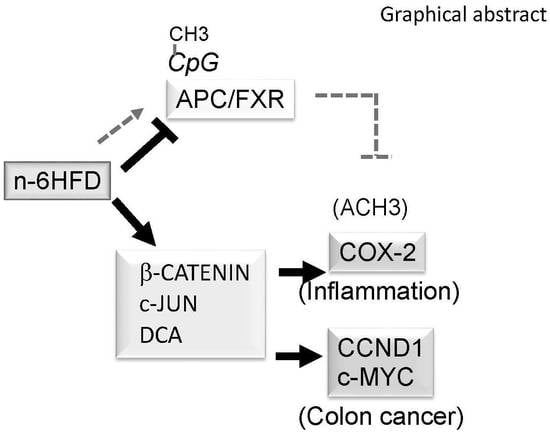
n-6 Linoleic Acid Induces Epigenetics Alterations Associated with Colonic Inflammation and Cancer
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animal Models
2.2. Cell Lines and Reagents
2.3. Real-Time PCR
2.4. Genomic DNA Methylation
2.5. Statistical Analysis
3. Results
3.1. n-6HFD Induces Fxr Gene Promoter Hypomethylation and Expression in Proximal Colonic Mucosa
3.2. n-6HFD Induces Ptsg-2 Gene Promoter Hypomethylation and Expression in Proximal Colonic Mucosa
3.3. n-6HFD Lowers the Expression of Apc and Activates Downstream Targets for the β-Catenin/Wnt Pathway
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Owczarek, D.; Rodacki, T.; Domagała-Rodacka, R.; Cibor, D.; Mach, T. Diet and nutritional factors in inflammatory bowel diseases. World J. Gastroenterol. 2016, 22, 895–905. [Google Scholar] [CrossRef]
- Mills, S.C.; Windsor, A.C.; Knight, S.C. The potential interactions between polyunsaturated fatty acids and colonic inflammatory processes. Clin. Exp. Immunol. 2005, 142, 216–228. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tjonneland, A.; Overvad, K.; Bergmann, M.M.; Nagel, G.; Linseisen, J.; Hallmans, G.; Palmqvist, R.; Sjodin, H.; Hagglund, G.; Berglund, G.; et al. Linoleic acid, a dietary n-6 polyunsaturated fatty acid, and the aetiology of ulcerative colitis: A nested case-control study within a European prospective cohort study. Gut 2009, 58, 1606–1611. [Google Scholar] [PubMed]
- Ueda, Y.; Kawakami, Y.; Kunii, D.; Okada, H.; Azuma, M.; Le, D.S.; Yamamoto, S. Elevated concentrations of linoleic acid in erythrocyte membrane phospholipids in patients with inflammatory bowel disease. Nutr. Res. 2008, 28, 239–244. [Google Scholar] [CrossRef] [PubMed]
- Caderni, G.; Bianchini, F.; Dolara, P.; Kriebel, D. Proliferative activity in the colon of the mouse and its modulation by dietary starch, fat, and cellulose. Cancer Res. 1989, 49, 1655–1659. [Google Scholar] [PubMed]
- Caderni, G.; Stuart, E.W.; Bruce, W.R. Dietary factors affecting the proliferation of epithelial cells in the mouse colon. Nutr. Cancer 1988, 11, 147–153. [Google Scholar] [CrossRef]
- Reddy, B.S.; Mangat, S.; Sheinfil, A.; Weisburger, J.H.; Wynder, E.L. Effect of type and amount of dietary fat and 1,2-dimethylhydrazine on biliary bile acids, fecal bile acids, and neutral sterols in rats. Cancer Res. 1977, 37, 2132–2137. [Google Scholar]
- Reddy, B.S.; Watanabe, K.; Weisburger, J.H.; Wynder, E.L. Promoting effect of bile acids in colon carcinogenesis in germ-free and conventional F344 rats. Cancer Res. 1977, 37, 3238–3242. [Google Scholar]
- Imray, C.H.; Radley, S.; Davis, A.; Barker, G.; Hendrickse, C.W.; Donovan, I.A.; Lawson, A.M.; Baker, P.R.; Neoptolemos, J.P. Faecal unconjugated bile acids in patients with colorectal cancer or polyps. Gut 1992, 33, 1239–1245. [Google Scholar] [CrossRef]
- Bayerdörffer, E.; Mannes, G.A.; Richter, W.O.; Ochsenkühn, T.; Wiebecke, B.; Köpcke, W.; Paumgartner, G. Increased serum deoxycholic acid levels in men with colorectal adenomas. Gastroenterology 1993, 104, 145–151. [Google Scholar] [CrossRef]
