Comprehensive Preterm Breast Milk Metabotype Associated with Optimal Infant Early Growth Pattern
Abstract
:1. Introduction
2. Materials and Methods
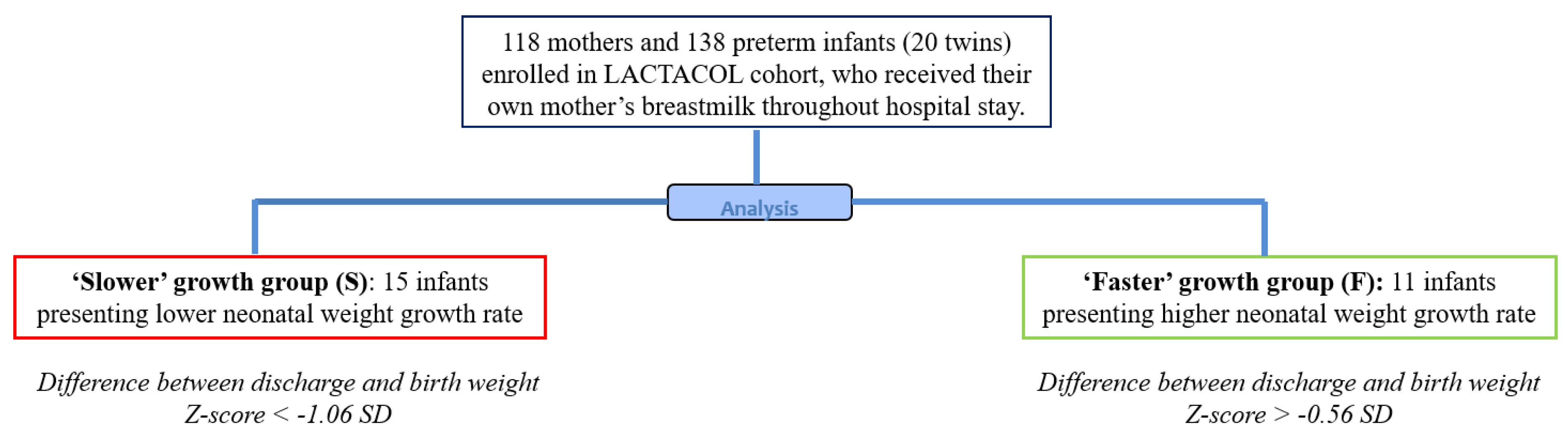
2.1. Study Design and Population
2.2. Ranking Infants according to Early Growth Trajectory
2.3. Ethics
2.4. Human Milk Collection and Targeted Free Amino Acid (FAA) Analysis
2.5. Breast Milk Liquid Chromatography-High-Resolution-Mass Spectrometry (LC-HRMS)–Based Glycomic Profiling
2.6. Breast Milk Liquid Chromatography-High-Resolution-Mass Spectrometry (LC-HRMS)–Based Lipidomic and Metabolomic Profiling
2.7. Lipidomic, Metabolomic and Glycomic Data Treatment and Metabolites Annotation
2.8. Statistical Analyses
3. Results
3.1. Subject Characteristics
3.2. Targeted Free Amino Acid Quantification
3.3. Lipidomics and Metabolomcs Profiling
3.4. Glycomics Profiling
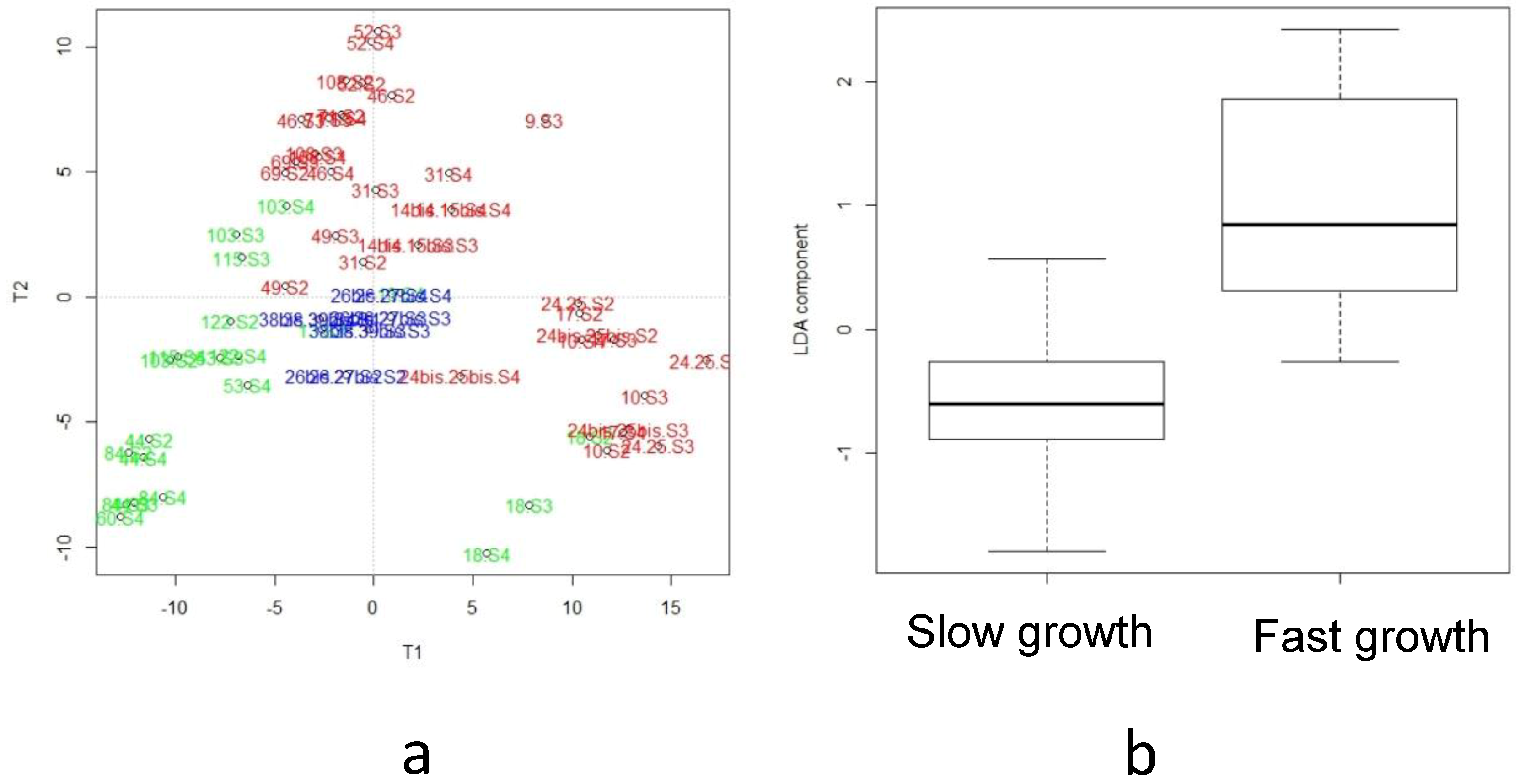
3.5. Integration of Multi-Omics Data sets
4. Discussion
4.1. Higher Breast Milk Content in Branched-Chain and Insulino-Trophic Amino Acid and in Tyrosine Associated to Optimal Infant Growth
4.2. Enhanced Milk Fat Availability by Infants with an Early “Faster” Growth Velocity
4.3. Di-Fucosylated HMOs Associated to Early Preterm Infant Growth
5. Conclusions
Supplementary Materials
Author Contributions
Acknowledgments
Conflicts of Interest
Abbreviations
| AoV-PLS | analysis of variance combined to partial least squares regression |
| BCFA | branched-chain fatty acids |
| ESI | electrospray ionization |
| FDR | false discovery rate |
| GA | gestational age |
| HMO | human milk oligosaccharides |
| LC-HR-MS | Liquid-Chromatography-High-Resolution-Mass-Spectrometry |
| MG PLS-DA | multi-group partial least squares discriminant analysis |
| MLR | multiple linear regression |
| MCSAT | medium-chain saturated fatty acid |
| U-PCA | unfold principal component analysis |
| SD | standard deviation |
| W4M | Workflow4Metabolomics® |
References
- Victora, C.G.; Bahl, R.; Barros, A.J.; França, G.V.; Horton, S.; Krasevec, J.; Murch, S.; Sankar, M.J.; Walker, N.; Rollins, N.C.; et al. Breastfeeding in the 21st century: Epidemiology, mechanisms, and lifelong effect. Lancet 2016, 387, 475–490. [Google Scholar] [CrossRef]
- Mills, S.; Ross, R.P.; Hill, C.; Fitzgerald, G.F.; Stanton, C. Milk intelligence: Mining milk for bioactive substances associated with human health. Int. Dairy J. 2011, 21, 377–401. [Google Scholar] [CrossRef]
- Andreas, N.J.; Kampmann, B.; Le-Doare, K.M. Human breast milk: A review on its composition and bioactivity. Early Hum. Dev. 2015, 91, 629–635. [Google Scholar] [CrossRef] [PubMed]
- Mosca, F.; Giannì, M.L. Human milk: Composition and health benefits. La Pediatria Medica e Chirurgica 2017, 39. [Google Scholar] [CrossRef] [PubMed]
- Boquien, C.Y. Human milk: An ideal food for nutrition of preterm newborn. Front. Pediatr. 2018, 6. [Google Scholar] [CrossRef] [PubMed]
- Johnson, T.J.; Patel, A.L.; Bigger, H.R.; Engstrom, J.L.; Meier, P.P. Economic benefits and costs of human milk feedings: A strategy to reduce the risk of prematurity-related morbidities in very-low-birth-weight infants. Adv. Nutr. Int. Rev. J. 2014, 5, 207–212. [Google Scholar] [CrossRef] [PubMed]
- Sankar, M.J.; Sinha, B.; Chowdhury, R.; Bhandari, N.; Taneja, S.; Martines, J.; Bahl, R. Optimal breastfeeding practices and infant and child mortality: A systematic review and meta-analysis. Acta Paediatr. 2015, 104, 3–13. [Google Scholar] [CrossRef] [PubMed]
- Quigley, M.; McGuire, W. Formula versus donor breast milk for feeding preterm or low birth weight infants. Cochrane Database Syst. Rev. 2014. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Corpeleijn, W.E.; de Waard, M.; Christmann, V.; van Goudoever, J.B.; Jansen-van der Weide, M.C.; Kooi, E.M.; Koper, J.F.; Kouwenhoven, S.M.; Lafeber, H.N.; Mank, E.; et al. Effect of donor milk on severe infections and mortality in very low-birth-weight infants: The early nutrition study randomized clinical trial. JAMA Pediatr. 2016, 170, 654–661. [Google Scholar] [CrossRef] [PubMed]
- Boyd, C.A.Q.; Quigley, M.A.; Brocklehurst, P. Donor breast milk versus infant formula for preterm infants: Systematic review and meta-analysis. Arch. Dis. Child. Fetal Neonatal Ed. 2007, 92, F169–F175. [Google Scholar] [CrossRef] [PubMed]
- Rozé, J.C.; Darmaun, D.; Boquien, C.Y.; Flamant, C.; Picaud, J.C.; Savagner, C.; Claris, O.; Lapillonne, A.; Mitanchez, D.; Branger, B.; et al. The apparent breastfeeding paradox in very preterm infants: Relationship between breast feeding; early weight gain and neurodevelopment based on results from two cohorts; EPIPAGE and LIFT. BMJ Open 2012, 2, e000834. [Google Scholar] [CrossRef] [PubMed]
- Curtis, M.; Rigo, J. Extrauterine growth restriction in very-low-birthweight infants. Acta Paediatr. 2004, 93, 1563–1568. [Google Scholar] [CrossRef] [PubMed]
