef] [PubMed]
- Li, W.X.; Dai, S.X.; Zheng, J.J.; Liu, J.Q.; Huang, J.F. Homocysteine metabolism gene polymorphisms (MTHFR C677T, MTHFR A1298C, MTR A2756G and MTRR A66G) jointly elevate the risk of folate deficiency. Nutrients 2015, 7, 6670–6687. [Google Scholar] [CrossRef] [PubMed]
- Lucock, M.; Yates, Z.; Martin, C.; Choi, J.H.; Beckett, E.; Boyd, L.; LeGras, K.; Ng, X.; Skinner, V.; Wai, R.; et al. Methylation diet and methyl group genetics in risk for adenomatous polyp occurrence. BBA Clin. 2015, 3, 107–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, D.; Mei, Q.; Luo, H.; Tang, B.; Yu, P. The polymorphisms in methylenetetrahydrofolate reductase, methionine synthase, methionine synthase reductase, and the risk of colorectal cancer. Int. J. Biol. Sci. 2012, 8, 819–830. [Google Scholar] [CrossRef] [PubMed]
- Inamura, K. Colorectal Cancers: An Update on Their Molecular Pathology. Cancers 2018, 10, 26. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Nishihara, R.; Qian, Z.R.; Tabung, F.K.; Nevo, D.; Zhang, X.; Song, M.; Cao, Y.; Mima, K.; Masugi, Y.; et al. Association Between Inflammatory Diet Pattern and Risk of Colorectal Carcinoma Subtypes Classified by Immune Responses to Tumor. Gastroenterology 2017, 153. [Google Scholar] [CrossRef] [PubMed]
- How AICR Recommendations Cuts Colorectal Cancer Risk for Both Men and Women. Available online: http://www.aicr.org/cancer-research-update/2016/11_02/cru-how-AICR-recommendations-cuts-colorectal-cancer-risk-for-men-and-women.html (accessed on 4 April 2018).
- Hastert, T.A.; White, E. Association between meeting the WCRF/AICR cancer prevention recommendations and colorectal cancer incidence: Results from the VITAL cohort. Cancer Causes Control 2016, 27, 1347–1359. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.-Q.; Li, F.; Dong, R.-H.; Chen, J.-S.; He, G.-S.; Li, S.G.; Chen, B. The Development of a Chinese Healthy Eating Index and Its Application in the General Population. Nutrients 2017, 9, 977. [Google Scholar] [CrossRef] [PubMed]
- United States (U.S.) Department of Health and Human Services and U.S. Department of Agriculture. 2015–2020 Dietary Guidelines for Americans. 8th Edition; December 2015. Available online: http://health.gov/dietaryguidelines/2015/guidelines/ (accessed on 31 March 2018).
- United States Department of Agriculture (USDA). Healthy Eating Index (HEI). November 2016. Available online: https://www.cnpp.usda.gov/healthyeatingindex (accessed on 31 March 2018).
- National Institute of Health (NIH). Nutrient Recommendations: Dietary Reference Intakes (DRI). (n.d.). Available online: https://ods.od.nih.gov/Health_Information/Dietary_Reference_Intakes.aspx (accessed on 31 March 2018).