- Hill, M.J.; Drasar, B.S.; Hawksworth, G.; Aries, V.; Crowther, J.S.; Williams, R.E. Bacteria and aetiology of cancer of large bowel. Lancet 1971, 1, 95–100. [Google Scholar] [CrossRef]
- Hill, M.J.; Drasar, B.S.; Williams, R.E.; Meade, T.W.; Cox, A.G.; Simpson, J.E.; Morson, B.C. Faecal bile-acids and clostridia in patients with cancer of the large bowel. Lancet 1975, 305, 535–539. [Google Scholar] [CrossRef]
- American Cancer Society. Cancer Facts & Figures 2018; American Cancer Society: Atlanta, GA, USA, 2018. [Google Scholar]
- Silviera, M.L.; Smith, B.P.; Powell, J.; Sapienza, C. Epigenetic differences in normal colon mucosa of cancer patients suggest altered dietary metabolic pathways. Cancer Prev. Res. 2012, 5, 374–384. [Google Scholar] [CrossRef] [PubMed]
- Willett, W.C. Diet and cancer. Oncology 2000, 5, 393–404. [Google Scholar]
- Khan, N.; Afaq, F.; Mukhtar, H. Lifestyle as risk factor for cancer: Evidence from human studies. Cancer Lett. 2010, 293, 133–143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Modica, S.; Gadaleta, R.M.; Moschetta, A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl. Recept. Signal. 2010, 8, e005. [Google Scholar] [CrossRef] [PubMed]
- De Gottardi, A.; Touri, F.; Maurer, C.A.; Perez, A.; Maurhofer, O.; Ventre, G.; Bentzen, C.L.; Niesor, E.J.; Dufour, J.F. The bile acid nuclear receptor FXR and the bile acid binding protein IBABP are differently expressed in colon cancer. Dig. Dis. Sci. 2004, 49, 982–989. [Google Scholar] [CrossRef] [PubMed]
- Lax, S.; Schauer, G.; Prein, K.; Kapitan, M.; Silbert, D.; Berghold, A.; Berger, A.; Trauner, M. Expression of the nuclear bile acid receptor/farnesoid X receptor is reduced inhuman colon carcinoma compared to nonneoplastic mucosa independent from site and may be associated with adverse prognosis. Int. J. Cancer 2012, 130, 2232–2239. [Google Scholar] [CrossRef]
- Modica, S.; Murzilli, S.; Salvatore, L.; Schmidt, D.R.; Moschetta, A. Nuclear Bile Acid Receptor FXR protects against intestinal tumorigenesis. Cancer Res. 2008, 68, 9589–9594. [Google Scholar] [CrossRef]
- Maran, R.R.; Thomas, A.; Roth, M.; Sheng, Z.; Esterly, N.; Pinson, D.; Gao, X.; Zhang, Y.; Ganapathy, V.; Gonzalez, F.J.; et al. Farnesoid X receptor deficiency in mice leads to increased intestinal epithelial cell proliferation and tumor development. J. Pharm. Exp. 2009, 328, 469–477. [Google Scholar] [CrossRef]
- Liu, Y.; Sun, H.; Hu, M.; Zhang, Y.; Chen, S.; Tighe, S.; Zhu, Y. The role of cyclooxygenase-2 in colorectal carcinogenesis. Clin. Colorectal Cancer 2017, 16, 165–172. [Google Scholar] [CrossRef] [PubMed]
- Yang, K.; Li, H.; Dong, J.; Dong, Y.; Wang, C.Z. Expression profile of polyunsaturated fatty acids in colorectal cancer. World J. Gastroenterol. 2015, 21, 2405–2412. [Google Scholar] [CrossRef] [PubMed]
- Dimberg, J.; Hugander, A.; Sirsjö, A.; Söderkvist, P. Enhanced expression of cyclooxygenase-2 and nuclear beta-catenin are related to mutations in the APC gene in human colorectal cancer. Anticancer Res. 2001, 21, 911–915. [Google Scholar] [PubMed]
- Suzuki, R.; Miyamoto, S.; Yasui, Y.; Sugie, S.; Tanaka, T. Global gene expression analysis of the mouse colonic mucosa treated with azoxymethane and dextran sodium sulfate. BMC Cancer 2007, 7, 84. [Google Scholar] [CrossRef] [PubMed]
