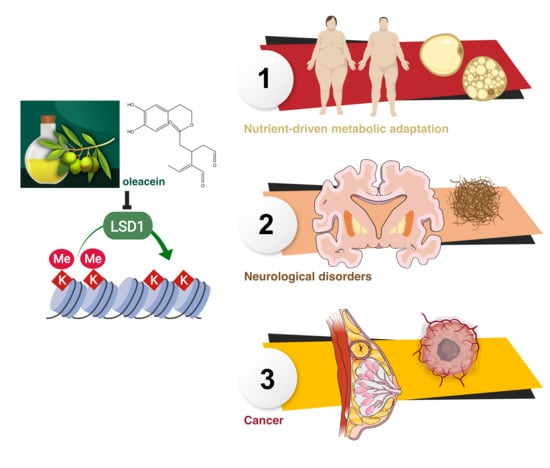
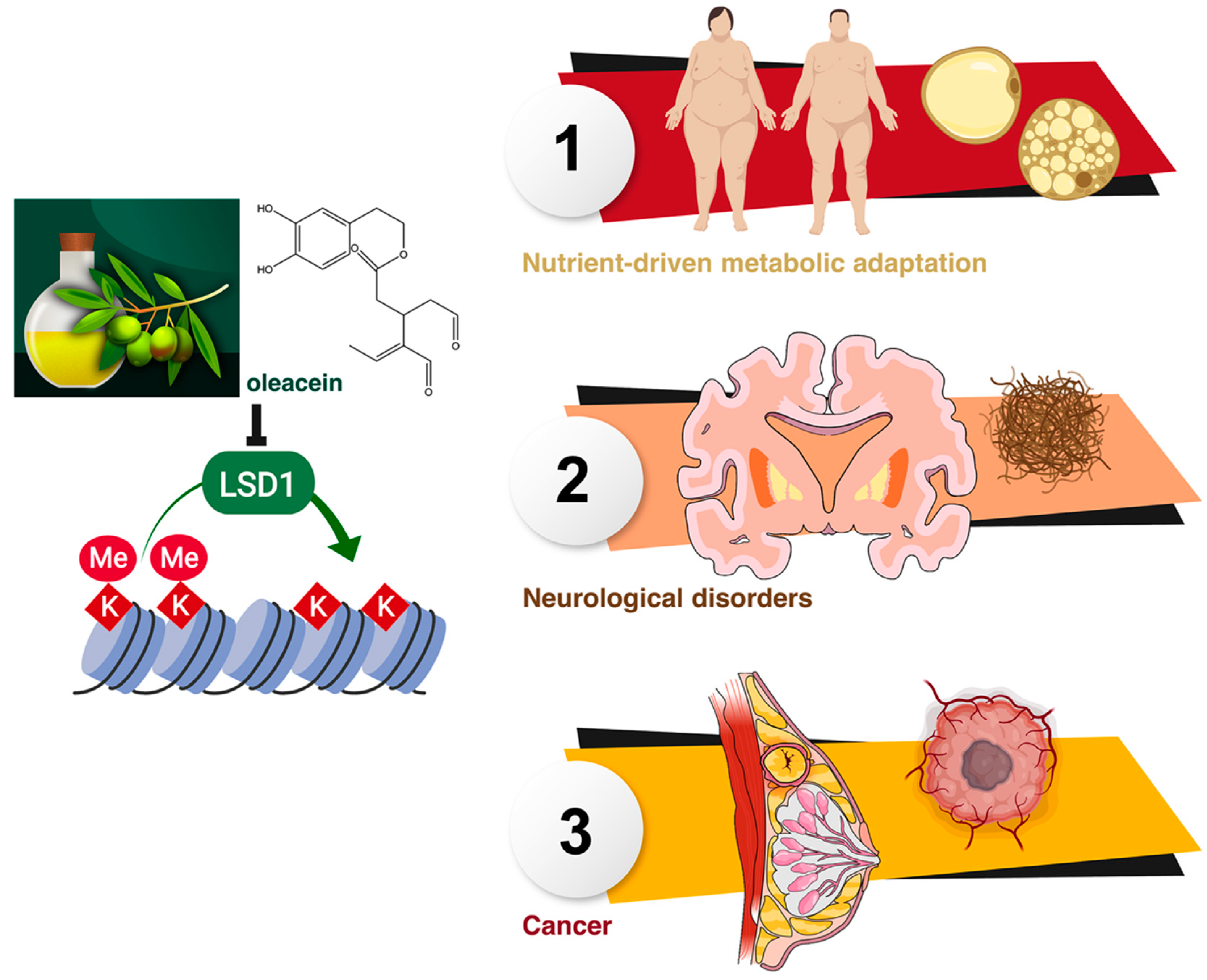
Extra Virgin Olive Oil Contains a Phenolic Inhibitor of the Histone Demethylase LSD1/KDM1A
Abstract
:1. Introduction
2. Materials and Methods
2.1. Docking Calculations, Molecular Dynamics Simulations, and Binding Free Energy Analysis
2.2. Oleacein Isolation and Purification
2.3. LSD1 Enzymatic Activity
2.4. Cell Lines and Culture Conditions
2.5. SOX2 Enhancer Reporter Assay
2.6. SOX2 Protein Expression
2.7. SOX2 Gene Expression
2.8. Statistical Analysis
3. Results
3.1. Computational Prediction of LSD1 as a Putative Target of Oleacein
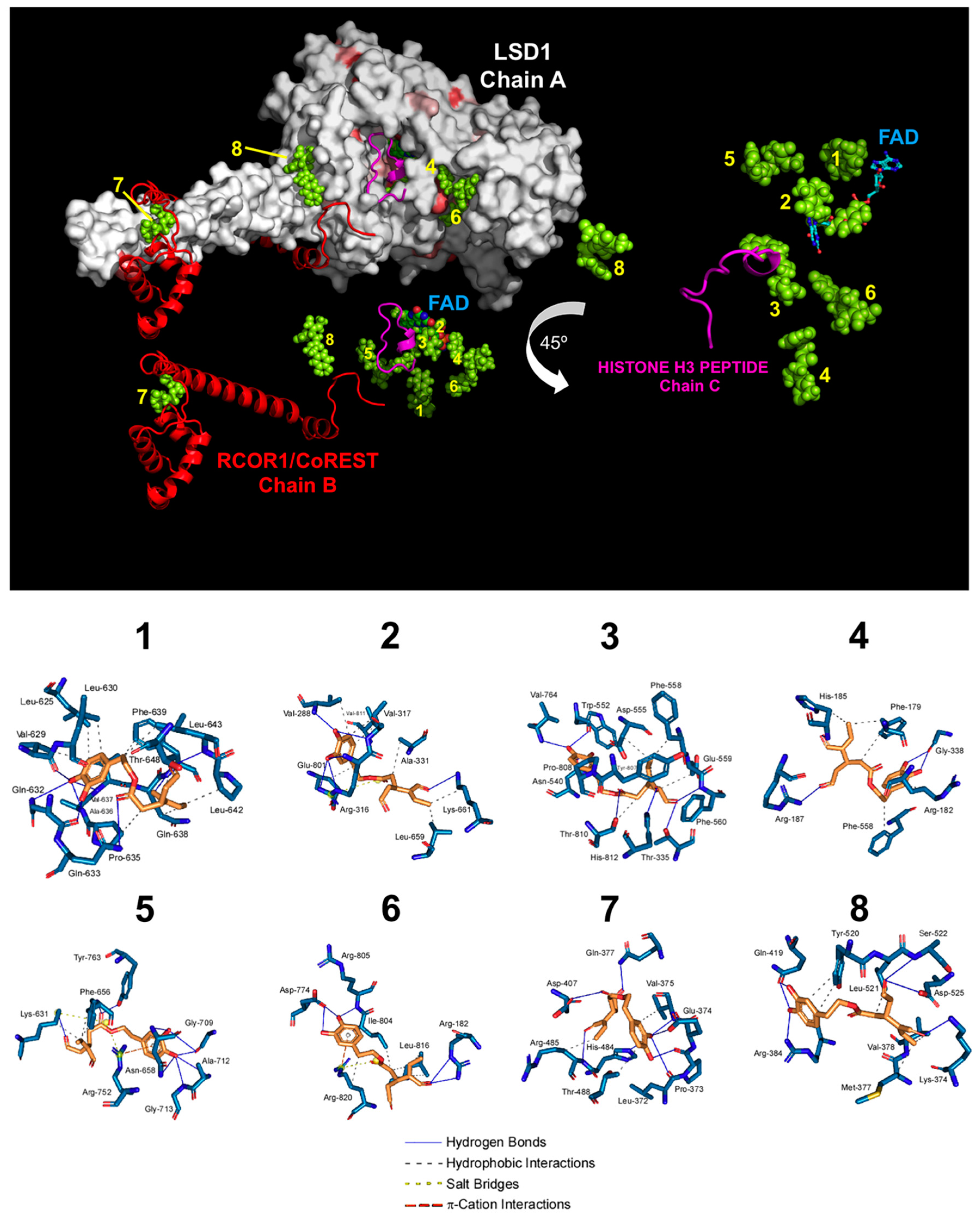
3.2. Computational Deconstruction of the Binding Mode of Oleacein to the LSD1-CoREST-histone H3 Complex
3.3. Oleacein Inhibits the In Vitro Enzymatic Activity of LSD1
3.4. Oleacein Suppresses LSD1-Targeted Enhancer-Driven Activation of the Stemness Transcription Factor SOX2 in Stem-Like Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Availability of Data and Materials
References
- Menendez, J.A.; Joven, J.; Aragonès, G.; Barrajón-Catalán, E.; Beltrán-Debón, R.; Borrás-Linares, I.; Camps, J.; Corominas-Faja, B.; Cufí, S.; Fernández-Arroyo, S.; et al. Xenohormetic and anti-aging activity of secoiridoid polyphenols present in extra virgin olive oil: A new family of gerosuppressant agents. Cell Cycle 2013, 12, 555–578. [Google Scholar] [CrossRef] [PubMed]
- Rigacci, S.; Stefani, M. Nutraceutical Properties of Olive Oil Polyphenols. An Itinerary from Cultured Cells through Animal Models to Humans. Int. J. Mol. Sci. 2016, 17, 843. [Google Scholar] [CrossRef] [PubMed]
- Angeloni, C.; Malaguti, M.; Barbalace, M.C.; Hrelia, S. Bioactivity of Olive Oil Phenols in Neuroprotection. Int. J. Mol. Sci. 2017, 18, 2230. [Google Scholar] [CrossRef] [PubMed]
- Peyrol, J.; Riva, C.; Amiot, M.J. Hydroxytyrosol in the Prevention of the Metabolic Syndrome and Related Disorders. Nutrients 2017, 9, 306. [Google Scholar] [CrossRef] [PubMed]