- Panizza, C.E.; Shvetsov, Y.B.; Harmon, B.E.; Wilkens, L.R.; Le Marchand, L.; Haiman, C.; Reedy, J.; Boushey, C.J. Testing the Predictive Validity of the Healthy Eating Index-2015 in the Multiethnic Cohort: Is the Score Associated with a Reduced Risk of All-Cause and Cause-Specific Mortality? Nutrients 2018, 10, 452. [Google Scholar] [CrossRef] [PubMed]
- Djuric, Z.; Severson, R.K.; Kato, I. Association of dietary quercetin with reduced risk of proximal colon cancer. Nutr. Cancer 2012, 64, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Miller, P.E.; Lazarus, P.; Lesko, S.M.; Muscat, J.E.; Harper, G.; Cross, A.J.; Sinha, R.; Ryczak, K.; Escobar, G.; Mauger, D.T.; et al. Diet index-based and empirically derived dietary patterns are associated with colorectal cancer risk. J. Nutr. 2010, 140, 1267–1273. [Google Scholar] [CrossRef] [PubMed]
- Reedy, J.; Wirfält, E.; Flood, A.; Mitrou, P.N.; Krebs-Smith, S.M.; Kipnis, V.; Midthune, D.; Leitzmann, M.; Hollenbeck, A.; Schatzkin, A.; et al. Comparing 3 dietary pattern methods—Cluster analysis, factor analysis, and index analysis—With colorectal cancer risk: The NIH-AARP Diet and Health Study. Am. J. Epidemiol. 2010, 171, 479–487. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.; Chen, K.; Ma, X.; Li, Q.; Yu, W.; Shu, G.; Yao, K. Diets, polymorphisms of Methylenetetrahydro- folate reductase, and the susceptibility of colon cancer and rectal cancer. Cancer Detect. Prev. 2005, 29, 146–154. [Google Scholar] [CrossRef] [PubMed]
- Guerreiro, C.S.; Cravo, M.L.; Brito, M.; Vidal, P.M.; Fidalgo, P.O.; Leitão, C.N. The D1822V APC polymorphism interacts with fat, calcium, and fiber intakes in modulating the risk of colorectal cancer in Portuguese persons. Am. J. Clin. Nutr. 2007, 85, 1592–1597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- May-Wilson, S.; Sud, A.; Law, P.J.; Palin, K.; Tuupanen, S.; Gylfe, A.; Hänninen, U.A.; Cajuso, T.; Tanskanen, T.; Kondelin, J.; et al. Pro-inflammatory fatty acid profile and colorectal cancer risk: A Mendelian randomization analysis. Eur. J. Cancer 2017, 84, 228–238. [Google Scholar] [CrossRef] [PubMed]
- De Vogel, S.; Wouters, K.A.; Gottschalk, R.W.; van Schooten, F.J.; de Goeij, A.F.; de Bruïne, A.P.; Goldbohm, R.A.; van den Brandt, P.A.; van Engeland, M.; Weijenberg, M.P. Dietary methyl donors, methyl metabolizing enzymes, and epigenetic regulators: Diet-gene interactions and promoter CpG island hypermethylation in colorectal cancer. Cancer Causes Control 2011, 22, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharp, L.; Little, J.; Brockton, N.T.; Cotton, S.C.; Masson, L.F.; Haites, N.E.; Cassidy, J. Polymorphisms in the methylenetetrahydrofolate reductase (MTHFR) gene, intakes of folate and related B vitamins and colorectal cancer: A case-control study in a population with relatively low folate intake. Br. J. Nutr. 2008, 99, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Shiao, S.P.K.; Grayson, J.; Yu, C.H.; Wasek, B.; Bottiglieri, T. Gene Environment Interactions and Predictors of Colorectal Cancer in Family-Based, Multi-Ethnic Groups. J. Pers. Med. 2018, 8, 10. [Google Scholar] [CrossRef] [PubMed]
- Simidjievski, N.; Todorovski, L.; Džeroski, S. Modeling dynamic systems with efficient ensembles of process-based models. PLoS ONE 2016, 11, e0153507. [Google Scholar] [CrossRef] [PubMed]
- Khalilia, M.; Chakraborty, S.; Popescu, M. Predicting disease risks from highly imbalanced data using random forest. BMC Med. Inform. Decis. Mak. 2011, 11, 51. [Google Scholar] [CrossRef] [PubMed]
- Islam, M.M.; Yao, X.; Shahriar Nirjon, S.M.; Islam, M.A.; Murase, K. Bagging and boosting negatively correlated neural networks. IEEE Trans. Syst. Man Cybern. B Cybern. 2008, 38, 771–784. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.W. New ensemble machine learning method for classification and prediction on gene expression data. Conf. Proc. IEEE Eng. Med. Biol. Soc. 2006, 1, 3478–3481. [Google Scholar] [PubMed]