- Ehrenkranz, R.A.; Dusick, A.M.; Vohr, B.R.; Wright, L.L.; Wrage, L.A.; Poole, W.K. Growth in the neonatal intensive care unit influences neurodevelopmental and growth outcomes of extremely low birth weight infants. Pediatrics 2006, 117, 1253–1261. [Google Scholar] [CrossRef] [PubMed]
- Larroque, B.; Ancel, P.Y.; Marret, S.; Marchand, L.; André, M.; Arnaud, C.; Pierrat, V.; Rozé, J.C.; Messer, J.; Thiriez, G.; et al. Neurodevelopmental disabilities and special care of 5-year-old children born before 33 weeks of gestation (the EPIPAGE study): A longitudinal cohort study. Lancet 2008, 371, 813–820. [Google Scholar] [CrossRef]
- Frondas-Chauty, A.; Simon, L.; Branger, B.; Gascoin, G.; Flamant, C.; Ancel, P.Y.; Darmaun, D.; Rozé, J.C. Early growth and neurodevelopmental outcome in very preterm infants: Impact of gender. Arch. Dis. Child. Fetal Neonatal Ed. 2014, 99, F366–F372. [Google Scholar] [CrossRef] [PubMed]
- Agostoni, C.; Buonocore, G.; Carnielli, V.P.; De Curtis, M.; Darmaun, D.; Decsi, T.; Domellöf, M.; Embleton, N.D.; Fusch, C.; Genzel-Boroviczeny, O.; et al. Enteral nutrient supply for preterm infants: Commentary from the European Society of Paediatric Gastroenterology; Hepatology and Nutrition Committee on Nutrition. J. Pediatr. Gastr. Nutr. 2010, 50, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Henriksen, C.; Westerberg, A.C.; Rønnestad, A.; Nakstad, B.; Veierød, M.B.; Drevon, C.A.; Iversen, P.O. Growth and nutrient intake among very-low-birth-weight infants fed fortified human milk during hospitalisation. Brit. J. Nutr. 2009, 102, 1179–1186. [Google Scholar] [CrossRef] [PubMed]
- Alexandre-Gouabau, M.C.; Courant, F.; Le Gall, G.; Moyon, T.; Darmaun, D.; Parnet, P.; Coupé, B.; Antignac, J.P. Offspring metabolomic response to maternal protein restriction in a rat model of intrauterine growth restriction (IUGR). J. Proteome Res. 2011, 10, 3292–3302. [Google Scholar] [CrossRef] [PubMed]
- Alexandre-Gouabau, M.C.; Courant, F.; Moyon, T.; Küster, A.; Le Gall, G.; Tea, I.; Antignac, J.P.; Darmaun, D. Maternal and cord blood LC-HRMS metabolomics reveal alterations in energy and polyamine metabolism, and oxidative stress in very-low birth weight infants. J. Proteome Res. 2013, 12, 2764–2778. [Google Scholar] [CrossRef] [PubMed]
- Alexandre-Gouabau, M.-C.; Moyon, T.; Cariou, V.; Antignac, J.-P.; Qannari, E.M.; Croyal, M.; Soumah, M.; Guitton, Y.; Billard, H.; Legrand, A.; et al. Breast milk lipidome is associated with early growth trajectory in preterm infants. Nutrients 2018, 10, 164. [Google Scholar] [CrossRef] [PubMed]
- Fanos, V.; Atzori, L.; Makarenko, K.; Melis, G.B.; Ferrazzi, E. Metabolomics application in maternal-fetal medicine. BioMed Res. Int. 2013, 9. [Google Scholar] [CrossRef] [PubMed]
- Demmelmair, H.; Koletzko, B. Variation of metabolite and hormone contents in human milk. Clin. Perinatol. 2017, 44, 151–164. [Google Scholar] [CrossRef] [PubMed]
- Carraro, S.; Baraldi, E.; Giordano, G.; Pirillo, P.; Stocchero, M.; Houben, M.; Bont, L. Metabolomic Profile of Amniotic Fluid and Wheezing in the First Year of Life—A Healthy Birth Cohort Study. J. Pediatr. 2018, 196, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Marincola, F.C.; Noto, A.; Caboni, P.; Reali, A.; Barberini, L.; Lussu, M.; Murgia, F.; Santoru, M.L.; Atzori, L.; Fanos, V.J. A metabolomic study of preterm human and formula milk by high resolution NMR and GC/MS analysis: Preliminary results. Matern. Fetal Neonatal Med. 2012, 25, 62–67. [Google Scholar] [CrossRef] [PubMed]
- Wu, J.; Domellöf, M.; Zivkovic, A.M.; Larsson, G.; Öhman, A.; Nording, M.L. NMR-based metabolite profiling of human milk: A pilot study of methods for investigating compositional changes during lactation. Biochem. Biophys. Res. Comm. 2016, 469, 626–632. [Google Scholar] [CrossRef] [PubMed]
- George, A.; Gay, M.; Trengove, R.; Geddes, D. Human Milk Lipidomics: Current Techniques and Methodologies. Nutrients 2018, 10, 1169. [Google Scholar] [CrossRef] [PubMed]
- Spevacek, A.R.; Smilowitz, J.T.; Chin, E.L.; Underwood, M.A.; German, J.B.; Slupsky, C.M. Infant Maturity at Birth Reveals Minor Differences in the Maternal Milk Metabolome in the First Month of Lactation. J. Nutr. 2015, 45, 1698–1708. [Google Scholar] [CrossRef] [PubMed]
- Sundekilde, U.K.; Downey, E.; O’Mahony, J.A.; O’Shea, C.A.; Ryan, C.A.; Kelly, A.L.; Bertram, H.C. The effect of gestational and lactational age on the human milk metabolome. Nutrients 2016, 8, 304. [Google Scholar] [CrossRef] [PubMed]
- Maffei, D.; Schanler, R.J. Human milk is the feeding strategy to prevent necrotizing enterocolitis! Semin. Perinatol. 2017, 41, 36–40. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Human Milk Oligosaccharides in the Prevention of Necrotizing Enterocolitis: A journey from in vitro and in vivo models to mother-infant cohort studies. Front Pediatr. 2018, 6, 385. [Google Scholar] [CrossRef] [PubMed]