- Selmin, O.I.; Fang, C.; Lyon, A.M.; Doetschman, T.C.; Thompson, P.A.; Martinez, J.D.; Smith, J.W.; Lance, P.M.; Romagnolo, D.F. Inactivation of Adenomatous Polyposis Coli Reduces Bile Acid/Farnesoid X Receptor Expression through Fxr gene CpG Methylation in Mouse Colon Tumors and Human Colon Cancer Cells. J. Nutr. 2016, 146, 236–242. [Google Scholar] [CrossRef] [PubMed]
- Kellermayer, R.; Dowd, S.E.; Harris, R.A.; Balasa, A.; Schaible, T.D.; Wolcott, R.D.; Tatevian, N.; Szigeti, R.; Li, Z.; Versalovic, J.; et al. Colonic mucosal DNA methylation, immune response, and microbiome patterns in Toll-like receptor 2-knockout mice. FASEB J. 2011, 25, 1449–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Wit, N.J.; Bosch-Vermeulen, H.; de Groot, P.J.; Hooiveld, G.J.; Bromhaar, M.M.; Jansen, J.; Müller, M.; van der Meer, R. The role of the small intestine in the development of dietary fat-induced obesity and insulin resistance in C57BL/6J mice. BMC Med. Genom. 2008, 1, 14. [Google Scholar] [CrossRef] [PubMed]
- Renga, B.; Mencarelli, A.; Migliorati, M.; Cipriani, S.; D’Amore, C.; Distrutti, E.; Fiorucci, S. SHP-dependent and -independent induction of peroxisome proliferator-activated receptor-γ by the bile acid sensor farnesoid X receptor counter-regulates the pro-inflammatory phenotype of liver myofibroblasts. Inflamm. Res. 2011, 60, 577–587. [Google Scholar] [CrossRef]
- Stenman, L.K.; Holma, R.; Korpela, R. High-fat-induced intestinal permeability dysfunction associated with altered fecal bile acids. World J. Gastroenterol. 2012, 18, 923–929. [Google Scholar] [CrossRef]
- William, C.S.; Mann, M.; Dubois, R.N. The role of cycloxygenases in inflammation, cancer and development. Oncogene 1999, 18, 7906–7916. [Google Scholar]
- Romagnolo, D.F.; Papoutsis, A.J.; Selmin, O. Nutritional targeting of cyclooxygenase-2 for colon cancer prevention. Inflamm. Allergy Drug Targets 2010, 9, 181–191. [Google Scholar] [CrossRef] [PubMed]
- Reddy, K.V.; Naidu, K.A. Maternal and neonatal dietary intake of balanced n-6/n-3 fatty acids modulates experimental colitis in young adult rats. Eur. J. Nutr. 2016, 55, 1875–1890. [Google Scholar] [CrossRef] [PubMed]
- Sano, H.; Kawahito, Y.; Wilder, R.L.; Hashiramoto, A.; Mukai, S.; Asai, K.; Kimura, S.; Kato, H.; Kondo, M.; Hla, T. Expression of cyclooxygenase-1 and -2 in human colorectal cancer. Cancer Res. 1995, 55, 3785–3789. [Google Scholar] [PubMed]
- DuBois, R.N.; Radhika, A.; Reddy, B.S.; Entingh, A.J. Increased cyclooxygenase-2 levels in carcinogen-induced rat colonic tumors. Gastroenterology 1996, 110, 1259–1262. [Google Scholar] [CrossRef] [PubMed]
- Kulendran, M.; Stebbing, J.F.; Marks, C.G.; Rockall, T.A. Predictive and prognostic factors in colorectal cancer: A personalized approach. Cancers 2011, 3, 1622–1638. [Google Scholar] [CrossRef]
- Markowitz, S.D.; Bertagnolli, M.M. Molecular origins of cancer: Molecular basis of colorectal cancer. N. Engl. J. Med. 2009, 361, 2449–2460. [Google Scholar] [CrossRef]
- Phelps, R.A.; Chidester, S.; Dehghanizadeh, S.; Phelps, J.; Sandoval, I.T.; Rai, K.; Broadbent, T.; Sarkar, S.; Burt, R.W.; Jones, D.A. A two-step model for colon adenoma initiation and progression caused by APC loss. Cell 2009, 137, 623–634. [Google Scholar] [CrossRef]