- Friedman, J.; Hastie, T.; Tibshirani, R. Regularization paths for generalized linear models via coordinate descent. J. Stat. Softw. 2010, 33, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Song, L.; Langfelder, P.; Horvath, S. Random generalized linear model: A highly accurate and interpretable ensemble predictor. BMC Bioinform. 2013, 14, 5. [Google Scholar] [CrossRef] [PubMed]
- Witten, D.M.; Tibshirani, R. Covariance-regularized regression and classification for high-dimensional problems. J. R. Stat. Soc. Ser. B Stat. Methodol. 2009, 71, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y. Elastic Net for Cox’s proportional hazards model with a solution path algorithm. Stat. Sin. 2012, 22, 27–294. [Google Scholar] [CrossRef] [PubMed]
- Krist, A.H.; Glenn, B.A.; Glasgow, R.E.; Balasubramanian, B.A.; Chambers, D.A.; Fernandez, M.E.; Heurtin-Roberts, S.; Kessler, R.; Ory, M.G.; Phillips, S.M.; et al. Designing a valid randomized pragmatic primary care implementation trial: The my own health report (MOHR) project. Implement. Sci. 2013, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Center for Disease Control and Prevention (CDC). 2012. National Health and Nutrition Examination Survey. Center for Disease Control and Prevention. Available online: http://www.cdc.gov/nchs/nhanes/nhanes_questionnaires.htm (accessed on 15 December 2012).
- National Coalition for Health Professional Education in Genetics. Family History Educational Aids. NCHPEG. Available online: http://www.nchpeg.org/index.php?option=com_content&view=article&id=145&Itemid=64 (accessed on 10 October 2016).
- Lievers, K.J.; Boers, G.H.; Verhoef, P.; Heijer, M.; Kluijtmans, L.A.; Put, N.M.; Trijbels, F.J.; Blom, H.J. A second common variant in the methylenetetrahydrofolate reductase (MTHFR) gene and its relationship to MTHFR enzyme activity, homocysteine, and cardiovascular disease risk. J. Mol. Med. 2001, 79, 522–528. [Google Scholar] [CrossRef] [PubMed]
- Wren, M.E.; Shirtcliff, E.A.; Drury, S.S. Not all biofluids are created equal: Chewing over salivary diagnostics and the epigenome. Clin. Ther. 2015, 37, 529–539. [Google Scholar] [CrossRef] [PubMed]
- Torres-Sánchez, L.; Chen, J.; Díaz-Sánchez, Y.; Palomeque, C.; Bottiglieri, T.; López-Cervantes, M.; López-Carrillo, L. Dietary and genetic determinants of homocysteine levels among Mexican women of reproductive age. Eur. J. Clin. Nutr. 2006, 60, 691–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Neuhouser, M.L.; Kristal, A.R.; McLerran, D.; Patterson, R.E.; Atkinson, J. Validity of short food frequency questionnaires used in cancer chemoprevention trials: Results from the Prostate Cancer Prevention Trial. Cancer Epidemiol. Biomark. Prev. 1999, 8, 721–725. [Google Scholar]
- Patterson, R.E.; Kristal, A.R.; Tinker, L.F.; Carter, R.A.; Bolton, M.P.; Agurs-Collins, T. Measurement characteristics of the Women’s Health Initiative food frequency questionnaire. Ann. Epidemiol. 1999, 9, 178–187. [Google Scholar] [CrossRef]
- Schakel, S.F.; Sievert, Y.A.; Buzzard, I.M. Sources of data for developing and maintaining a nutrient database. J. Am. Diet. Assoc. 1988, 88, 1268–1271. [Google Scholar] [PubMed]
- Harnack, L.; Lee, S.; Schakel, S.F.; Duval, S.; Luepker, R.V.; Arnett, D.K. Trends in the trans-fatty acid composition of the diet in a metropolitan area: The Minnesota Heart Survey. J. Am. Diet Assoc. 2003, 103, 1160–1166. [Google Scholar] [CrossRef]
- Young, L.; Shiao, S.P.K. Validation of Methyl Donors between Two Food Measurements in a Colorectal Cancer Study. Available online: http://www.aacr.org/Documents/AACR2017_Proceedings.pdf (accessed on 20 May 2018).