- De Leoz, M.L.A.; Gaerlan, S.C.; Strum, J.S.; Dimapasoc, L.M.; Mirmiran, M.; Tancredi, D.J.; Smilowitz, J.T.; Kalanetra, K.M.; Mills, D.A.; German, J.B.; et al. Lacto-N-tetraose; fucosylation; and secretor status are highly variable in human milk oligosaccharides from women delivering preterm. J. Proteome Res. 2012, 11, 4662–4672. [Google Scholar] [CrossRef] [PubMed]
- Alderete, T.L.; Autran, C.; Brekke, B.E.; Knight, R.; Bode, L.; Goran, M.I.; Fields, D.A. Associations between human milk oligosaccharides and infant body composition in the first 6 mo of life. Am. J. Clin. Nutr. 2015, 102, 1381–1388. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charbonneau, M.R.; O’Donnell, D.; Blanton, L.V.; Totten, S.M.; Davis, J.C.; Barratt, M.J.; Cheng, J.; Guruge, J.; Talcott, M.; Bain, J.R.; et al. Sialylated milk oligosaccharides promote microbiota-dependent growth in models of infant undernutrition. Cell 2016, 164, 859–871. [Google Scholar] [CrossRef] [PubMed]
- Rezaei, R.; Wu, Z.; Hou, Y.; Bazer, F.W.; Wu, G. Amino acids and mammary gland development: Nutritional implications for milk production and neonatal growth. J. Anim. Sci. Biotechnol. 2016, 7, 20. [Google Scholar] [CrossRef] [PubMed]
- Metrustry, S.J.; Karhunen, V.; Edwards, M.H.; Menni, C.; Geisendorfer, T.; Huber, A.; Reichel, C.; Dennison, E.M.; Cooper, C.; Spector, T.; et al. Metabolomic signatures of low birthweight: Pathways to insulin resistance and oxidative stress. PLoS ONE 2018, 13, e0194316. [Google Scholar] [CrossRef] [PubMed]
- Ferchaud-Roucher, V.; Desnots, E.; Naël, C.; Martin Agnoux, A.; Alexandre-Gouabau, M.C.; Darmaun, D.; Boquien, C.Y. Use of UPLC-ESI-MS/MS to quantitate free amino acid concentrations in micro-samples of mammalian milk. SpringerPlus 2013, 2, 622. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oursel, S.; Junot, C.; Fenaille, F. Comparative analysis of native and permethylated human milk oligosaccharides by liquid chromatography coupled to high resolution mass spectrometry. J. Chromatogr. B 2017, 1071, 49–57. [Google Scholar] [CrossRef] [PubMed]
- Bligh, E.G.; Dyer, W.J. A rapid method of total lipid extraction and purification. Can. J. Biochem. Physiol. 1959, 37, 911–917. [Google Scholar] [CrossRef] [PubMed]
- Gallart-Ayala, H.; Courant, F.; Severe, S.; Antignac, J.P.; Morio, F.; Abadie, J.; Le Bizec, B. Versatile lipid profiling by liquid chromatography-high resolution mass spectrometry using all ion fragmentation and polarity switching. Preliminary application for serum samples phenotyping related to canine mammary cancer. Anal. Chim. Acta 2013, 796, 75–83. [Google Scholar] [CrossRef] [PubMed]
- Courant, F.; Royer, A.L.; Chéreau, S.; Morvan, M.L.; Monteau, F.; Antignac, J.P.; Le Bizec, B. Implementation of a semi-automated strategy for the annotation of metabolomic fingerprints generated by liquid chromatography-high resolution mass spectrometry from biological samples. Analyst 2012, 137, 4958–4967. [Google Scholar] [CrossRef] [PubMed]
- Kessner, D.; Chambers, M.; Burke, R.; Agus, D.; Mallick, P. ProteoWizard: Open source software for rapid proteomics tools development. Bioinformatics 2008, 24, 2534–2536. [Google Scholar] [CrossRef] [PubMed]
- Smith, C.A.; Want, E.J.; O’Maille, G.; Abagyan, R.; Siuzdak, G. XCMS: Processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal. Chem. 2006, 78, 779–787. [Google Scholar] [CrossRef] [PubMed]
- Giacomoni, F.; Le Corguillé, G.; Monsoor, M.; Landi, M.; Pericard, P.; Pétéra, M.; Duperier, C.; Tremblay-Franco, M.; Martin, J.F.; Jacob, D.; et al. Workflow4Metabolomics: A collaborative research infrastructure for computational metabolomics. Bioinformatics 2014, 31, 1493–1495. [Google Scholar] [CrossRef] [PubMed]
- Kuhl, C.; Tautenhahn, R.; Bottcher, C.; Larson, T.R.; Neumann, S. CAMERA: An integrated strategy for compound spectra extraction and annotation of liquid chromatography/mass spectrometry data sets. Anal. Chem. 2011, 84, 283–289. [Google Scholar] [CrossRef] [PubMed]
- Van Der Kloet, F.M.; Bobeldijk, I.; Verheij, E.R.; Jellema, R.H. Analytical error reduction using single point calibration for accurate and precise metabolomic phenotyping. J. Prot. Res. 2009, 8, 5132–5141. [Google Scholar] [CrossRef] [PubMed]
- Dunn, W.B.; Broadhurst, D.; Begley, P.; Zelena, E.; Francis-McIntyre, S.; Anderson, N.; Brown, M.; Knowles, J.D.; Halsall, A.; Haselden, J.N.; et al. Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat. Protoc. 2011, 6, 1060. [Google Scholar] [CrossRef] [PubMed]
- Ferchaud-Roucher, V.; Croyal, M.; Krempf, M.; Ouguerram, K. Plasma lipidome characterization using UHPLC-HRMS and ion mobility of hypertriglyceridemic patients on nicotinic acid. Atherosclerosis 2015, 241, e123–e124. [Google Scholar] [CrossRef]
- Van den Berg, R.A.; Hoefsloot, H.C.; Westerhuis, J.A.; Smilde, A.K.; van der Werf, M.J. Centering, scaling, and transformations: Improving the biological information content of metabolomics data. BMC Genom. 2006, 7, 142. [Google Scholar] [CrossRef] [PubMed]