- Saibandith, B.; Spencer, J.P.E.; Rowland, I.R.; Commane, D.M. Olive Polyphenols and the Metabolic Syndrome. Molecules 2017, 22, 1082. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, G.E.; Lepore, S.M.; Morittu, V.M.; Arcidiacono, B.; Colica, C.; Procopio, A.; Maggisano, V.; Bulotta, S.; Costa, N.; Mignogna, C.; et al. Effects of Oleacein on High-Fat Diet-Dependent Steatosis, Weight Gain, and Insulin Resistance in Mice. Front. Endocrinol. 2018, 9, 116. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Reboredo-Rodríguez, P.; Varela-López, A.; Forbes-Hernández, T.Y.; Gasparrini, M.; Afrin, S.; Cianciosi, D.; Zhang, J.; Manna, P.P.; Bompadre, S.; Quiles, J.L.; et al. Phenolic Compounds Isolated from Olive Oil as Nutraceutical Tools for the Prevention and Management of Cancer and Cardiovascular Diseases. Int. J. Mol. Sci. 2018, 19, 2305. [Google Scholar] [CrossRef] [PubMed]
- Pedret, A.; Fernández-Castillejo, S.; Valls, R.M.; Catalán, Ú.; Rubió, L.; Romeu, M.; Macià, A.; López de Las Hazas, M.C.; Farràs, M.; Giralt, M.; et al. Cardiovascular Benefits of Phenol-Enriched Virgin Olive Oils: New Insights from the Virgin Olive Oil and HDL Functionality (VOHF) Study. Mol. Nutr. Food Res. 2018, 62, 1800456. [Google Scholar] [CrossRef]
- Celano, M.; Maggisano, V.; Lepore, S.M.; Russo, D.; Bulotta, S. Secoiridoids of olive and derivatives as potential coadjuvant drugs in cancer: A critical analysis of experimental studies. Pharmacol. Res. 2019, 142, 77–86. [Google Scholar] [CrossRef]
- Corominas-Faja, B.; Cuyàs, E.; Lozano-Sánchez, J.; Cufí, S.; Verdura, S.; Fernández-Arroyo, S.; Borrás-Linares, I.; Martin-Castillo, B.; Martin, Á.G.; Lupu, R.; et al. Extra-virgin olive oil contains a metabolo-epigenetic inhibitor of cancer stem cells. Carcinogenesis 2018, 39, 601–613. [Google Scholar] [CrossRef] [Green Version]
- Verdura, S.; Cuyàs, E.; Lozano-Sánchez, J.; Bastidas-Velez, C.; Llorach-Parés, L.; Fernández-Arroyo, S.; Hernández-Aguilera, A.; Joven, J.; Nonell-Canals, A.; Bosch-Barrera, J.; et al. An olive oil phenolic is a new chemotype of mutant isocitrate dehydrogenase 1 (IDH1) inhibitors. Carcinogenesis 2019, 40, 27–40. [Google Scholar] [CrossRef] [PubMed]
