Structural Insights into Phylloquinone (Vitamin K1), Menaquinone (MK4, MK7), and Menadione (Vitamin K3) Binding to VKORC1
Abstract
1. Introduction
2. Materials and Methods
2.1. Molecular Modelling, Docking, and Dynamics of Vitamin K and VKORC1 Human Protein
2.2. MD Analysis and Figures
2.3. VKOR Activity Assays and Kinetics
3. Results
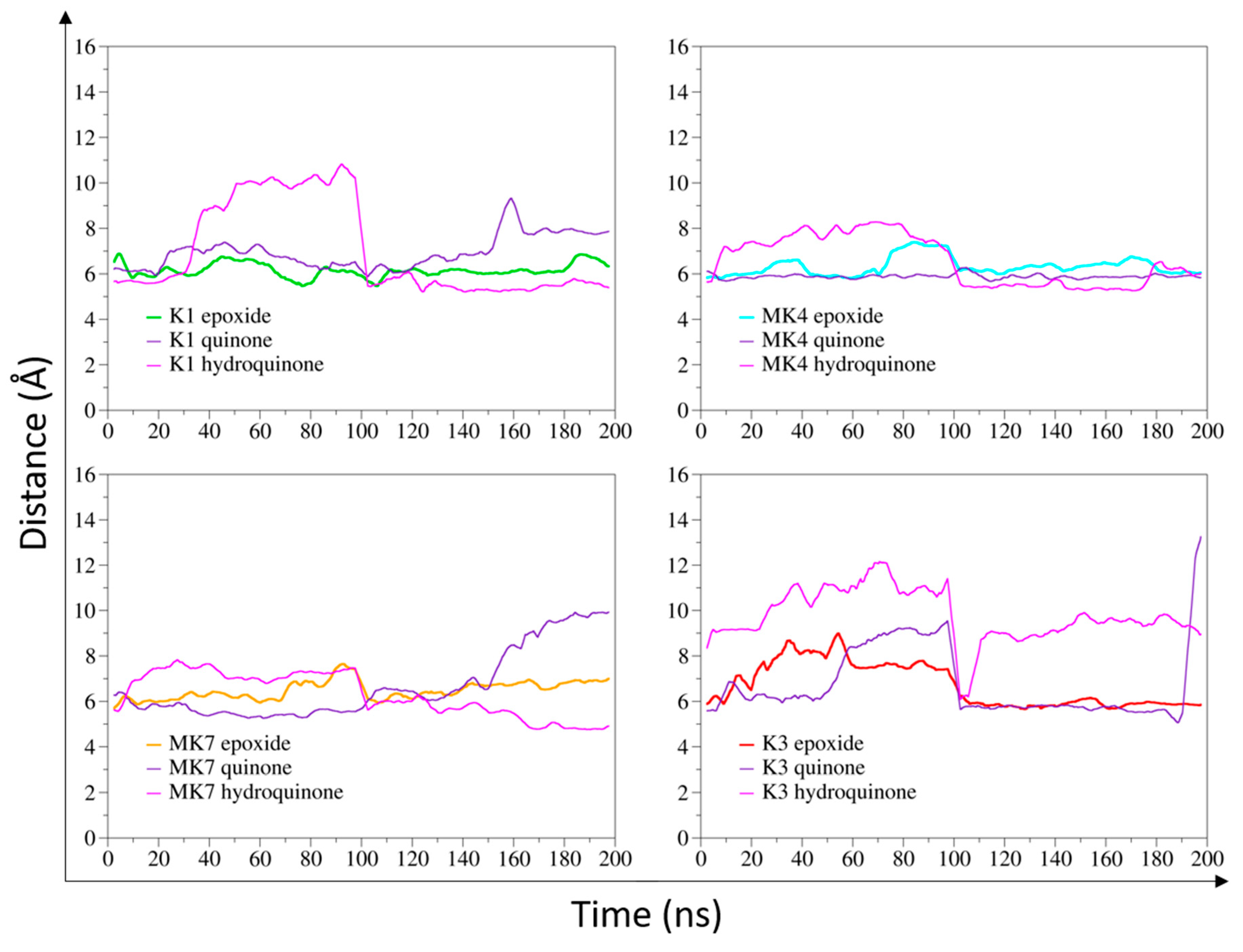
3.1. Calculated Distance between Vitamin K and the VKORC1 Catalytic Site in MD Simulations
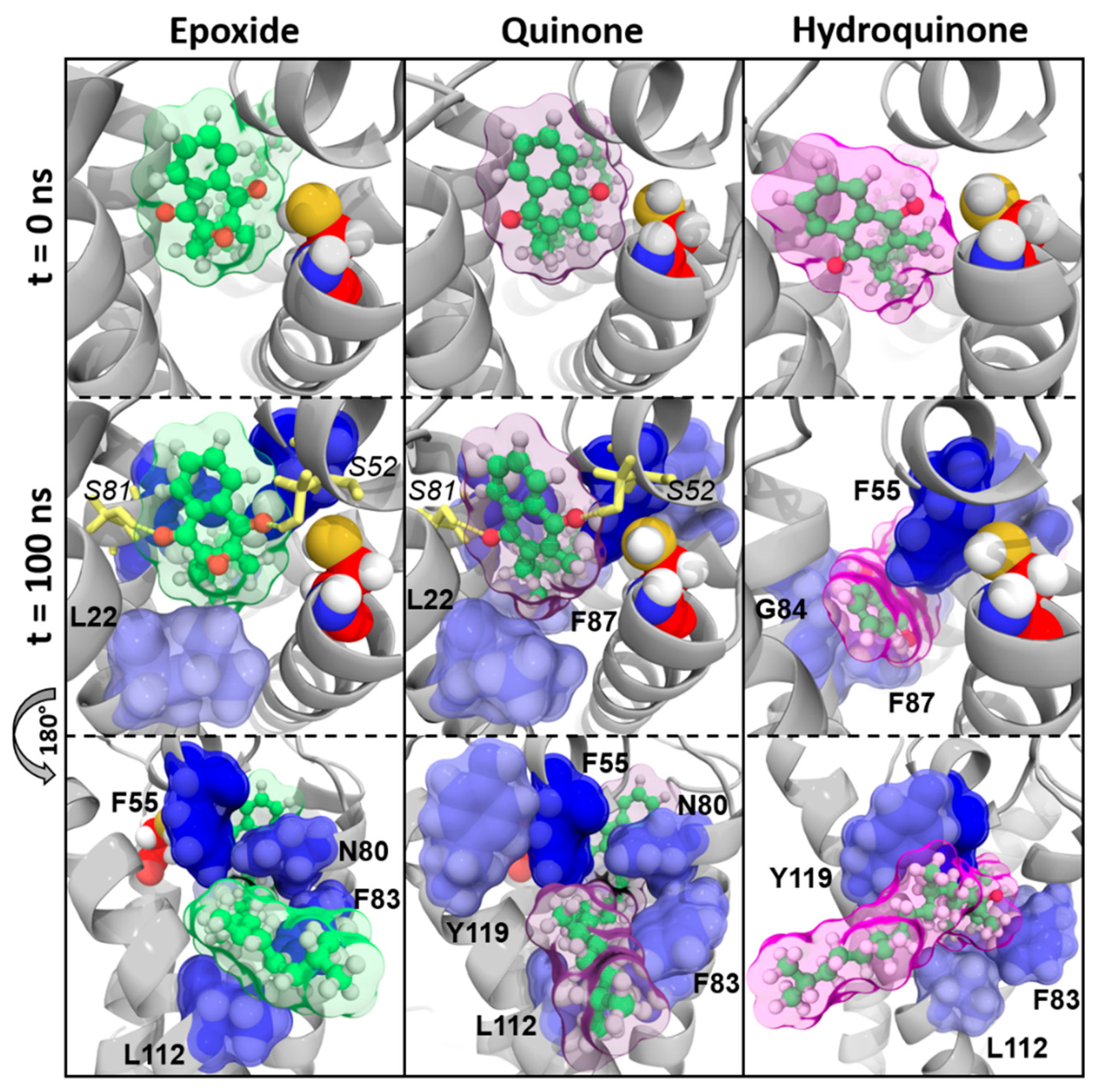
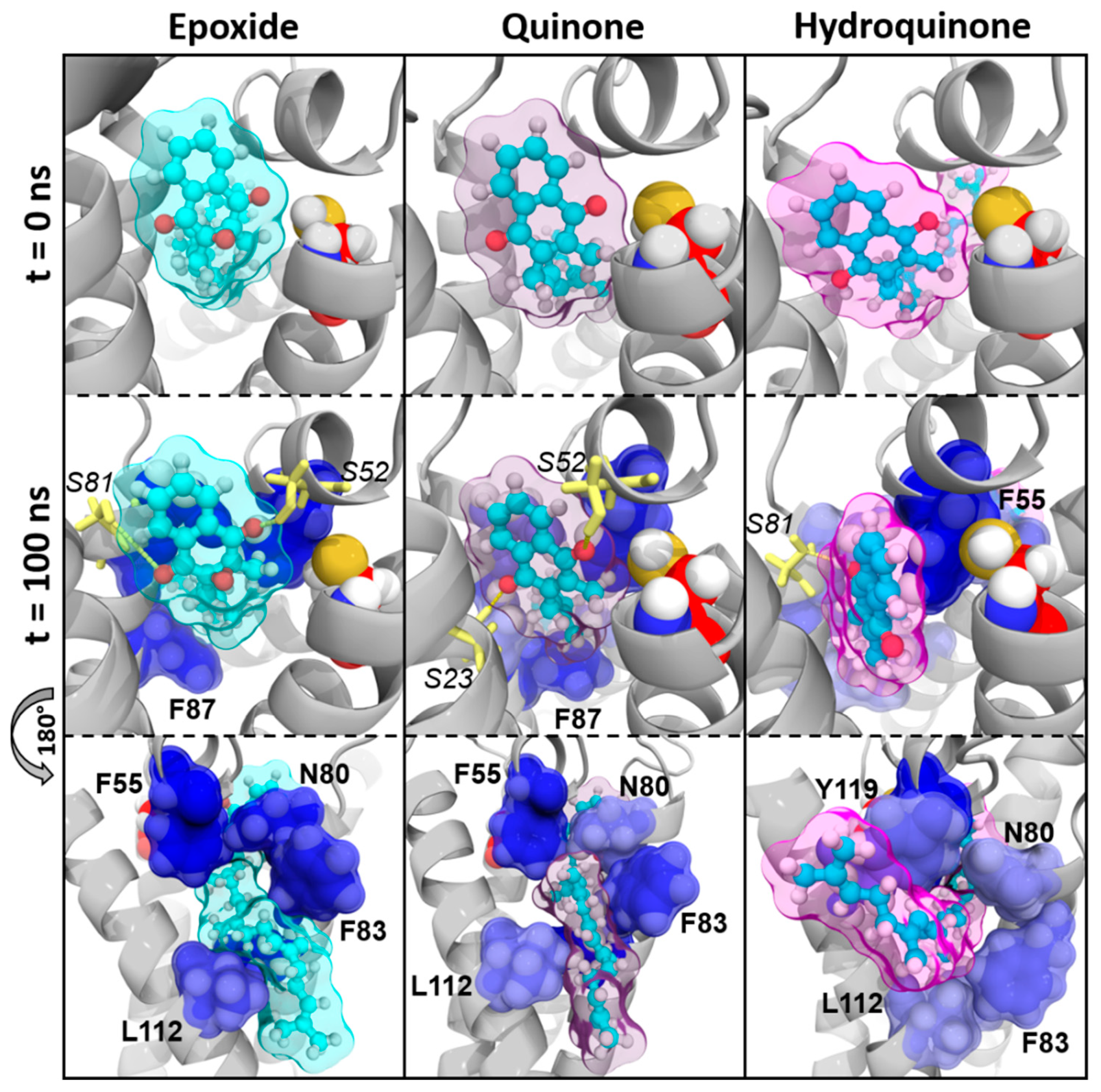
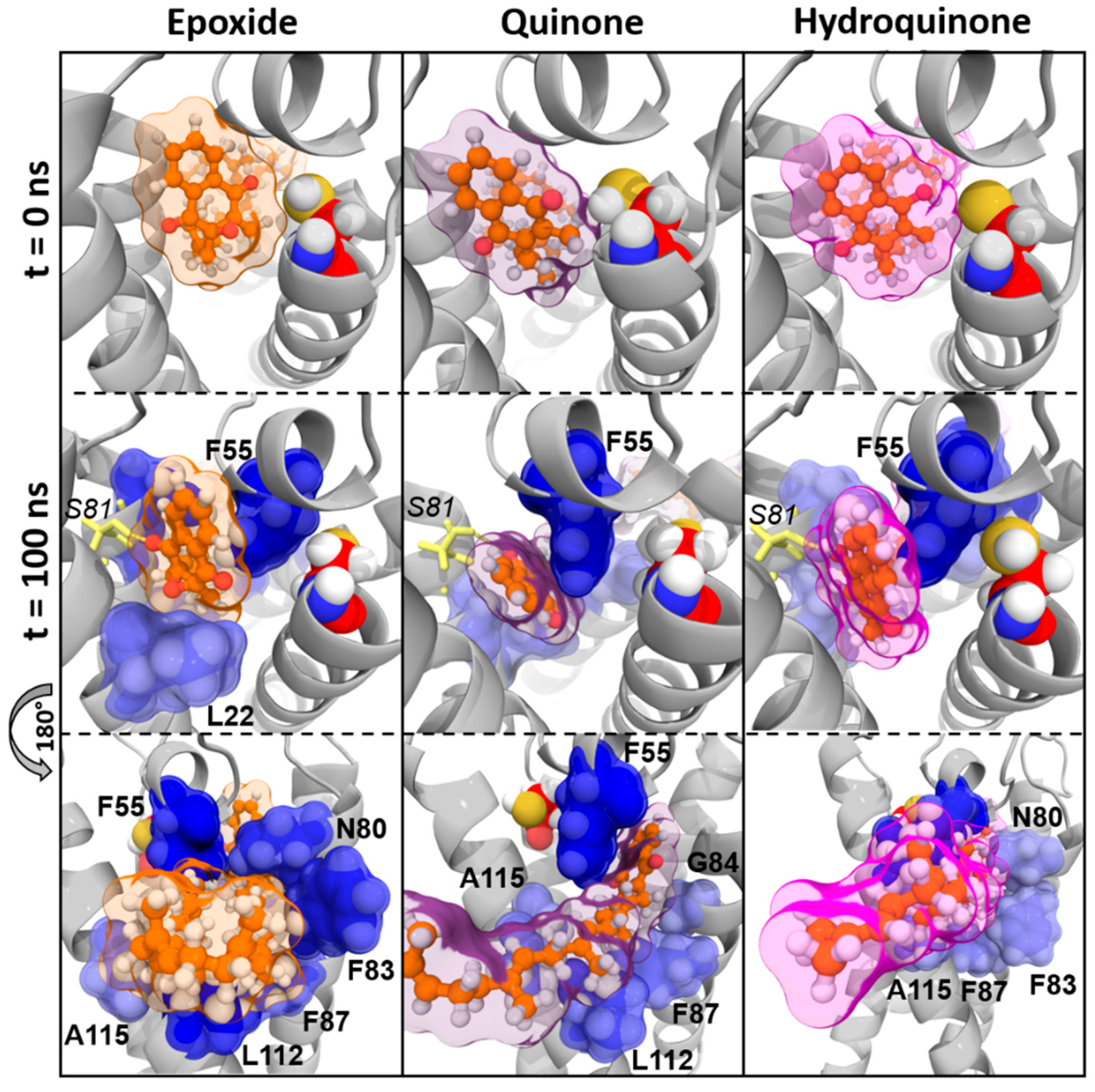
3.2. Structural Characterization of Vitamin K Binding to VKORC1
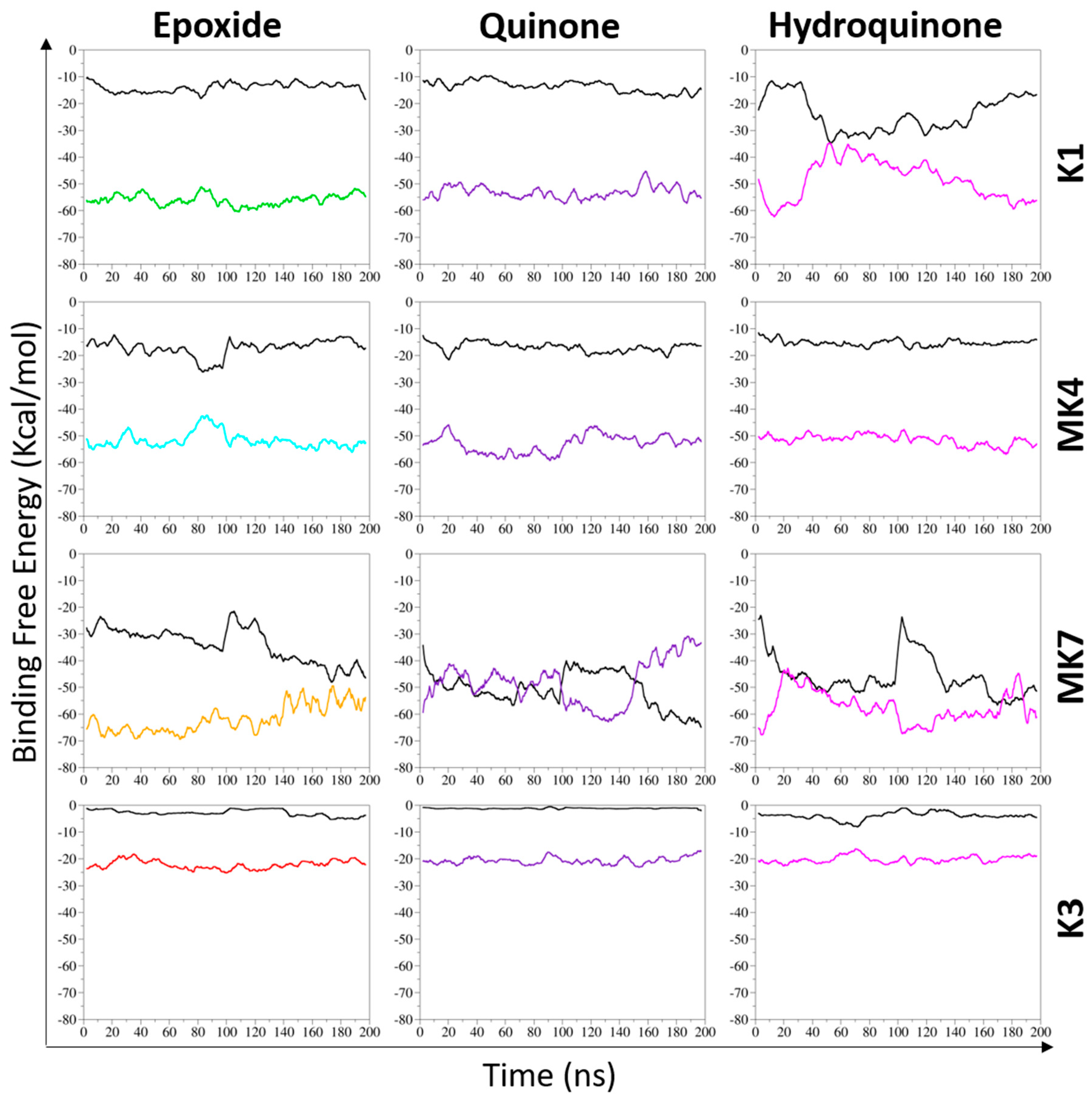
3.2.1. Overall Binding Free Energy of Vitamin K–VKORC1 Complexes
3.2.2. The VKORC1 Amino Acid Contribution to Vitamin K Binding
3.2.3. Vitamins K–VKORC1 Binding Overview
Vitamin K1
Vitamin MK4
Vitamin MK7
Vitamin K3
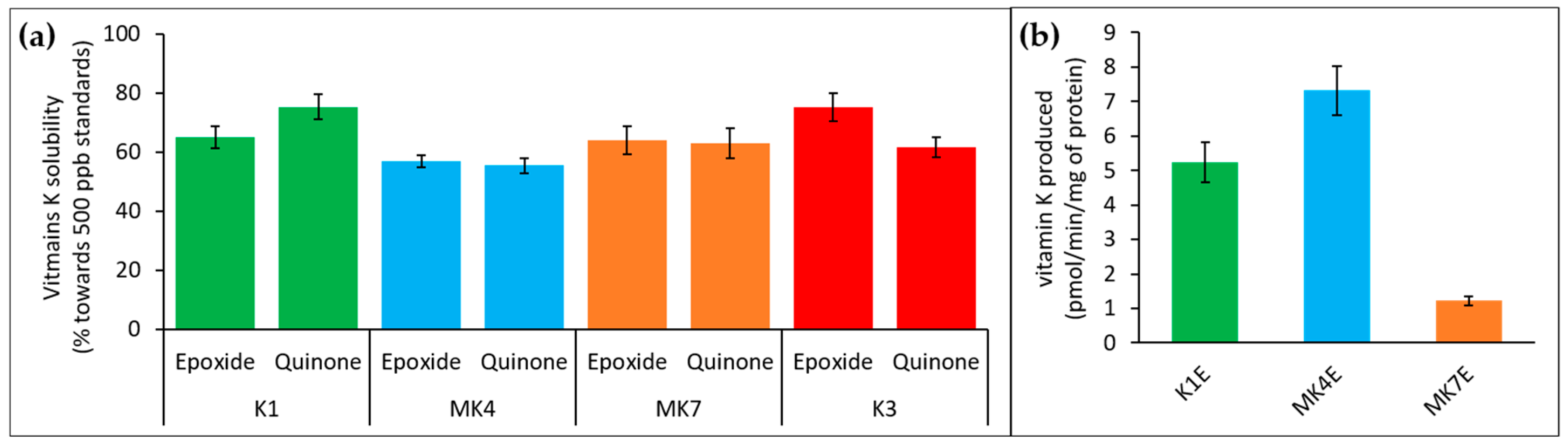
3.3. Membrane Effect of Vitamins K Binding to VKORC1
3.4. VKORC1 Activity Towards Vitamins K1, MK4, MK7, and K3
4. Discussion
4.1. Vitamin K1
4.2. Vitamin MK4
4.3. Vitamin MK7
4.4. Vitamin K3
5. Conclusions
Author Contributions
Conflicts of Interest
Appendix A


References
- Koshihara, Y.; Hoshi, K.; Shiraki, M. Vitamin K2 (menatetrenone) inhibits prostaglandin synthesis in cultured human osteoblast-like periosteal cells by inhibiting prostaglandin H synthase activity. Biochem. Pharmacol. 1993, 46, 1355–1362. [Google Scholar] [CrossRef]
- Hara, K.; Akiyama, Y.; Nakamura, T.; Murota, S.; Morita, I. The inhibitory effect of vitamin K2 (menatetrenone) on bone resorption may be related to its side chain. Bone 1995, 16, 179–184. [Google Scholar] [CrossRef]