- El Ghaziri, A.; Qannari El, M.; Moyon, T.; Alexandre-Gouabau, M.-C. ANOVA-PLS: A new method for the analysis of multivariate data depending on several factors. Electron. J. Appl. Stat. Anal. 2015, 8, 214–235. [Google Scholar] [CrossRef]
- Henrion, R. N-way principal component analysis theory; algorithms and applications. Chemometr. Intell. Lab. 1994, 25, 1–23. [Google Scholar] [CrossRef]
- Westerhuis, J.A.; Kourti, T.; MacGregor, J.F. Analysis of multiblock and hierarchical PCA and PLS models. J. Chemometr. 1998, 12, 301–321. [Google Scholar] [CrossRef]
- Simon, L.; Frondas-Chauty, A.; Senterre, T.; Flamant, C.; Darmaun, D.; Rozé, J.C. Determinants of body composition in preterm infants at the time of hospital discharge. Am. J. Clin. Nutr. 2014, 100, 98–104. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Steward, D.K.; Pridham, K.F. Growth patterns of extremely low-birth-weight hospitalized preterm infants. J. Obstet. Gynaecol. Neonat. Nurs. 2002, 31, 57–65. [Google Scholar] [CrossRef]
- Sprenger, N.; De Castro, C.A.; Steenhout, P.; Thakkar, S.K. Longitudinal change of selected human milk oligosaccharides and association to infants’ growth, an observatory, single center, longitudinal cohort study. PLoS ONE 2017, 12, e0171814. [Google Scholar] [CrossRef] [PubMed]
- Lynch, C.J.; Adams, S.H. Branched-chain amino acids in metabolic signalling and insulin resistance. Nat. Rev. Endocrinol. 2014, 10, 723. [Google Scholar] [CrossRef] [PubMed]
- De Luca, A.; Hankard, R.; Alexandre-Gouabau, M.C.; Ferchaud-Roucher, V.; Darmaun, D.; Boquien, C.Y. Higher concentrations of branched-chain amino acids in breast milk of obese mothers. Nutr. J. 2016, 32, 1295–1298. [Google Scholar] [CrossRef] [PubMed]
- McCormack, S.E.; Shaham, O.; McCarthy, M.A.; Deik, A.A.; Wang, T.J.; Gerszten, R.E.; Clish, C.B.; Mootha, V.K.; Grinspoon, S.K.; Fleischman, A. Circulating branched-chain amino acid concentrations are associated with obesity and future insulin resistance in children and adolescents. Pediatr. Obes. 2013, 8, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Kirchberg, F.F.; Harder, U.; Weber, M.; Grote, V.; Demmelmair, H.; Peissner, W.; Rzehak, P.; Xhonneux, A.; Carlier, C.; Ferre, N.; et al. Dietary protein intake affects amino acid and acylcarnitine metabolism in infants aged 6 months. J. Clin. Endocr. Metab. 2015, 100, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Hu, S.; Bannai, M.; Wu, G. L-Arginine regulates protein turnover in porcine mammary epithelial cells to enhance milk protein synthesis. Amino Acids 2018, 50, 621–628. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.W.; Wu, G. Regulatory role for amino acids in mammary gland growth and milk synthesis. Amino Acids 2009, 37, 89–95. [Google Scholar] [CrossRef] [PubMed]
- Tan, C.; Zhai, Z.; Ni, X.; Wang, H.; Ji, Y.; Tang, T.; Ren, W.; Long, H.; Deng, B.; Deng, J.; et al. Metabolomic profiles reveal potential factors that correlate with lactation performance in sow milk. Sci. Rep. UK 2018, 8. [Google Scholar] [CrossRef] [PubMed]
- Bode, L. Recent advances on structure; metabolism; and function of human milk oligosaccharides. J. Nutr. 2006, 136, 2127–2130. [Google Scholar] [CrossRef] [PubMed]
- Plaza-Zamora, J.; Sabater-Molina, M.M.; Rodríguez-Palmero, M.; Rivero, M.; Bosch, V.; Nadal, J.M.; Zamora, S.; Larqué, E. Polyamines in human breast milk for preterm and term infants. Br. J. Nutr. 2013, 3, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Shah, P.S.; Shah, V.S.; Kelly, L.E. Arginine supplementation for prevention of necrotising enterocolitis in preterm infants. Cochrane Database Syst. Rev. 2017. [Google Scholar] [CrossRef] [PubMed]
- Blazquez, A.M.; Cives-Losada, C.; Iglesia, A.; Marin, J.J.; Monte, M.J. Lactation during cholestasis: Role of ABC proteins in bile acid traffic across the mammary gland. Sci. Rep. UK 2017, 7, 7475. [Google Scholar] [CrossRef] [PubMed]
- Luan, H.; Liu, L.F.; Tang, Z.; Zhang, M.; Chua, K.K.; Song, J.X.; Mok, V.C.; Li, M.; Cai, Z. Comprehensive urinary metabolomic profiling and identification of potential noninvasive marker for idiopathic Parkinson’s disease. Sci. Rep. UK 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Puchalska, P.; Crawford, P.A. Multi-dimensional roles of ketone bodies in fuel metabolism; signaling; and therapeutics. Cell Metab. 2017, 25, 262–284. [Google Scholar] [CrossRef] [PubMed]
- Dingess, K.A.; Valentine, C.J.; Ollberding, N.J.; Davidson, B.S.; Woo, J.G.; Summer, S.; Peng, Y.M.; Guerrero, M.L.; Ruiz-Palacios, G.M.; Ran-Ressler, R.R.; et al. Branched-chain fatty acid composition of human milk and the impact of maternal diet: The Global Exploration of Human Milk (GEHM) Study. Am. J. Clin. Nutr. 2016, 105, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Ran-Ressler, R.R.; Khailova, L.; Arganbright, K.M.; Adkins-Rieck, C.K.; Jouni, Z.E.; Koren, O.; Ley, R.E.; Brenna, J.T.; Dvorak, B. Branched chain fatty acids reduce the incidence of necrotizing enterocolitis and alter gastrointestinal microbial ecology in a neonatal rat model. PLoS ONE 2011, 6. [Google Scholar] [CrossRef] [PubMed]