- Cuyàs, E.; Castillo, D.; Llorach-Parés, L.; Lozano-Sánchez, J.; Verdura, S.; Nonell-Canals, A.; Brunet, J.; Bosch-Barrera, J.; Joven, J.; Valdés, R.; et al. Computational de-orphanization of the olive oil biophenols oleacein: Discovery of new metabolic and epigenetic targets. Food Chem. Toxicol. 2019, 131, 110529. [Google Scholar] [CrossRef] [PubMed]
- Shi, Y.; Lan, F.; Matson, C.; Mulligan, P.; Whetstine, J.R.; Cole, P.A.; Casero, R.A.; Shi, Y. Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 2004, 119, 941–953. [Google Scholar] [CrossRef] [PubMed]
- Lan, F.; Nottke, A.C.; Shi, Y. Mechanisms involved in the regulation of histone lysine demethylases. Curr. Opin. Cell Biol. 2008, 20, 316–325. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Metzger, E.; Wissmann, M.; Yin, N.; Müller, J.M.; Schneider, R.; Peters, A.H.; Günther, T.; Buettner, R.; Schüle, R. LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 2005, 437, 436–439. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Bassets, I.; Kwon, Y.S.; Telese, F.; Prefontaine, G.G.; Hutt, K.R.; Cheng, C.S.; Ju, B.G.; Ohgi, K.A.; Wang, J.; Escoubet-Lozach, L.; et al. Histone methylation-dependent mechanisms impose ligand dependency for gene activation by nuclear receptors. Cell 2007, 128, 505–518. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Scully, K.; Zhu, X.; Cai, L.; Zhang, J.; Prefontaine, G.G.; Krones, A.; Ohgi, K.A.; Zhu, P.; Garcia-Bassets, I.; et al. Opposing LSD1 complexes function in developmental gene activation and repression programmes. Nature 2007, 446, 882–887. [Google Scholar] [CrossRef]
- Forneris, F.; Binda, C.; Battaglioli, E.; Mattevi, A. LSD1: Oxidative chemistry for multifaceted functions in chromatin regulation. Trends Biochem. Sci. 2008, 33, 181–189. [Google Scholar] [CrossRef]
- Abdulla, A.; Zhang, Y.; Hsu, F.N.; Xiaoli, A.M.; Zhao, X.; Yang, E.S.; Ji, J.Y.; Yang, F. Regulation of lipogenic gene expression by lysine-specific histone demethylase-1 (LSD1). J. Biol. Chem. 2014, 289, 29937–29947. [Google Scholar] [CrossRef]
- Duteil, D.; Metzger, E.; Willmann, D.; Karagianni, P.; Friedrichs, N.; Greschik, H.; Günther, T.; Buettner, R.; Talianidis, I.; Metzger, D.; et al. LSD1 promotes oxidative metabolism of white adipose tissue. Nat. Commun. 2014, 5, 4093. [Google Scholar] [CrossRef]