- Reddi, K.; Henderson, B.; Meghji, S.; Wilson, M.; Poole, S.; Hopper, C.; Harris, M.; Hodges, S.J. Interleukin 6 production by lipopolysaccharide-stimulated human fibroblasts is potently inhibited by naphthoquinone (vitamin K) compounds. Cytokine 1995, 7, 287–290. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, J.C.; Wang, H.; Peterson, J.W.; Furie, B.C.; Furie, B.; Booth, S.L.; Volpe, J.J.; Rosenberg, P.A. Novel role of vitamin k in preventing oxidative injury to developing oligodendrocytes and neurons. J. Neurosci. 2003, 23, 5816–5826. [Google Scholar] [CrossRef] [PubMed]
- Lev, M.; Milford, A.F. Effect of vitamin K depletion and restoration on sphingolipid metabolism in Bacteroides melaninogenicus. J. Lipid Res. 1972, 13, 364–370. [Google Scholar] [PubMed]
- Thomas, M.W.; Gebara, B.M. Difficulty breathing. Late onset vitamin K deficiency bleeding. Clin. Pediatr. 2004, 43, 499–502. [Google Scholar] [CrossRef]
- Dam, H. The antihaemorrhagic vitamin of the chick. Biochem. J. 1935, 29, 1273–1285. [Google Scholar] [CrossRef]
- van Summeren, M.J.; Braam, L.A.; Lilien, M.R.; Schurgers, L.J.; Kuis, W.; Vermeer, C. The effect of menaquinone-7 (vitamin K2) supplementation on osteocalcin carboxylation in healthy prepubertal children. Br. J. Nutr. 2009, 102, 1171–1178. [Google Scholar] [CrossRef]
- Willems, B.A.; Vermeer, C.; Reutelingsperger, C.P.; Schurgers, L.J. The realm of vitamin K dependent proteins: Shifting from coagulation toward calcification. Mol. Nutr. Food Res 2014, 58, 1620–1635. [Google Scholar] [CrossRef]
- Oldenburg, J.; Marinova, M.; Müller-Reible, C.; Watzka, M. The vitamin K cycle. Vitam. Horm. 2008, 78, 35–62. [Google Scholar]
- Bell, R.G.; Matschiner, J.T. Vitamin K activity of phylloquinone oxide. Arch. Biochem. Biophys. 1970, 141, 473–476. [Google Scholar] [CrossRef]