- Wongtangtintharn, S.; Oku, H.; Iwasaki, H.; TODA, T. Effect of branched-chain fatty acids on fatty acid biosynthesis of human breast cancer cells. J. Nutr. Sci. Vitaminol. 2004, 50, 137–143. [Google Scholar] [CrossRef]
- Vlaeminck, B.; Fievez, V.; Cabrita, A.R.J.; Fonseca, A.J.M.; Dewhurst, R.J. Factors affecting odd-and branched-chain fatty acids in milk: A review. Anim. Feed Sci. Tech. 2006, 131, 389–417. [Google Scholar] [CrossRef]
- Jenkins, B.; West, J.; Koulman, A. A review of odd-chain fatty acid metabolism and the role of pentadecanoic acid (C15: 0) and heptadecanoic acid (C17: 0) in health and disease. Molecules 2015, 20, 2425–2444. [Google Scholar] [CrossRef] [PubMed]
- Gill, B.D.; Indyk, H.E. Development and application of a liquid chromatographic method for analysis of nucleotides and nucleosides in milk and infant formulas. Int. Dairy J. 2007, 17, 596–605. [Google Scholar] [CrossRef]
- Gianni, M.L.; Roggero, P.; Mosca, F. Human milk protein vs. formula protein and their use in preterm infants. Cur. Opin. Clin. Nutr. 2019, 22. [Google Scholar] [CrossRef] [PubMed]
- Zeisel, S.H. Choline: Critical role during fetal development and dietary requirements in adults. Annu. Rev. Nutr. 2006, 26, 229–250. [Google Scholar] [CrossRef] [PubMed]
- Newburg, D.S.; Morelli, L. Human milk and infant intestinal mucosal glycans guide succession of the neonatal intestinal microbiota. Pediatr. Res. 2015, 77, 115–120. [Google Scholar] [CrossRef] [PubMed]

| Characteristics | “Faster” Growth Rate | “Slower” Growth Rate | p-Value |
|---|---|---|---|
| Maternal characteristics | 11 | 11 | |
| Age (years) | 29.00 ± 4.52 (25.00; 35.00) | 30.00 ± 4.12 (26.00; 33.00) | 0.908 |
| BMI before gestation (kg/m2) | 22.32 ± 5.26 (19.14; 28.91) | 24.00 ± 5.11 (20.83; 30.80) | 0.789 |
| Infants characteristics at birth | 11 (7 males and 4 females) | 15 (10 males and 5 females) | |
| Neonatal Morbidity (number of events) * | 0 | 0 | |
| Gestational age (weeks) | 31.00 ± 1.37 (30.0; 32.00) | 30.00 ± 1.68 (29.00; 32.00) | 0.288 |
| Length of hospital stay (days) | 51.50 ± 3.16 (37.25; 56.25) | 49.50 ± 4.21 (36.75; 54.75) | 0.849 |
| Birth weight (kg) | 1.200 ± 0.293 (1.020; 1.445) | 1.605 ± 0.211 (1.465; 1.705) | 0.005 |
| Birth weight Z-score (SD) | −1.592 ± 0.958 (−2.079; −0.571) | 0.564 ± 0.718 (−0.290; 0.842) | 0.000 |
| BMI at birth (kg/m2) | 7.694 ± 1.573 (7.139; 9.884) | 9.455 ± 0.857 (8.843; 9.900) | 0.161 |
| Discharge weight (kg) | 2.340 ± 0.320 (2.029; 2.520) | 2.565 ± 0.270 (2.355; 2.720) | 0.041 |
| Discharge weight Z-score (SD) | −1.878 ± 0.857 (−2.264; −1.127) | −1.142 ± 0.682 (−1.552; −0.953) | 0.146 |
| BMI at Discharge (kg/m2) | 11.98 ± 0.485 (11.66; 12.28) | 12.67 ± 0.955 (11.78; 13.36) | 0.047 |
| Difference between discharge and birth weight Z-score (SD) | −0.479 ± 0.189 (−0.668; −0.294) | −1.538 ± 0.417 (−1.953; −1.230) | <0.001 |
| Free Amino Acids (µM) | W2 to W4 Median (25% and 75% Percentile) | Mann-Whitney p-Value from W2 to W4 | FDR Corrected MW q-Value from W2 to W4 | MLR p-Value From W2 to W4 | FDR Corrected MLR q-Value from W2 to W4 | |
|---|---|---|---|---|---|---|
| “Slower” Growth (n = 38) | “Faster” Growth (n = 29) | |||||
| EAA | 234.4 (216.9–278.5) | 277.1 (221.4–343.5) | 0.0675 t | 0.18 | 0.06 # | 0.08 # |
| Arginine | 11.02 (7.56–21.84) | 18.34 (11.45–28.13) | 0.0079 ** | 0.05 * | 0.10 # | 0.08 # |
| Isoleucine | 10.12 (7.56–14.51) | 10.70 (8.12–18.01) | 0.2224 | 0.29 | 0.08 # | 0.44 |
| Leucine | 30.23 (19.93–35.46) | 32.25 (24.00–39.02) | 0.2407 | 0.29 | 0.05 # | 0.40 |
| Proline | 30.21 (26.56–37.13) | 30.32 (26.16–38.80) | 0.9425 | 0.49 | 0.85 | 0.83 |
| Methionine | 5.29 (3.11–8.35) | 6.27 (3.88–8.90) | 0.6038 | 0.43 | 0.44 | 0.74 |
| Phenylalanine | 12.43 (8.81–15.47) | 12.42 (7.25–16.43) | 0.9277 | 0.48 | 0.48 | 0.77 |
| Threonine | 86.71 (74.34–115.7) | 78.62 (66.76–107.5) | 0.3197 | 0.31 | 0.39 | 0.71 |
| Tryptophan | 2.48 (1.89–4.23) | 2.73 (1.97–4.22) | 0.7676 | 0.45 | 0.59 | 0.21 |
| Valine | 51.13 (38.90–55.06) | 52.09 (37.75–56.30) | 0.8034 | 0.45 | 0.41 | 0.71 |
| NEAA | 2610 (2146–3280) | 2512 (1753–3182) | 0.2592 | 0.32 | 0.78 | 0.25 |
| Alanine | 206.7 (186.9–332.4) | 201.0 (166.5–254.3) | 0.5466 | 0.43 | 0.78 | 0.83 |
| Aspartic acid & asparagine | 66.93 (39.53–90.83) | 57.39 (29.92–80.49) | 0.3382 | 0.31 | 0.86 | 0.83 |
| Glutamine | 455.9 (211.4–902.3) | 375.0 (134.4–572.0) | 0.0838 t | 0.17 | 0.14 | 0.52 |
| Glutamic acid | 1319 (898.5–1449) | 1220 (906.3–1480) | 0.6567 | 0.43 | 0.27 | 0.62 |
| Glx | 1838 (1381–2259) | 1754 (1177–2098) | 0.0613 t | 0.14 | 0.52 | 0.21 |
| Glycine | 89.25 (68.99–105.2) | 69.60 (54.38–103.09) | 0.0126 * | 0.05 * | 0.18 | 0.56 |
| Serine | 86.01 (68.78–116.2) | 86.01 (63.81–112.3) | 0.8034 | 0.46 | 0.23 | 0.59 |
| Tyrosine | 11.64 (6.70–15.56) | 14.49 (11.47–21.00) | 0.0349 * | 0.10 | 0.11 | 0.49 |
| Taurine | 313.9 (275.5–428.2) | 270.0 (174.0–313.2) | 0.0031 ** | 0.03 * | 0.14 | 0.51 |