- Health Information. Nutrient recommendations: Dietary reference intakes. U. S. Department of Health and Human Services, National Institutes of Health, Office of Dietary Supplements. Available online: https://ods.od.nih.gov/Health_Information/Dietary_Reference_Intakes.aspx (accessed on 19 June 2018).
- Zhao, L.P.; Le Marchand, L. An analytical method for assessing patterns of familial aggregation in case-control studies. Genet. Epidemiol. 1992, 9, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Grayson, J.; Gardner, S.; Stephens, M. Building Better Models with JMP® Pro. 2015; SAS Press: Cary, NC, USA, 2015. [Google Scholar]
- Klimberg, R.; McCullough, B.D. Fundamentals of Predictive Analytics with JMP, 2nd ed.; SAS Press: Cary, NC, USA, 2016. [Google Scholar]
- Yu, C.H. Resampling: A Conceptual and Procedural Introduction. In Best Practices in Quantitative Methods; Osborne, J., Ed.; Sage Publications: Thousand Oaks, CA, USA, 2007; pp. 283–298. [Google Scholar]
- Faraway, J.J. Extending the Linear Model with R: Generalized Linear, Mixed Effects and Nonparametric Regression Models (Texts in Statistical Science); Chapman & Hall/CRC: Boca Raton, FL, USA, 2005. [Google Scholar]
- Meir, R.; Rätsch, G. An Introduction to Boosting and Leveraging. In Advanced Lectures on Machine Learning. Lecture Notes in Computer Science; Mendelson, S., Smola, A.J., Eds.; Springer: Berlin, Germany, 2003; Volume 2600, pp. 118–183. [Google Scholar] [CrossRef]
- Zaman, M.F.; Hirose, H. Classification performance of bagging and boosting type ensemble methods with small training sets. New Gener. Comput. 2011, 29, 277–292. [Google Scholar] [CrossRef]
- Burnham, K.P.; Anderson, D.R. Model Selection and Multimodel Inference: A Practical Information-Theoretic Approach, 2nd ed.; Springer-Verlag: New York, NY, USA, 2002. [Google Scholar]
- Burnham, K.P.; Anderson, D.R. Multimodel inference: Understanding AIC and BIC in model selection. Sociol. Meth. Res. 2004, 33, 261–304. [Google Scholar] [CrossRef]
- Yang, Y. Can the strengths of AIC and BIC be shared? Biometrika 2005, 92, 937–950. [Google Scholar] [CrossRef]
- Akaike, H. A new look at the statistical model identification. IEEE Trans. Autom. Control 1974, 19, 716–723. [Google Scholar] [CrossRef]
- Akaike, H. A Bayesian analysis of the minimum AIC procedure. Ann. Inst. Stat. Math. 1978, 30, 9–14. [Google Scholar] [CrossRef]
- SAS Institute Inc. JMP 13 Fitting Linear Models, 2nd ed.; SAS Institute Inc.: Cary, NC, USA, 2016. [Google Scholar]
- Cheng, H.; Garrick, D.J.; Fernando, R.L. Efficient strategies for leave-one-out cross validation for genomic best linear unbiased prediction. J. Anim. Sci. Biotechnol. 2017, 8, 38. [Google Scholar] [CrossRef] [PubMed]
- Zou, H.; Hastie, T. Regularization and variable selection via the elastic net. J. R. Stat. Soc. B 2005, 67, 301–320. Available online: https://web.stanford.edu/~hastie/Papers/B67.2%20(2005)%20301-320%20Zou%20&%20Hastie.pdf (accessed on 30 May 2018). [CrossRef]
- Efron, B.; Hastie, T.; Johnstone, I.; Tibshirani, R. Least angle regression. Ann. Stat. 2004, 32, 407–499. Available online: http://statweb.stanford.edu/~tibs/ftp/lars.pdf (accessed on 30 May 2018).[Green Version]
- SAS Institute. Overview of the Generalized Regression Personality; SAS Institute: Cary, NC, USA, 2017; Available online: https://www.jmp.com/support/help/14/overview-of-the-generalized-regression-personali.shtml (accessed on 30 May 2018).