- Rost, S.; Fregin, A.; Ivaskevicius, V.; Conzelmann, E.; Hörtnagel, K.; Pelz, H.J.; Lappegard, K.; Seifried, E.; Scharrer, I.; Tuddenham, E.G.; et al. Mutations in VKORC1 cause warfarin resistance and multiple coagulation factor deficiency type 2. Nature 2004, 427, 537–541. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Chang, C.Y.; Jin, D.Y.; Lin, P.J.; Khvorova, A.; Stafford, D.W. Identification of the gene for vitamin K epoxide reductase. Nature 2004, 427, 541–544. [Google Scholar] [CrossRef]
- Tie, J.K.; Stafford, D.W. Structure and function of vitamin K epoxide reductase. Vitam. Horm. 2008, 78, 103–130. [Google Scholar]
- Jin, D.Y.; Tie, J.K.; Stafford, D.W. The conversion of vitamin K epoxide to vitamin K quinone and vitamin K quinone to vitamin K hydroquinone uses the same active site cysteines. Biochemistry 2007, 46, 7279–7283. [Google Scholar] [CrossRef]
- Goodstadt, L.; Ponting, C.P. Vitamin K epoxide reductase: Homology, active site and catalytic mechanism. Trends Biochem. Sci. 2004, 29, 289–292. [Google Scholar] [CrossRef]
- Oldenburg, J.; Bevans, C.G.; Müller, C.R.; Watzka, M. Vitamin K epoxide reductase complex subunit 1 (VKORC1): The key protein of the vitamin K cycle. Antioxid. Redox. Signal 2006, 8, 347–353. [Google Scholar] [CrossRef] [PubMed]
- Rost, S.; Fregin, A.; Hünerberg, M.; Bevans, C.G.; Müller, C.R.; Oldenburg, J. Site-directed mutagenesis of coumarin-type anticoagulant-sensitive VKORC1: Evidence that highly conserved amino acids define structural requirements for enzymatic activity and inhibition by warfarin. Thromb. Haemost. 2005, 94, 780–786. [Google Scholar]