| BCAA | 85.15 (71.48–93.3) | 101.1 (84.99–121.5) | 0.0075 ** | 0.04 * | 0.06 # | 0.08 # |
| Insulino-trophic amino acid | 182.0 (166.6–219.2) | 224.9 (175.7–275.0) | 0.0427 * | 0.10 t | 0.07 # | 0.08 # |
| SAA | 327.9 (297.0–430.7) | 227.9 (168.6–355.1) | 0.0019 ** | 0.03 * | 0.13 | 0.09 # |
| Abundance of Metabolites (106) | ||||||
|---|---|---|---|---|---|---|
| Metabolites (Annotation Level) | a, b, c | mz | Median (25% and 75% Percentile), W2 to W4 | |||
| “Slower“ Growth (n = 38) | “Faster“ Growth (n = 29) | Mann-Whitney p-Value (FDR-Corrected MW q-Value in Exposant) | MLR p-Value (FDR-Corrected MLR q-Value in Exposant) | |||
| Amino acid | ||||||
| Hippuric acid 1 | a | 180.0654 (M + H)+ | 1.36 (0.79–1.81) | 1.36 (0.88–2.20) | 0.86 | 0.33 |
| 2-hydroxyhippuric acid 2 | a, b, c | 194.0459 (M – H)− | 0.07 (0.05–0.126) | 0.08 (0.05–0.15) | 0.25 | 0.04 |
| Valine 1 | a | 118.0865 (M + H)+ | 2.81 (1.72–3.23) | 2.67 (0.91–3.17) | 0.79 | 0.17 |
| Leucine 1 | a, c | 130.0872 (M – H)− | 1.92 (1.45–2.87) | 2.99 (1.61–4.66) | 0.02 ** | 0.92 |
| N-Carbamoylsarcosine 2 | a, c | 133.0609 (M + H)+ | 0.96 (0.66–1.45) | 1.69 (1.18–2.38) | 0.0003 ** | 0.96 |
| Tryptophan metabolism | ||||||
| Tryptophan 1 | a, c | 205.0970 (M + H)+ | 4.20 (3.59–4.77) | 4.76 (3.51–7.95) | 0.18 | 0.79 |
| Kynurenine 1 | a, c | 192.0653 (M–NH3 + H)+ | 0.97 (0.64–1.55) | 0.72 (0.59–0.93) | 0.06 t | 0.80 |
| 1H-Indole-3-carboxaldehyde 2 | a, b | 146.0599 (M + H)+ | 2.18 (1.55–2.81) | 2.26 (1.66–3.63) | 0.27 | 0.98 |
| Indole-3-ethanol 2 | a | 184.0732 (M + Na)+ | 9.66 (6.11–13.01) | 9.59 (6.62–12.06) | 0.82 | 0.14 |
| 3-Methylindole 2 | a | 132.0806 (M + H)+ | 0.43 (0.35–0.55) | 0.47 (0.39–0.63) | 0.10 | 0.70 |
| Tyrosine metabolism | ||||||
| hydroxyphenylacetic acid 1 | a, c | 151.0399 (M – H)− | 0.32 (0.25–0.37) | 0.30 (0.21–0.37) | 0.61 | 0.08 |
| p-Cresol (4-methylphenol) 2 | a, b, c | 107.0501 (M – H)− | 1.84 (1.16–2.85) | 1.37 (0.09–2.21) | 0.04 * | 0.20 |
| p-Cresol sulfate 2 | a | 187.0070 (M – H)− | 5.28 (4.14–9.27) | 4.89 (2.67–6.79) | 0.13 t | 0.17 |
| Sulphur metabolism | ||||||
| Cystathionine 2 | a | 240.1015 (M + NH4)+ | 0.67 (0.51–0.80) | 0.87 (0.66–1.35) | 0.02 * | 0.90 |
| Methionin 1 | a, c | 150.0580 (M + H)+ | 1.82 (1.49–2.19) | 1.81 (1.63–2.69) | 0.36 | 0.89 |
| Se-Adenosylselenohomocysteine 2 | a | 228.0314 (M + H + Na)+ | 0.22 (0.17–0.25) | 0.19 (0.13–0.24) | 0.15 t | 0.09 |
| S-Adenosylhomocysteine 2 | a | 365.1048 (M – H2O– H)+ | 6.53 (5.02–7.63) | 4.48 (3.92–6.84) | 0.007 * | 0.20 |
| Hydrogen sulfite 2 | a, b, c | 79.9573 (M – H)− | 0.52 (0.38–0.85) | 0.44 (0.31–0.67) | 0.07 t | 0.23 |
| Thiocyanic acid 2 | a, c | 150.0018 (M – H)− | 0.75 (0.64–0.83) | 0.57 (0.51–0.76) | 0.02 * | 0.69 |
| Aromatic compound | ||||||
| Benzoic acid 1 | a | 121.0294 (M – H)− | 3.14 (2.27–4.18) | 2.49 (2.09–2.95) | 0.01 * | 0.16 |
| Hydroxyphenyllactic acid 2 | a | 241.0730 (M + Hac-H)− | 0.14 (0.11–0.16) | 0.11 (0.08–0.18) | 0.17 | 0.96 |
| Pyridines and Derivatives/Nucleosides | ||||||
| Niacinamide 1 | a | 123.0554 (M + H)+ | 6.45 (5.27–10.82) | 11.00 (6.17–13.84) | 0.029 * | 0.16 |
| Energy metabolism | ||||||
| Hydroxyhexanoylcarnitine 2 | a | 276.1803 (M + H)+ | 0.11 (0.04–0.22) | 0.08 (0.05–0.21) | 0.84 | 0.52 |
| Oxoicosanoyl-CoA 2 | a, b | 547.2129 (M + H+ NH4)+ | 0.66 (0.53–0.89) | 0.81 (0.63–1.20) | 0.04 * | 0.87 |
| 3-Hydroxypimelyl-CoA 2 | a | 943.2103 (M + NH4)+ | 1.77 (1.66–2.02) | 1.79 (1.61–2.04) | 0.98 | 0.91 |
| Hexanoylglycine 2 | a, b | 174.1123 (M + H)+ | 0.20 (0.12–0.26) | 0.13 (0.08–0.19) | 0.02 * | 0.15 |
| Heptanoylglycine 2 | a | 229.1544 (M + H)+ | 0.64 (0.50–0.97) | 0.65 (0.49–0.81) | 0.58 | 0.69 |
| Gamma-Butyrolactone/ | a | 85.0293 (M – H)− | 42.9 (25.08–58.13) | 48.37 (34.67–65.61) | 0.32 | 0.68 |
| But-2-enoic/Isocrotonic acid 2 | a, b | 631.3089 (M – H)− | 0.01 (0.00–0.03) | 0.02 (0.01–0.06) | 0.01 * | 0.63 |
| butyl 2-dodecanoic acid/ 5-Tetra dodecanoic acid 2 | a | 225.1859 (M – H)− | 0.09 (0.04–0.19) | 0.06 (0.01–0.10) | 0.05 t | 0.81 |
| caproic acid 1 | a | 115.0763 (M – H)− | 0.13 (0.11–0.15) | 0.14 (0.11–0.18) | 0.46 | 0.26 |
| 3-hydroxycapric acid 2 | a, b, c | 187.1339 (M – H)− | 0.43 (0.26–0.81) | 0.62 (0.50–1.11) | 0.01 * | 0.97 |
| Geranic acid 2 | 167.1077 (M – H)− | 0.09 (0.07–0.22) | 0.08 (0.06–0.16) | 0.13 | 0.03 | |
| Sebacic acid 1 | a, c | 261.1345 (M-CH3COO)− | 0.10 (0.07–0.18) | 0.13 (0.08–0.19) | 0.18 t | 0.72 |
| 3-Hydroxysebacic acid 2 | a, c | 217.1081 (M – H)− | 0.04 (0.03–0.06) | 0.06 (0.03–0.07) | 0.07 t | 0.68 |
| 3,4-Methylenesebacic acid 2 | a | 225.1132 (M – H)− | 0.04 (0.03–0.06) | 0.03 (0.02–0.04) | 0.12 t | 0.82 |
| 2-Hydroxybutyric acid 1 | a, b, c | 103.0399 (M – H)− | 2.93 (2.35–3.72) | 3.85 (3.05–4.81) | 0.005 * | 0.77 |
| 2-hydroxy-3-methylbutyric acid 1 | a, b, c | 117.0555 (M – H)− | 0.99 (0.81–1.82) | 1.52 (1.07–2.07) | 0.01 * | 0.06 |
| pyridosine 2 | a, c | 253.1195 (M – H)− | 0.20 (0.09–0.28) | 0.11 (0.07–0.22) | 0.06 t | 0.17 |