- Crotty, M.; Barker, C. Penalizing Your Models: An Overview of the Generalized Regression Platform; SAS Institute: Cary, NC, USA, 2014; Available online: https://analytics.ncsu.edu/sesug/2014/RIV-08.pdf (accessed on 30 May 2018).
- Tibshirani, R. Regression shrinkage and selection via the lasso: A retrospective. J. R. Stat. Soc. Ser. B Stat. Methodol. 2011, 73, 273–282. [Google Scholar] [CrossRef]
- Gonzales, G.B.; de Saeger, S. Elastic net regularized regression for time-series analysis of plasma metabolome stability under sub-optimal freezing condition. Sci. Rep. 2018, 8, 3659. [Google Scholar] [CrossRef] [PubMed]
- Shmueli, G. To Explain or to Predict? Stat. Sci. 2010, 25, 289–310. [Google Scholar] [CrossRef]
- Diaconis, P.; Efron, B. Computer-intensive methods in statistics. Sci. Am. 1983, 248, 116–130. [Google Scholar] [CrossRef]
- Cenit, M.C.; Olivares, M.; Codoñer-Franch, P.; Sanz, Y. Intestinal microbiota and celiac disease: Cause, consequence or co-evolution? Nutrients 2015, 7, 6900–6923. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.; Liu, Y.; Yin, L.; Zhang, K. Confidence intervals and regions for the lasso by using stochastic variational inequality techniques in optimization. J. R. Stat. Soc. 2017, 79, 1–23. Available online: https://www.unc.edu/~zhangk/2017%20Confidence%20Intervals%20and%20Regions%20for%20the%20LASSO%20Using%20Stochastic%20Variational%20Inequality%20Techniques%20in%20Optimization.pdf (accessed on 30 May 2018). [CrossRef]




| Parameters | Control (N = 53) n (%) | Cancer (N = 53) n (%) | p | |
|---|---|---|---|---|
| Gender | Male | 14 (26%) | 25 (47%) | 0.027 |
| Female | 39 (74%) | 28 (53%) | ||
| Age (years) | Mean ± SD | 47 ± 17 | 61 ± 11 | <0.0001 |
| Range | 18–80 | 37–79 | ||
| Ethnicity | Asian | 22 (42%) | 18 (34%) | 0.88 |
| Caucasian | 16 (30%) | 18 (34%) | ||
| Hispanic | 11 (21%) | 12 (23%) | ||
| African American | 4 (7.5%) | 5 (9.4%) | ||
| BMI status | Obese | 11 (21%) | 15 (28%) | 0.37 |
| Alcohol drinker | Yes | 25 (47%) | 32 (60%) | 0.17 |
| Smoker | Yes | 5 (9.4%) | 4 (7.6%) | 0.73 |
| Total polymorphisms (0–6) | ≥4 | 16 (30%) | 27 (51%) | 0.03 |
| Parameters (Amount, Score) | Control (N = 53) Mean ± SD | Cancer (N = 53) Mean ± SD | p |
|---|---|---|---|
| Calorie (per day) | 1640 ± 1021 | 1603 ± 784 | 0.84 |
| Total Fruit (≥0.8 cup, 5 points) | 1.6 ± 1.5 | 1.6 ± 1.4 | 0.98 |
| Whole Fruit (≥0.4 cup, 5 points) | 1.2 ± 1.1 | 1.2 ± 1.0 | 0.95 |
| Vegetables (≥1.1 cups, 5 points) | 1.6 ± 1.1 | 1.5 ± 1.3 | 0.86 |
| Dark Green (≥0.4 cup, 5 points) | 0.9 ± 0.7 | 0.8 ± 0.7 | 0.66 |