- Li, W.; Schulman, S.; Dutton, R.J.; Boyd, D.; Beckwith, J.; Rapoport, T.A. Structure of a bacterial homologue of vitamin K epoxide reductase. Nature 2010, 463, 507–512. [Google Scholar] [CrossRef] [PubMed]
- Rishavy, M.A.; Usubalieva, A.; Hallgren, K.W.; Berkner, K.L. Novel insight into the mechanism of the vitamin K oxidoreductase (VKOR): Electron relay through Cys43 and Cys51 reduces VKOR to allow vitamin K reduction and facilitation of vitamin K-dependent protein carboxylation. J. Biol. Chem. 2011, 286, 7267–7278. [Google Scholar] [CrossRef]
- Silverman, R.B.; Nandi, D.L. Reduced thioredoxin: A possible physiological cofactor for vitamin K epoxide reductase. Further support for an active site disulfide. Biochem. Biophys. Res. Commun. 1988, 155, 1248–1254. [Google Scholar] [CrossRef]
- Beulens, J.W.; Booth, S.L.; van den Heuvel, E.G.; Stoecklin, E.; Baka, A.; Vermeer, C. The role of menaquinones (vitamin K(2)) in human health. Br. J. Nutr. 2013, 110, 1357–1368. [Google Scholar] [CrossRef] [PubMed]
- Thijssen, H.H.; Drittij-Reijnders, M.J. Vitamin K status in human tissues: Tissue-specific accumulation of phylloquinone and menaquinone-4. Br. J. Nutr. 1996, 75, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Benton, M.E.; Price, P.A.; Suttie, J.W. Multi-site-specificity of the vitamin K-dependent carboxylase: In vitro carboxylation of des-gamma-carboxylated bone Gla protein and Des-gamma-carboxylated pro bone Gla protein. Biochemistry 1995, 34, 9541–9551. [Google Scholar] [CrossRef]
- Kawashima, H.; Nakajima, Y.; Matubara, Y.; Nakanowatari, J.; Fukuta, T.; Mizuno, S.; Takahashi, S.; Tajima, T.; Nakamura, T. Effects of vitamin K2 (menatetrenone) on atherosclerosis and blood coagulation in hypercholesterolemic rabbits. Jpn. J. Pharmacol. 1997, 75, 135–143. [Google Scholar] [CrossRef] [PubMed]