- Duteil, D.; Tosic, M.; Lausecker, F.; Nenseth, H.Z.; Müller, J.M.; Urban, S.; Willmann, D.; Petroll, K.; Messaddeq, N.; Arrigoni, L.; et al. Lsd1 Ablation Triggers Metabolic Reprogramming of Brown Adipose Tissue. Cell Rep. 2016, 17, 1008–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maes, T.; Mascaró, C.; Ortega, A.; Lunardi, S.; Ciceri, F.; Somervaille, T.C.; Buesa, C. KDM1 histone lysine demethylases as targets for treatments of oncological and neurodegenerative disease. Epigenomics 2015, 7, 609–626. [Google Scholar] [CrossRef]
- Sakamoto, A.; Hino, S.; Nagaoka, K.; Anan, K.; Takase, R.; Matsumori, H.; Ojima, H.; Kanai, Y.; Arita, K.; Nakao, M. Lysine Demethylase LSD1 Coordinates Glycolytic and Mitochondrial Metabolism in Hepatocellular Carcinoma Cells. Cancer Res. 2015, 75, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Sun, H.; Liang, L.; Li, Y.; Feng, C.; Li, L.; Zhang, Y.; He, S.; Pei, D.; Guo, Y.; Zheng, H. Lysine-specific histone demethylase 1 inhibition promotes reprogramming by facilitating the expression of exogenous transcriptional factors and metabolic switch. Sci. Rep. 2016, 6, 30903. [Google Scholar] [CrossRef] [Green Version]
- Abdulla, A.; Zhao, X.; Yang, F. Natural Polyphenols Inhibit Lysine-Specific Demethylase-1 in vitro. J. Biochem. Pharmacol. Res. 2013, 1, 56–63. [Google Scholar] [PubMed]
- Duan, Y.C.; Guan, Y.Y.; Zhai, X.Y.; Ding, L.N.; Qin, W.P.; Shen, D.D.; Liu, X.Q.; Sun, X.D.; Zheng, Y.C.; Liu, H.M. Discovery of resveratrol derivatives as novel LSD1 inhibitors: Design, synthesis and their biological evaluation. Eur. J. Med. Chem. 2017, 126, 246–258. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.C.; Shen, D.D.; Ren, M.; Liu, X.Q.; Wang, Z.R.; Liu, Y.; Zhang, Q.N.; Zhao, L.J.; Zhao, L.J.; Ma, J.L.; et al. Baicalin, a natural LSD1 inhibitor. Bioorg. Chem. 2016, 69, 129–131. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, G.F.S.; Silva, G.D.B.; Pavan, A.R.; Chiba, D.E.; Chin, C.M.; Dos Santos, J.L. Epigenetic Regulatory Mechanisms Induced by Resveratrol. Nutrients 2017, 9, 1201. [Google Scholar] [CrossRef]