- Mei, J.M.; Hord, N.G.; Winterstein, D.F.; Donald, S.P.; Phang, J.M. Differential expression of prostaglandin endoperoxide H synthase-2 and formation of activated beta-catenin-Lef-1 transcription complex in mouse colonic epithelial cells contrasting in APC. Carcinogenesis 1999, 20, 737–740. [Google Scholar] [CrossRef]
- Tetsu, O.; McCormick, F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature 1999, 398, 422–426. [Google Scholar] [CrossRef]
- Shtutman, M.; Zhurinsky, J.; Simcha, I.; Albanese, C.; D’Amico, M.; Pestell, R.; Ben-Ze’ev, A. The cyclin D1 gene is a target of the beta-catenin/LEF-1 pathway. Proc. Natl. Acad. Sci. USA 1999, 96, 5522–5527. [Google Scholar] [CrossRef]
- Mann, B.; Gelos, M.; Siedow, A.; Hanski, M.L.; Gratchev, A.; Ilyas, M.; Bodmer, W.F.; Moyer, M.P.; Riecken, E.O.; Buhr, H.J.; et al. Target genes of beta-catenin-T cell-factor/lymphoid-enhancer-factor signaling in human colorectal carcinomas. Proc. Natl. Acad. Sci. USA 1999, 96, 1603–1608. [Google Scholar] [CrossRef] [PubMed]
- Degner, S.C.; Kemp, M.Q.; Bowden, G.T.; Romagnolo, D.F. Conjugated linoleic acid attenuates cyclooxygenase-2 transcriptional activity via an anti-AP-1 mechanism in MCF-7 breast cancer cells. J. Nutr. 2006, 136, 421–427. [Google Scholar] [CrossRef] [PubMed]
- Day, S.D.; Enos, R.T.; McClellan, J.L.; Steiner, J.L.; Velázquez, K.T.; Murphy, E.A. Linking inflammation to tumorigenesis in a mouse model of high-fat-diet-enhanced colon cancer. Cytokine 2013, 64, 454–462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, F.Y.; Pai, M.H.; Chiang, E.P. Consumption of high-fat diet induces tumor progression and epithelial-mesenchymal transition of colorectal cancer in a mouse xenograft model. J. Nutr. Biochem. 2012, 23, 1302–1313. [Google Scholar] [CrossRef] [PubMed]
- Abdelkarim, M.; Caron, S.; Duhem, C.; Prawitt, J.; Dumont, J.; Lucas, A.; Bouchaert, E.; Briand, O.; Brozek, J.; Kuipers, F.; et al. The farnesoid X receptor regulates adipocyte differentiation and function by promoting peroxisome proliferator-activated receptor-gamma and interfering with the Wnt/beta-catenin pathways. J. Biol. Chem. 2010, 285, 36759–36767. [Google Scholar] [CrossRef]
- Panza, A.; Pazienza, V.; Ripoli, M.; Benegiamo, G.; Gentile, A.; Valvano, M.R.; Augello, B.; Merla, G.; Prattichizzo, C.; Tavano, F.; et al. Interplay between SOX9, β-catenin and PPARγ activation in colorectal cancer. Biochim. Biophys. Acta 2013, 1833, 1853–1865. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Girnun, G.D.; Smith, W.M.; Drori, S.; Sarraf, P.; Mueller, E.; Eng, C.; Nambiar, P.; Rosenberg, D.W.; Bronson, R.T.; Edelmann, W.; et al. APC-dependent suppression of colon carcinogenesis by PPARgamma. Proc. Natl. Acad. Sci. USA 2002, 99, 13771–13776. [Google Scholar] [CrossRef]
- Looby, E.; Abdel-Latif, M.M.; Athié-Morales, V.; Duggan, S.; Long, A.; Kelleher, D. Deoxycholate induces COX-2 expression via Erk1/2-, p38-MAPK and AP-1-dependent mechanisms in esophageal cancer cells. BMC Cancer 2009, 9, 190. [Google Scholar] [CrossRef]