- Suttie, J.W. The importance of menaquinones in human nutrition. Annu. Rev. Nutr. 1995, 15, 399–417. [Google Scholar] [CrossRef] [PubMed]
- Buitenhuis, H.C.; Soute, B.A.; Vermeer, C. Comparison of the vitamins K1, K2 and K3 as cofactors for the hepatic vitamin K-dependent carboxylase. Biochim. Biophys. Acta 1990, 1034, 170–175. [Google Scholar] [CrossRef]
- Akiyama, Y.; Hara, K.; Matsumoto, A.; Takahashi, S.; Tajima, T. Comparison of intestinal absorption of vitamin K2 (menaquinone) homologues and their effects on blood coagulation in rats with hypoprothrombinaemia. Biochem. Pharmacol. 1995, 49, 1801–1807. [Google Scholar] [CrossRef]
- Hirota, Y.; Tsugawa, N.; Nakagawa, K.; Suhara, Y.; Tanaka, K.; Uchino, Y.; Takeuchi, A.; Sawada, N.; Kamao, M.; Wada, A.; et al. Menadione (vitamin K3) is a catabolic product of oral phylloquinone (vitamin K1) in the intestine and a circulating precursor of tissue menaquinone-4 (vitamin K2) in rats. J. Biol. Chem. 2013, 288, 33071–33080. [Google Scholar] [CrossRef]
- Shen, G.; Cui, W.; Zhang, H.; Zhou, F.; Huang, W.; Liu, Q.; Yang, Y.; Li, S.; Bowman, G.R.; Sadler, J.E.; et al. Warfarin traps human vitamin K epoxide reductase in an intermediate state during electron transfer. Nat. Struct. Mol. Biol. 2017, 24, 69–76. [Google Scholar] [CrossRef]
- Czogalla, K.J.; Biswas, A.; Höning, K.; Hornung, V.; Liphardt, K.; Watzka, M.; Oldenburg, J. Warfarin and vitamin K compete for binding to Phe55 in human VKOR. Nat. Struct. Mol. Biol. 2017, 24, 77–85. [Google Scholar] [CrossRef]
- Chatron, N.; Chalmond, B.; Trouvé, A.; Benoît, E.; Caruel, H.; Lattard, V.; Tchertanov, L. Identification of the functional states of human vitamin K epoxide reductase from molecular dynamics simulations. RSC Adv. 2017, 7, 52071–52090. [Google Scholar] [CrossRef]