- Nettore, I.C.; Rocca, C.; Mancino, G.; Albano, L.; Amelio, D.; Grande, F.; Puoci, F.; Pasqua, T.; Desiderio, S.; Mazza, R.; et al. Quercetin and its derivative Q2 modulate chromatin dynamics in adipogenesis and Q2 prevents obesity and metabolic disorders in rats. J. Nutr. Biochem. 2019, 69, 151–162. [Google Scholar] [CrossRef]
- Cuyàs, E.; Verdura, S.; Llorach-Parés, L.; Fernández-Arroyo, S.; Joven, J.; Martin-Castillo, B.; Bosch-Barrera, J.; Brunet, J.; Nonell-Canals, A.; Sanchez-Martinez, M.; et al. Metformin Is a Direct SIRT1-Activating Compound: Computational Modeling and Experimental Validation. Front. Endocrinol. 2018, 9, 657. [Google Scholar] [CrossRef] [Green Version]
- Cuyàs, E.; Verdura, S.; Lozano-Sánchez, J.; Viciano, I.; Llorach-Parés, L.; Nonell-Canals, A.; Bosch-Barrera, J.; Brunet, J.; Segura-Carretero, A.; Sanchez-Martinez, M.; et al. The extra virgin olive oil phenolic oleacein is a dual substrate-inhibitor of catechol-O-methyltransferase. Food Chem. Toxicol. 2019, 128, 35–45. [Google Scholar] [CrossRef]
- Salentin, S.; Schreiber, S.; Haupt, V.J.; Adasme, M.F.; Schroeder, M. PLIP: Fully automated protein-ligand interaction profiler. Nucleic Acids Res. 2015, 43, W443–W447. [Google Scholar] [CrossRef]
- Leis, O.; Eguiara, A.; Lopez-Arribillaga, E.; Alberdi, M.J.; Hernandez-Garcia, S.; Elorriaga, K.; Pandiella, A.; Rezola, R.; Martin, A.G. Sox2 expression in breast tumours and activation in breast cancer stem cells. Oncogene 2012, 31, 1354–1365. [Google Scholar] [CrossRef]
- Selga, E.; Sendfeld, F.; Martinez-Moreno, R.; Medine, C.N.; Tura-Ceide, O.; Wilmut, S.I.; Pérez, G.J.; Scornik, F.S.; Brugada, R.; Mills, N.L. Sodium channel current loss of function in induced pluripotent stem cell-derived cardiomyocytes from a Brugada syndrome patient. J. Mol. Cell. Cardiol. 2018, 114, 10–19. [Google Scholar] [CrossRef]
- Takanaga, H.; Tsuchida-Straeten, N.; Nishide, K.; Watanabe, A.; Aburatani, H.; Kondo, T. Gli2 is a novel regulator of sox2 expression in telencephalic neuroepithelial cells. Stem Cells 2009, 27, 165–174. [Google Scholar] [CrossRef]
- Yang, M.; Gocke, C.B.; Luo, X.; Borek, D.; Tomchick, D.R.; Machius, M.; Otwinowski, Z.; Yu, H. Structural basis for CoREST-dependent demethylation of nucleosomes by the human LSD1 histone demethylase. Mol. Cell 2006, 23, 377–387. [Google Scholar] [CrossRef]