| Total Grains (≥3 oz, 5 points) | 4.6 ± 3.3 | 4.6 ± 2.7 | 0.95 |
| Whole Grains (≥1.5 oz, 5 points) | 1.5 ± 1.4 | 2.0 ± 1.9 | 0.16 |
| Dairy (≥1.3 cups, 10 points) | 1.8 ± 4.3 | 1.0 ± 1.2 | 0.19 |
| Protein (≥2.5 oz, 10 points) | 6.3 ± 5.0 | 5.3 ± 3.2 | 0.22 |
| Oil and Nuts (≥12 g. 10 points) | 37 ± 25 | 36 ± 19 | 0.72 |
| Saturated Fat (g, ≤8% energy) | 18 ± 9.6 | 19 ± 13 | 0.82 |
| Sodium (≤1.1 g. 10 points) | 3.3 ± 2.1 | 3.0 ± 1.8 | 0.34 |
| Empty Calories (≤19% energy) | 348 ± 235 | 353 ± 216 | 0.91 |
| HEI score (≤50, 51–79, ≥80) | 75 ± 10 | 76 ± 8.3 | 0.43 |
| HEI score (≥77) | 24 (45%) | 30 (57%) | 0.24 |
| HEI score (≥80) | 20 (38%) | 21 (40%) | 0.84 |
| Parameters, Unit, RDI | Control (N = 53) n (%) | Cancer (N = 53) n (%) | p | |
|---|---|---|---|---|
| Carbohydrates, g, 45–65% calorie | ≥45% | 38 (73%) | 37 (70%) | 0.83 |
| Protein, g, 10–35% calorie | ≥20% | 21 (40%) | 17 (32%) | 0.42 |
| Total Fat, g, 20–35% calorie | <35% | 35 (66%) | 34 (64%) | 0.84 |
| Saturated Fat, g, <10% calorie | <10% | 28 (53%) | 23 (43%) | 0.33 |
| Cholesterol, <300 mg | <100% | 39 (74%) | 39 (74%) | 1.00 |
| Sodium, <2300 mg | <100% | 19 (36%) | 21 (40%) | 0.69 |
| Fiber, ≥25 g | ≥100% | 9 (17%) | 7 (13%) | 0.59 |
| Total Folate, 400 mcg | ≥100% | 13 (25%) | 21 (40%) | 0.10 |
| Vitamin B1 (Thiamine), 1.1 mg | ≥100% | 30 (57%) | 35 (66%) | 0.32 |
| Vitamin B2 (Riboflavin), 1.1 mg | ≥100% | 37 (70%) | 41 (77%) | 0.38 |
| Vitamin B6, 1.3 mg | ≥100% | 35 (66%) | 33 (62%) | 0.69 |
| Vitamin B12, 2.4 mcg | <150% | 25 (47%) | 19 (36%) | 0.24 |
| Niacin, 14 mg | ≥100% | 35 (66%) | 37 (70%) | 0.68 |
| Calcium, 1000 mg | ≥75% | 24 (45%) | 22 (42%) | 0.70 |
| Magnesium, 320 mg | ≥75% | 27 (51%) | 25 (47%) | 0.70 |
| Iron, 8 mg | ≥100% | 19 (36%) | 25 (47%) | 0.24 |
| Zinc, 8 mg | ≥100% | 27 (51%) | 26 (49%) | 0.85 |
| Methionine, 13 mg/kg | <150% | 22 (42%) | 23 (43%) | 0.84 |
| Term | Number of Splits | G2 | Column Contribution | Portion |
|---|---|---|---|---|
| Age (≤ or >56 years) | 61 | 3.12 |  | 0.28 |
| Gender | 44 | 1.35 |  | 0.12 |
| Total Polymorphisms (≥4) | 49 | 1.30 |  | 0.11 |
| Total Vegetable Intake 10 oz | 43 | 1.24 |  | 0.11 |
| Total Folate Intake 100% | 49 | 1.05 |  | 0.09 |
| HEI 77 | 42 | 0.72 |  | 0.06 |
| Overweight BMI | 44 | 0.70 |  | 0.06 |
| Vitamin B12 150% | 35 | 0.66 |  | 0.06 |
| Thiamine 100% | 38 | 0.65 |  | 0.06 |
| MTHFR 677 Polymorphism | 39 | 0.52 |  | 0.05 |
| Parameters | Logistic Regression with Validation | Generalized Regression Elastic Net Model | |||||
|---|---|---|---|---|---|---|---|