- Abdel-Latif, M.M.; Inoue, H.; Kelleher, D.; Reynolds, J.V. Factors regulating nuclear factor-kappa B activation in esophageal cancer cells: Role of bile acids and acid. J. Cancer Res. Ther. 2016, 12, 364–373. [Google Scholar]
- Glinghammar, B.; Rafter, J. Colonic luminal contents induce cyclooxygenase 2 transcription in human colon carcinoma cells. Gastroenterology 2001, 120, 401–410. [Google Scholar] [CrossRef]
- Kim, Y.S.; Han, C.Y.; Kim, S.W.; Kim, J.H.; Lee, S.K.; Jung, D.J.; Park, S.Y.; Kang, H.; Choi, H.S.; Lee, J.W.; et al. The orphan nuclear receptor small heterodimer partner as a novel coregulator of nuclear factor-kappa b in oxidized low density lipoprotein-treated macrophage cell line RAW 264.7. J. Biol. Chem. 2001, 276, 33736–33740. [Google Scholar] [CrossRef] [PubMed]
- Park, M.J.; Kim, K.H.; Kim, H.Y.; Kim, K.; Cheong, J. Bile acid induces expression of COX-2 through the homeodomain transcription factor CDX1 and orphan nuclear receptor SHP in human gastric cancer cells. Carcinogenesis 2008, 29, 2385–2393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.D.; Chen, W.D.; Wang, M.; Yu, D.; Forman, B.M.; Huang, W. Farnesoid X receptor antagonizes nuclear factor kappa B in hepatic inflammatory response. Hepatology 2008, 48, 1632–1643. [Google Scholar] [CrossRef]
- Hsu, H.H.; Lin, Y.M.; Shen, C.Y.; Shibu, M.A.; Li, S.Y.; Chang, S.H.; Lin, C.C.; Chen, R.J.; Viswanadha, V.P.; Shih, H.N.; et al. Prostaglandin E2-Induced COX-2 Expressions via EP2 and EP4 Signaling Pathways in Human LoVo Colon Cancer Cells. Int. J. Mol. Sci. 2017, 18, 1132. [Google Scholar] [CrossRef] [PubMed]
- Lim, K.; Han, C.; Xu, L.; Isse, K.; Demetris, A.J.; Wu, T. Cyclooxygenase-2-derived prostaglandin E2 activates beta-catenin in human cholangiocarcinoma cells: Evidence for inhibition of these signaling pathways by omega 3 polyunsaturated fatty acids. Cancer Res. 2008, 68, 553–560. [Google Scholar] [CrossRef] [PubMed]
- Behrens, A.; Jochum, W.; Sibilia, M.; Wagner, E.F. Oncogenic transformation by ras and fos is mediated by c-Jun N-terminal phosphorylation. Oncogene 2000, 19, 2657–2663. [Google Scholar] [CrossRef]
- Geerling, B.J.; Dagnelie, P.C.; Badart-Smook, A.; Russel, M.G.; Stockbrügger, R.W.; Brummer, R.J. Diet as a risk factor for the development of ulcerative colitis. Am. J. Gastroenterol. 2000, 95, 1008–1013. [Google Scholar] [CrossRef]
- Bailey, A.M.; Zhan, L.; Maru, D.; Shureiqi, I.; Pickering, C.R.; Kiriakova, G.; Izzo, J.; He, N.; Wei, C.; Baladandayuthapani, V.; et al. FXR silencing in human colon cancer by DNA methylation and KRAS signaling. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 306, G48–G58. [Google Scholar] [CrossRef] [Green Version]
- Zeng, H.; Ishaq, S.L.; Zhao, F.Q.; Wright, A.G. Colonic inflammation accompanies an increase of β-catenin signaling and Lachnospiraceae/Streptococcaceae bacteria in the hind gut of high-fat diet-fed mice. J. Nutr. Biochem. 2016, 35, 30–36. [Google Scholar] [CrossRef]
- Penrose, H.M.; Heller, S.; Cable, C.; Nakhoul, H.; Baddoo, M.; Flemington, E.; Crawford, S.E.; Savkovic, S.D. High-fat diet induced leptin and Wnt expression: RNA-sequencing and pathway analysis of mouse colonic tissue and tumors. Carcinogenesis 2017, 38, 302–311. [Google Scholar] [CrossRef] [Green Version]