- Tie, J.K.; Nicchitta, C.; von Heijne, G.; Stafford, D.W. Membrane topology mapping of vitamin K epoxide reductase by in vitro translation/cotranslocation. J. Biol. Chem. 2005, 280, 16410–16416. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.K.; Jin, D.Y.; Stafford, D.W. Human vitamin K epoxide reductase and its bacterial homologue have different membrane topologies and reaction mechanisms. J. Biol. Chem. 2012, 287, 33945–33955. [Google Scholar] [CrossRef] [PubMed]
- Tie, J.K.; Stafford, D.W. Structural and functional insights into enzymes of the vitamin K cycle. J. Thromb. Haemost. 2016, 14, 236–247. [Google Scholar] [CrossRef]
- Wu, S.; Tie, J.K.; Stafford, D.W.; Pedersen, L.G. Membrane topology for human vitamin K epoxide reductase. J. Thromb. Haemost. 2014, 12, 112–114. [Google Scholar] [CrossRef]
- Liu, S.; Cheng, W.; Fowle Grider, R.; Shen, G.; Li, W. Structures of an intramembrane vitamin K epoxide reductase homolog reveal control mechanisms for electron transfer. Nat. Commun. 2014, 5, 3110. [Google Scholar] [CrossRef]
- Czogalla, K.J.; Biswas, A.; Wendeln, A.C.; Westhofen, P.; Müller, C.R.; Watzka, M.; Oldenburg, J. Human VKORC1 mutations cause variable degrees of 4-hydroxycoumarin resistance and affect putative warfarin binding interfaces. Blood 2013, 122, 2743–2750. [Google Scholar] [CrossRef]
- Cao, Z.; van Lith, M.; Mitchell, L.J.; Pringle, M.A.; Inaba, K.; Bulleid, N.J. The membrane topology of vitamin K epoxide reductase is conserved between human isoforms and the bacterial enzyme. Biochem. J. 2016, 473, 851–858. [Google Scholar] [CrossRef]