| Glycerophosphorylcholine 2 | a, c | 292.0724 (M – H)− | 0.68 (0.49–0.96) | 0.58 (0.41–0.71) | 0.07 t | 0.40 |
| N-Heptanoylglycine 2 | a, b | 186.1135 (M – H)− | 0.93 (0.47–1.83) | 0.61 (0.40–1.16) | 0.07 t | 0.14 |
| Butyryl glycine/Saccharopine 2 | a | 335.1455 * (M + Fa − H)− | 0.22 (0.11–0.53) | 0.28 (0.08–0.51) | 0.58 | 0.62 |
| 2-Phenylglycine 2 | a, c | 150.0559 (M – H)− | 0.14 (0.09–0.22) | 0.17 (0.10–0.29) | 0.11 t | 0.08 |
| Cis-aconitic acid 1 | a | 154.9983 (M-H2O – H) − | 1.33 (0.58–2.27) | 1.73 (1.28–2.49) | 0.09 t | 0.85 |
| Pyruvic acid 1 | a, b, c | 147.0297 (M-CH3COO)− | 2.15 (1.27–3.49) | 3.55 (1.94–6.10) | 0.03 * | 0.95 |
| Citraconic 1 | a, b, c | 129.0192 (M – H)− | 13.82 (9.49–26.19) | 21.43 (13.82–29.91) | 0.02 * | 0.95 |
| 2-Keto-glutaramic acid 2 | a, c | 144.0302 (M – H)− | 0.38 (0.30–0.44) | 0.37 (0.31–0.58) | 0.21 | 0.44 |
| Panthothenic acid 1 | a, c | 200.0929 (M-H2O – H)− | 0.09 (0.06–0.11) | 0.10 (0.08–0.11) | 0.51 | 0.007 |
| 4-Heptenal 2 | a, b | 111.0814 (M – H)− | 0.43 (0.33–0.50) | 0.35 (0.26–0.45) | 0.04 * | 0.55 |
| 2-Methylpentanal 2 | a | 99.0814 (M – H)− | 0.12 (0.09–0.13) | 0.10 (0.09–0.12) | 0.06 t | 0.98 |
| Undecenal 2 | a, b, c | 167.1440 (M – H)− | 0.08 (0.06–0.12) | 0.06 (0.04–0.13) | 0.006 * | 0.02 |
| Methyl 2-octynoate 2 | a | 153.0919 (M – H)− | 0.10 (0.07–0.13) | 0.07 (0.06–0.11) | 0.09 t | 0.06 |
| 4-Methylphenyl-acetaldehyde 2 | a | 133.0658 (M – H)− | 0.07 (0.05–0.11) | 0.06 (0.05–0.09) | 0.04 * | 0.14 |
| 4-Hydroxynonenal 2 | a, c | 155.1077 (M – H)− | 0.06 (0.04–0.09) | 0.07 (0.05–0.10) | 0.13 t | 0.79 |
| cis-4-Decenedioic acid 2 | a, b, c | 199.0973 (M – H)− | 0.07 (0.05–0.11) | 0.10 (0.07–0.16) | 0.01 * | 0.94 |
| Tetradecanedioic acid 1 | a, c | 257.1761 (M – H)− | 0.08 (0.05–0.12) | 0.12 (0.08–0.19) | 0.07 t | 0.62 |
| Dodecanedioic acid 2 | a, c | 229.1445 (M – H)− | 0.41 (0.30–0.56) | 0.35 (0.30–0.54) | 0.42 | 0.01 |
| Heptanoic acid 2 | a, c | 129.0920 (M – H)− | 0.17 (0.14–0.21) | 0.14 (0.10–0.19) | 0.06 t | 0.74 |
| 2-benzyloctanoic acid 2 | a, b, c | 233.1544 (M – H)− | 1.45 (0.78–2.31) | 0.87 (0.61–1.45) | 0.007 * | 0.81 |
| N-methylethanolaminium phosphate 2 | a, b, c | 136.0165 (M–H2O – H)− | 0.36 (0.27–0.51) | 0.26 (0.19–0.30) | <0.0001 ** | 0.81 |
| Phosphorylcholine 1 | a, | 206.0551 (M + Na)+ | 5.78 (4.26–6.63) | 4.03 (0.68–5.67) | 0.001 * | 0.21 |
| Glycerophosphocholin 2 | a | 280.0917 (M + Na)+ | 18.88 (9.66–30.18) | 16.98 (3.54–24.57) | 0.12 t | 0.57 |
| Choline 1 | a, | 105.11080 (M + H)+ | 2.67 (2.25–3.66) | 3.30 (2.74–4.65) | 0.008 * | 0.02 |
| Glucuronide/oligosides | ||||||
| Dihydrocaffeic acid 3-O-glucuronide 2 | a, b, c | 383.0763 (M + Na)+ | 1.15 (0.87–1.29) | 1.05 (0.86–1.23) | 0.49 | 0.51 |
| 2-Fucosyllactose 2 | a, c | 511.1629 (M + H)+ | 106.6 (82.1–152.5) | 122.6 (98.5–148.7) | 0.34 | 0.79 |
| N-acetyl-D-glucosamine 2 | a | 244.0788 (M + Na)+ | 2.94 (2.71–3.51) | 3.20 (2.55–3.81) | 0.74 | 0.68 |
| Lacto-N-fucopentaose-2 2 | a, b | 876.2936 (M + Na)+ | 9.29 (7.62–12.68) | 10.56 (8.07–15.04) | 0.15 t | 0.72 |
| Saccharopine 2 | a, c | 335.1455 (M-CH3COO)− | 0.22 (0.11–0.53) | 0.28 (0.08–0.51) | 0.58 | 0.62 |
| HMOs | Composition | Median (25% and 75% Percentile) from W2 to W4 (Secretors and Non Secretors Mothers) | Mann-Whitney p-Value (FDR-Corrected MW q-Value in Exposant) | VIP –PLS-DA (C1–C2) | MLR-p-value (FDR-Corrected MLR q-value in Exposant) | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| mz | RT | Hex | HexNac | Fuc | NeuAc | “Slower” Growth (n = 38) | “Faster” Growth (n = 29) | Secretors and Non Secretors | Secretors Only | Secretors and Non Secretors | Secretors Only | Secretors and Non Secretors | Secretors Only | |
| Fucosylated | 61.46 (50.28–65.13) | 62.82 (60.20–65.19) | 0.1847 t | 0.1370 | 0.73 | |||||||||
| Sialylated | 8.47 (6.97–9.40) | 7.45 (6.70–9.11) | 0.2545 | 0.2773 | 0.37 | |||||||||
| Fucosylated./Sialylated | 1.95 (1.50-2.32) | 1.63 (1.33–2.30) | 0.0973 t | 0.1422 | 0.35 | |||||||||
| Neutral | 28.30 (26.78–38.23) | 26.87 (25.92–29.57) | 0.0352 * | 0.6966 | 0.95 | |||||||||
| Mono Fucosylated | 28.49 (19.06–34.74) | 36.95 (31.83-39.21) | 0.0020 | 0.0669 | 0.19 | |||||||||
| Di Fucosylated | 26.64 (23.83–28.53) | 23.12 (19.14–26.77) | 0.0026 | 0.0054 * | 0.10 | |||||||||
| Tri Fucosylated | 3.52 (2.96–4.00) | 2.85 (2.63–3.26) | 0.0061 | 0.0883 | 0.43 | |||||||||
| Tetra Fucosylated | 3.20 (2.54–4.07) | 2.94 (1.77–3.83) | 0.1520 | 0.0243 * | 0.004 * | |||||||||
| LNFPI≠ | 856.3280 | 10.2 | 3 | 1 | 1 | 0 | 11.99 (0.36-18.65) | 19.86 (15.00–23.69) | 0.0003 ** | 0.0045 * | 1.22 | 1.26 | 0.63 | 0.03 * |
| pLNH≠ | 1075.4023 | 18 | 4 | 2 | 0 | 0 | 0.26 (0.14–0.35) | 0.38 (0.26–0.54) | 0.0014 ** | 0.0013 * | 1.44 | 1.75 | 0.50 | 0.10 * |
| 2’-FL≠ | 491.1958 | 8.5 | 2 | 0 | 1 | 0 | 11.22 (0.10–13.87) | 12.58 (10.29–15.09) | 0.0882 t | 0.9433 | 1.10 | 1.08 | 0.63 | 0.22 * |