- Arita, S.; Kinoshita, Y.; Ushida, K.; Enomoto, A.; Inagaki-Ohara, K. High-fat diet feeding promotes stemness and precancerous changes in murine gastric mucosa mediated by leptin receptor signaling pathway. Arch. Biochem. Biophys. 2016, 610, 16–24. [Google Scholar] [CrossRef] [PubMed]
- Mao, J.; Hu, X.; Xiao, Y.; Yang, C.; Ding, Y.; Hou, N.; Wang, J.; Cheng, H.; Zhang, X. Overnutrition stimulates intestinal epithelium proliferation through ß-catenin signaling in obese mice. Diabetes 2013, 62, 3736–3746. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.C.; Gao, R.Y.; Wu, W.; Guo, B.M.; Peng, J.Y.; Qin, H.L. Effect of a high-fat diet in development of colonic adenoma in an animal model. World. J. Gastroenterol. 2014, 20, 8119–8129. [Google Scholar] [CrossRef] [PubMed]
- Korinek, V.; Barker, N.; Morin, P.J.; van Wichen, D.; de Weger, R.; Kinzler, K.W.; Vogelstein, B.; Clevers, H. Constitutive transcriptional activation by a beta-catenin-Tcf complex in APC−/− colon carcinoma. Science 1997, 275, 1784–1787. [Google Scholar] [CrossRef]
- Morin, P.J.; Sparks, A.B.; Korinek, V.; Barker, N.; Clevers, H.; Vogelstein, B.; Kinzler, K.W. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science 1997, 275, 1787–1790. [Google Scholar] [CrossRef]
- Kinzler, K.W.; Nilbert, M.C.; Su, L.K.; Vogelstein, B.; Bryan, T.M.; Levy, D.B.; Smith, K.J.; Preisinger, A.C.; Hedge, P.; Mckechnie, D.; et al. Identification of FAP locus genes from chromosome 5q21. Science 1991, 253, 661–665. [Google Scholar] [CrossRef]
- Romagnolo, D.F.; Zempleni, J.; Selmin, O.I. Nuclear receptors and epigenetic regulation: Opportunities for nutritional targeting and disease prevention. Adv. Nutr. 2014, 5, 373–385. [Google Scholar] [CrossRef]
- Wang, H.; Liu, A.; Kuo, Y.; Chi, E.; Yang, X.; Zhang, L.; Yang, C.S. Obesity promotes PhIP-induced small intestinal carcinogenesis in hCYP1A-db/db mice: Involvement of mutations and DNA hypermethylation of Apc. Carcinogenesis 2016, 37, 723–730. [Google Scholar] [CrossRef]
- Li, R.; Grimm, S.A.; Chrysovergis, K.; Kosak, J.; Wang, X.; Du, Y.; Burkholder, A.; Janardhan, K.; Mav, D.; Shah, R.; et al. Obesity, rather than diet, drives epigenomic alterations in colonic epithelium resembling cancer progression. Cell Metab. 2014, 19, 702–711. [Google Scholar] [CrossRef]
- Esteller, M.; Sparks, A.; Toyota, M.; Sanchez-Cespedes, M.; Capella, G.; Peinado, M.A.; Gonzalez, S.; Tarafa, G.; Sidransky, D.; Meltzer, S.J.; et al. Analysis of adenomatous polyposis coli promoter hypermethylation in human cancer. Cancer Res. 2000, 60, 4366–4371. [Google Scholar]
- Lao, V.V.; Grady, W.M. Epigenetics and colorectal cancer. Nat. Rev. Gastroenterol. Hepatol. 2011, 8, 686–700. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.K.; Liu, J.; Liu, C.; Wang, F.Y.; Chen, C.Y.; Zhang, X.H. Hypermethylation of adenomatous polyposis coli gene promoter is associated with novel Wnt signaling pathway in gastric adenomas. J. Gastroenterol. Hepatol. 2012, 27, 1629–1634. [Google Scholar] [CrossRef] [PubMed]
- Deng, G.; Song, G.A.; Pong, E.; Sleisenger, M.; Kim, Y.S. Promoter methylation inhibits APC gene expression by causing changes in chromatin conformation and interfering with the binding of transcription factor CCAAT-binding factor. Cancer Res. 2004, 64, 2692–2698. [Google Scholar] [CrossRef] [PubMed]