- Morris, G.M.; Huey, R.; Lindstrom, W.; Sanner, M.F.; Belew, R.K.; Goodsell, D.S.; Olson, A.J. AutoDock4 and AutoDockTools4: Automated docking with selective receptor flexibility. J. Comput. Chem. 2009, 30, 2785–2791. [Google Scholar] [CrossRef]
- Van Der Spoel, D.; Lindahl, E.; Hess, B.; Groenhof, G.; Mark, A.E.; Berendsen, H.J. GROMACS: Fast, flexible, and free. J. Comput. Chem. 2005, 26, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Best, R.B.; Zhu, X.; Shim, J.; Lopes, P.E.; Mittal, J.; Feig, M.; Mackerell, A.D., Jr. Optimization of the additive CHARMM all-atom protein force field targeting improved sampling of the backbone phi, psi and side-chain chi(1) and chi(2) dihedral angles. J. Chem. Theory Comput. 2012, 8, 3257–3273. [Google Scholar] [CrossRef] [PubMed]
- Jambeck, J.P.; Lyubartsev, A.P. Derivation and systematic validation of a refined all-atom force field for phosphatidylcholine lipids. J. Phys. Chem. B 2012, 116, 3164–3179. [Google Scholar] [CrossRef] [PubMed]
- Hou, T.; Wang, J.; Li, Y.; Wang, W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J. Chem. Inf. Model. 2011, 51, 69–82. [Google Scholar] [CrossRef]
- Kumari, R.; Kumar, R.; Lynn, A. g_mmpbsa—A GROMACS tool for high-throughput MM-PBSA calculations. J. Chem. Inf. Model 2014, 54, 1951–1962. [Google Scholar] [CrossRef] [PubMed]
- Turner, P.J. XMGRACE; Version 5.1.19; Center for Coastal and Land-Margin Research, Oregon Graduate Institute of Science and Technology: Beaverton, OR, USA, 2005. [Google Scholar]
- The PyMOL Molecular Graphics System; Version 1.8; Schrödinger LLC: New York, NY, USA, 2015.
- Hodroge, A.; Matagrin, B.; Moreau, C.; Fourel, I.; Hammed, A.; Benoit, E.; Lattard, V. VKORC1 mutations detected in patients resistant to vitamin K antagonists are not all associated with a resistant VKOR activity. J. Thromb. Haemost. 2012, 10, 2535–2543. [Google Scholar] [CrossRef] [PubMed]