| 6’-SL | 636.2333 | 9.6 | 2 | 0 | 0 | 1 | 2.50 (2.30–3.06) | 2.47 (1.81–3.05] | 0.3219 | 0.9433 | 0.65 | 0.29 | 0.26 | 0.31 * |
| LNnH≠ | 1075.4023 | 15.1 | 4 | 2 | 0 | 0 | 0.82 (0.32–1.44) | 0.95 (0.58–2.31) | 0.0689 t | 0.8116 | 1.25 | 1.22 | 0.67 | 0.52 t |
| LNT/LNnT≠ | 710.2701 | 10.9 | 3 | 1 | 0 | 0 | 22.52 (20.29–35.65) | 22.46 (20.04–25.21) | 0.1991 t | 0.2591 | 1.00 | 0.81 | 0.48 | 0.62 t |
| LSTc/b | 1001.3655 | 18.4 | 3 | 1 | 0 | 1 | 2.59 (2.00–3.57) | 2.63 (2.25–3.41) | 0.7012 | 0.1422 | 0.89 | 0.87 | 0.78 | 0.65 t |
| LNDFH I | 1002.3859 | 5.7 | 3 | 1 | 2 | 0 | 4.26 (0.44–6.25) | 5.10 (0.51–6.96) | 0.3986 | 0.8903 | 0.55 | 0.74 | 0.22 | 0.50 t |
| 3’ FL | 491.1958 | 1.8 | 2 | 0 | 1 | 0 | 0.03 (0.00–0.08) | 0.00 (0.00–0.03) | 0.0133 * | 0.1697 | 0.94 | 0.70 | 0.12 | 0.09 * |
| 3’SL | 636.2333 | 18.9 | 2 | 0 | 0 | 1 | 0.15 (0.13–0.21) | 0.14 (0.11–0.18) | 0.1555 t | 0.9433 | 0.46 | 0.48 | 0.26 | 0.16 * |
| LNDFH ≠ | 1002.3859 | 9.6 | 3 | 1 | 2 | 0 | 0.26 (0.14-0.36) | 0.36 (0.27-0.45) | 0.0020 ** | 0.1050 | 1.12 | 1.05 | 0.43 | 0.04 * |
| LNDFHx | 1002.3859 | 6.8 | 3 | 1 | 2 | 0 | 0.26 (0.11–1.11) | 0.14 (0.07–0.24) | 0.0293 * | 0.0726 t | 0.97 | 1.04 | 0.15 | 0.06 * |
| 4230c≠ | 1513.5760 | 13.5 | 4 | 2 | 3 | 0 | 0.11 (0.00–0.18) | 0.07 (0.04–0.14) | 0.5405 | 0.0002 ** | 1.90 | 1.55 | 0.0062 * | 0.07 * |
| 4210d≠ | 1221.4602 | 17.4 | 4 | 2 | 1 | 0 | 0.18 (0.00–0.39) | 0.47 (0.27–1.31) | 0.0005 ** | 0.0142 * | 1.69 | 1.66 | 0.10 | 0.04 * |
| 4220e≠ | 1367.5181 | 13 | 4 | 2 | 2 | 0 | 1.59 (0.37–2.43) | 1.75 (1.35–2.77) | 0.1799 t | 0.6128 | 1.28 | 1.76 | 0.86 | 0.78 t |
| 4230b≠ | 1513.576 | 8.2 | 4 | 2 | 3 | 0 | 0.65 (0.00–1.49) | 0.61 (0.06–1.20) | 0.7967 | 0.0893 t | 1.38 | 1.33 | 0.0095 * | 0.09 * |
| 3000≠ | 507.1907 | 6.4 | 3 | 0 | 0 | 0 | 0.26 (0.19–0.31) | 0.16 (0.12–0.23) | <0.0001 *** | 0.0022 * | 2.06 | 0.90 | 0.53 | 0.22 * |
| 5300 (2+) | 720.7709 | 18.1 | 5 | 3 | 0 | 0 | 0.15 (0.10–0.23) | 0.16 (0.14–0.32) | 0.0771 t | 0.6305 | 1.41 | 1.39 | 0.72 | 0.37 * |
| 6420c (2+)≠ | 1049.3949 | 18.1 | 6 | 4 | 2 | 0 | 0.44 (0.09–0.64) | 0.50 (0.32–0.72) | 0.1893 t | 0.5220 | 1.11 | 0.67 | 0.30 | 0.76 t |
| 6430d (2+)≠ | 1122.4238 | 16.8 | 6 | 4 | 3 | 0 | 0.14 (0.00–0.26) | 0.15 (0.10–0.24) | 0.2102 t | 0.2663 | 1.18 | 1.00 | 0.34 | 0.87 t |
| 5310c | 1586.5924 | 18 | 5 | 3 | 1 | 0 | 0.29 (0.22–0.38) | 0.38 (0.25–0.46) | 0.0428 * | 0.4094 | 0.90 | 1.02 | 0.95 | 0.16 * |
| 2110a | 694.2752 | 3.6 | 2 | 1 | 1 | 0 | 0.01 (0.00–0.27) | 0.24 (0.00–0.51) | 0.0811 t | 0.6772 | 0.90 | 1.49 | 0.48 | 0.60 t |
| 4240b≠ | 1659.6339 | 13.9 | 4 | 2 | 4 | 0 | 0.04 (0.00–0.11) | 0.04 (0.00–0.08) | 0.9654 | 0.0457 * | 1.29 | 0.68 | 0.0007 ** | 0.05 * |
| 2020a≠ | 637.2537 | 11.1 | 2 | 0 | 2 | 0 | 0.49 (0.04–0.61) | 0.55 (0.45–0.66) | 0.0840 t | 0.9700 | 1.10 | 1.23 | 0.72 | 0.20 * |
| 5310b | 1586.5924 | 17 | 5 | 3 | 1 | 0 | 0.24 (0.13–0.31) | 0.16 (0.09–0.27) | 0.1091 t | 0.0420 * | 0.93 | 0.97 | 0.84 | 0.40 * |
| 4220a | 684.2627 | 7.7 | 4 | 2 | 2 | 0 | 0.20 (0.14–0.29) | 0.11 (0.07–0.25) | 0.0216 * | 0.0911 t | 0.49 | 0.52 | 0.10 | 0.82 t |
| 4220b≠ | 1367.5181 | 8.8 | 4 | 2 | 2 | 0 | 0.31 (0.09–1.78) | 0.23 (0.07–0.33) | 0.0895 t | 0.3352 | 1.02 | 0.99 | 0.15 | 0.14 * |
| 4210b≠ | 1221.4602 | 12.6 | 4 | 2 | 1 | 0 | 7.19 (5.03–8.90) | 3.75 (2.37–6.33) | <0.0001 *** | 0.0003 ** | 1.05 | 1.84 | 0.30 | 0.07 * |
| 5330b | 1878.7082 | 13.9 | 5 | 3 | 3 | 0 | 0.23 (0.00–0.39) | 0.29 (0.00–0.38) | 0.3749 | 0.4106 | 0.96 | 1.10 | 0.05 | 0.26 * |
| 5320a | 1732.6503 | 12.9 | 5 | 3 | 2 | 0 | 0.27 (0.13–0.70) | 0.13 (0.05–0.22) | 0.0004 ** | 0.0045 * | 1.06 | 1.37 | 0.47 | 0.16 * |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alexandre-Gouabau, M.-C.; Moyon, T.; David-Sochard, A.; Fenaille, F.; Cholet, S.; Royer, A.-L.; Guitton, Y.; Billard, H.; Darmaun, D.; Rozé, J.-C.; et al. Comprehensive Preterm Breast Milk Metabotype Associated with Optimal Infant Early Growth Pattern. Nutrients 2019, 11, 528. https://doi.org/10.3390/nu11030528
Alexandre-Gouabau M-C, Moyon T, David-Sochard A, Fenaille F, Cholet S, Royer A-L, Guitton Y, Billard H, Darmaun D, Rozé J-C, et al. Comprehensive Preterm Breast Milk Metabotype Associated with Optimal Infant Early Growth Pattern. Nutrients. 2019; 11(3):528. https://doi.org/10.3390/nu11030528
Chicago/Turabian StyleAlexandre-Gouabau, Marie-Cécile, Thomas Moyon, Agnès David-Sochard, François Fenaille, Sophie Cholet, Anne-Lise Royer, Yann Guitton, Hélène Billard, Dominique Darmaun, Jean-Christophe Rozé, and et al. 2019. "Comprehensive Preterm Breast Milk Metabotype Associated with Optimal Infant Early Growth Pattern" Nutrients 11, no. 3: 528. https://doi.org/10.3390/nu11030528
APA StyleAlexandre-Gouabau, M.-C., Moyon, T., David-Sochard, A., Fenaille, F., Cholet, S., Royer, A.-L., Guitton, Y., Billard, H., Darmaun, D., Rozé, J.-C., & Boquien, C.-Y. (2019). Comprehensive Preterm Breast Milk Metabotype Associated with Optimal Infant Early Growth Pattern. Nutrients, 11(3), 528. https://doi.org/10.3390/nu11030528