- Li, B.Q.; Liu, P.P.; Zhang, C.H. Correlation between the methylation of APC gene promoter and colon cancer. Oncol. Lett. 2017, 14, 2315–2319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mai, V.; Colbert, L.H.; Perkins, S.N.; Schatzkin, A.; Hursting, S.D. Intestinal microbiota: A potential diet-responsive prevention target in ApcMin mice. Mol. Carcinog. 2007, 46, 42–48. [Google Scholar] [CrossRef] [PubMed]






| Diet | Control | n-6HFD | ||
|---|---|---|---|---|
| Formula | (g/Kg) | (g/Kg) | ||
| Casein | 200.0 | 240.0 | ||
| L-Cystine | 3.0 | 3.6 | ||
| Corn Starch | 397.5 | 199.4 | ||
| Maltodextrin | 132.0 | 150.0 | ||
| Sucrose | 120.0 | 80.0 | ||
| Safflower Oil | 50.0 | 220.0 | ||
| Cellulose | 50.0 | 50.0 | ||
| Mineral Mix, AIN-93G-MX(94046) | 35.0 | 42.0 | ||
| Mineral Mix, AIN-93-VX(94047) | 10.0 | 12.0 | ||
| Choline Bitartrate | 2.5 | 3.0 | ||
| TBHQ, Antioxidant | 0.01 | 0.045 | ||
| Nutrient Composition | (% Weight) | (% Kcal) | (% Weight) | (% Kcal) |
| Protein | 17.7 | 19.3 | 21.2 | 18.7 |
| Carbohydrate | 62.1 | 67.9 | 42.3 | 37.3 |
| Fat | 5.2 | 12.8 | 22.2 | 44.0 |
| Energy (Kcal/g) | 3.7 | 4.5 | ||
| Target | Primer Sequence a |
|---|---|
| mRNA: | |
| Fxr | F: TTAGTCTTCACCACAGCCACC |
| R: ACCTGTATACATACATTCAGCCAAC | |
| Apc | F: CTGAGCCTGGATGAGCCATT |
| R: GTGAGTCCAAGGCGAACGTC | |
| Pparγ1 | F: GTGAGACCAACAGCCTGACG |
| R: ACAGACTCGGCACTCAATGG | |
| Cox-2 | F: GAAGTCTTTGGTCTGGTGCCT |
| R: GCTCCTGCTTGAGTATGTCG | |
| Gapdh | F: CACTTGAAGGGTGGAGCCAA |
| R: AGTGATGGCATGGACTGTGG | |
| Ibabp | F: CAGGAGACGTGATTGAAAGGG |
| R: GCCCCCAGAGTAAGACTGGG | |
| Shp | F: GTACCTGAAGGGCACGATCC |
| R: AGCCTCCTGTTGCAGGTGT | |
| Ccnd1 | F: CTAAACAAGCACCCCCTCCA |
| R: GGTAACAGGGCTGTAGGCAC | |
| Methylation-specific: | |
| Fxr | F: CGTTTAGCGATGGGGTTAATTAG |
| R: CGTCTTCTTTACTTATCTAAACCTCCTT | |
| Apc | F: GAGTGTGGTTGTCGGAAATTC |
| R: CAAAAAAACGTACATAAAAAACGCT | |
| Ptsg-2 | F: TTTTAGTTAGGATTTTAGATTTCGG |
| R: ATAATACCAAAAAAACTACACCGC | |
| β-Actin | F: AATAGTTATTTTAAGTATTTATGAAATAAG |
| R: TAACTACCTCAACACCTCAAC |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Romagnolo, D.F.; Donovan, M.G.; Doetschman, T.C.; Selmin, O.I. n-6 Linoleic Acid Induces Epigenetics Alterations Associated with Colonic Inflammation and Cancer. Nutrients 2019, 11, 171. https://doi.org/10.3390/nu11010171
Romagnolo DF, Donovan MG, Doetschman TC, Selmin OI. n-6 Linoleic Acid Induces Epigenetics Alterations Associated with Colonic Inflammation and Cancer. Nutrients. 2019; 11(1):171. https://doi.org/10.3390/nu11010171
Chicago/Turabian StyleRomagnolo, Donato F., Micah G. Donovan, Tom C. Doetschman, and Ornella I. Selmin. 2019. "n-6 Linoleic Acid Induces Epigenetics Alterations Associated with Colonic Inflammation and Cancer" Nutrients 11, no. 1: 171. https://doi.org/10.3390/nu11010171
APA StyleRomagnolo, D. F., Donovan, M. G., Doetschman, T. C., & Selmin, O. I. (2019). n-6 Linoleic Acid Induces Epigenetics Alterations Associated with Colonic Inflammation and Cancer. Nutrients, 11(1), 171. https://doi.org/10.3390/nu11010171