- Goulois, J.; Lambert, V.; Legros, L.; Benoit, E.; Lattard, V. Adaptative evolution of the Vkorc1 gene in Mus musculus domesticus is influenced by the selective pressure of anticoagulant rodenticides. Ecol. Evol. 2017, 7, 2767–2776. [Google Scholar] [CrossRef] [PubMed]



| Epoxide | Quinone | Hydroquinone | |
|---|---|---|---|
| K1 | −55.91 ± 3.73 | −53.22 ± 3.85 | −48.48 ± 7.79 |
| MK4 | −51.56 ± 4.35 | −52.99 ± 4.49 | −51.49 ± 3.35 |
| MK7 | −61.67 ± 7.24 | −47.96 ± 9.95 | −57.42 ± 7.49 |
| K3 | −22.19 ± 2.37 | −20.65 ± 2.36 | −20.19 ± 2.15 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatron, N.; Hammed, A.; Benoît, E.; Lattard, V. Structural Insights into Phylloquinone (Vitamin K1), Menaquinone (MK4, MK7), and Menadione (Vitamin K3) Binding to VKORC1. Nutrients 2019, 11, 67. https://doi.org/10.3390/nu11010067
Chatron N, Hammed A, Benoît E, Lattard V. Structural Insights into Phylloquinone (Vitamin K1), Menaquinone (MK4, MK7), and Menadione (Vitamin K3) Binding to VKORC1. Nutrients. 2019; 11(1):67. https://doi.org/10.3390/nu11010067
Chicago/Turabian StyleChatron, Nolan, Abdessalem Hammed, Etienne Benoît, and Virginie Lattard. 2019. "Structural Insights into Phylloquinone (Vitamin K1), Menaquinone (MK4, MK7), and Menadione (Vitamin K3) Binding to VKORC1" Nutrients 11, no. 1: 67. https://doi.org/10.3390/nu11010067
APA StyleChatron, N., Hammed, A., Benoît, E., & Lattard, V. (2019). Structural Insights into Phylloquinone (Vitamin K1), Menaquinone (MK4, MK7), and Menadione (Vitamin K3) Binding to VKORC1. Nutrients, 11(1), 67. https://doi.org/10.3390/nu11010067