| AICc Validation | Leave-One-Out Validation | ||||||
| Estimate (95% CI) | p (X2) | Estimate (95% CI) | p (X2) | Estimate (95% CI) | p (X2) | ||
| (Intercept) | −0.59 (−2.5, 1.3) | 0.54 | −0.56 (−2.2, 1.1) | 0.51 | −1.02 (−2.5, 0.49) | 0.19 | |
| Thiamine * HEI | −3.67 (−6.6, −0.79) | 0.01 | −2.80 (−5.1, −0.51) | 0.02 | −2.73 (−4.9, −0.56) | 0.01 | |
| Gender * BMI Overweight | −2.4 (−5.1, 0.15) | 0.06 | −3.49 (−5.6, −1.4) | 0.001 | −3.36 (−5.2, −1.5) | 0.0003 | |
| Gender | 1.86 (0.07, 3.6) | 0.04 | 2.50 (1.1, 3.9) | 0.0005 | 2.53 (1.3, 3.8) | <0.0001 | |
| Total Polymorphisms | −0.95 (−2.2, 0.33) | 0.15 | −1.54 (−2.7, −0.35) | 0.011 | −1.65 (−2.8, −0.53) | 0.004 | |
| HEI | 2.73 (0.41, 5.1) | 0.02 | 2.53 (0.35, 4.7) | 0.02 | 2.52 (0.49, 4.6) | 0.02 | |
| Thiamine | 1.75 (−0.08, 3.6) | 0.06 | 1.71 (0.18, 3.2) | 0.03 | 1.86 (0.42, 3.3) | 0.011 | |
| Age | −1.32 (−2.6, −0.08) | 0.04 | −1.48 (−2.5, −0.51) | 0.003 | −1.35 (−2.3, −0.41) | 0.005 | |
| Vegetable 10 oz | 1.20 (−0.19, 2.6) | 0.09 | 1.03 (−0.07, 2.1) | 0.07 | 1.02 (0.03, 2.0) | 0.04 | |
| MTHFR 677 * BMI | 1.42 (−1.2, 4.0) | 0.29 | 2.02 (−0.07, 4.1) | 0.06 | 1.43 (−0.29, 3.2) | 0.10 | |
| MTHFR 677 | −0.63 (−2.4, 1.1) | 0.48 | 0.63 (−1.9, 0.63) | 0.33 | −0.14 (−1.3, 1.1) | 0.82 | |
| BMI Overweight | −0.36 (−2.3, 1.5) | 0.71 | −0.33 (−1.9, 1.2) | 0.68 | 0 (0, 0) | 1.00 | |
| Misclassification Rate | 0.22 | 0.25 | 0.21 | ||||
| AICc | 71 | 130 | n/a | ||||
| Area Under the Curve | 0.85 | 0.85 | 0.86 | ||||
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Shiao, S.P.K.; Grayson, J.; Lie, A.; Yu, C.H. Personalized Nutrition—Genes, Diet, and Related Interactive Parameters as Predictors of Cancer in Multiethnic Colorectal Cancer Families. Nutrients 2018, 10, 795. https://doi.org/10.3390/nu10060795
Shiao SPK, Grayson J, Lie A, Yu CH. Personalized Nutrition—Genes, Diet, and Related Interactive Parameters as Predictors of Cancer in Multiethnic Colorectal Cancer Families. Nutrients. 2018; 10(6):795. https://doi.org/10.3390/nu10060795
Chicago/Turabian StyleShiao, S. Pamela K., James Grayson, Amanda Lie, and Chong Ho Yu. 2018. "Personalized Nutrition—Genes, Diet, and Related Interactive Parameters as Predictors of Cancer in Multiethnic Colorectal Cancer Families" Nutrients 10, no. 6: 795. https://doi.org/10.3390/nu10060795
APA StyleShiao, S. P. K., Grayson, J., Lie, A., & Yu, C. H. (2018). Personalized Nutrition—Genes, Diet, and Related Interactive Parameters as Predictors of Cancer in Multiethnic Colorectal Cancer Families. Nutrients, 10(6), 795. https://doi.org/10.3390/nu10060795