- Zibetti, C.; Adamo, A.; Binda, C.; Forneris, F.; Toffolo, E.; Verpelli, C.; Ginelli, E.; Mattevi, A.; Sala, C.; Battaglioli, E. Alternative splicing of the histone demethylase LSD1/KDM1 contributes to the modulation of neurite morphogenesis in the mammalian nervous system. J. Neurosci. 2010, 30, 2521–2532. [Google Scholar] [CrossRef]
- Wang, J.; Lu, F.; Ren, Q.; Sun, H.; Xu, Z.; Lan, R.; Liu, Y.; Ward, D.; Quan, J.; Ye, T.; et al. Novel histone demethylase LSD1 inhibitors selectively target cancer cells with pluripotent stem cell properties. Cancer Res. 2011, 71, 7238–7249. [Google Scholar] [CrossRef]
- Zhang, X.; Lu, F.; Wang, J.; Yin, F.; Xu, Z.; Qi, D.; Wu, X.; Cao, Y.; Liang, W.; Liu, Y.; et al. Pluripotent stem cell protein Sox2 confers sensitivity to LSD1 inhibition in cancer cells. Cell Rep. 2013, 5, 445–457. [Google Scholar] [CrossRef]
- Yin, F.; Lan, R.; Zhang, X.; Zhu, L.; Chen, F.; Xu, Z.; Liu, Y.; Ye, T.; Sun, H.; Lu, F.; et al. LSD1 regulates pluripotency of embryonic stem/carcinoma cells through histone deacetylase 1-mediated deacetylation of histone H4 at lysine 16. Mol. Cell. Biol. 2014, 34, 158–179. [Google Scholar] [CrossRef]
- Iglesias, J.M.; Leis, O.; Pérez Ruiz, E.; Gumuzio Barrie, J.; Garcia-Garcia, F.; Aduriz, A.; Beloqui, I.; Hernandez-Garcia, S.; Lopez-Mato, M.P.; Dopazo, J.; et al. The Activation of the Sox2 RR2 Pluripotency Transcriptional Reporter in Human Breast Cancer Cell Lines is Dynamic and Labels Cells with Higher Tumorigenic Potential. Front. Oncol. 2014, 4, 308. [Google Scholar] [CrossRef]
- Iglesias, J.M.; Gumuzio, J.; Martin, A.G. Linking Pluripotency Reprogramming and Cancer. Stem Cells Transl. Med. 2017, 6, 335–339. [Google Scholar] [CrossRef]
- Moreno-Manzano, V.; Rodríguez-Jiménez, F.J.; Aceña-Bonilla, J.L.; Fustero-Lardíes, S.; Erceg, S.; Dopazo, J.; Montaner, D.; Stojkovic, M.; Sánchez-Puelles, J.M. FM19G11, a new hypoxia-inducible factor (HIF) modulator, affects stem cell differentiation status. J. Biol. Chem. 2010, 285, 1333–1342. [Google Scholar] [CrossRef]
- Whyte, W.A.; Bilodeau, S.; Orlando, D.A.; Hoke, H.A.; Frampton, G.M.; Foster, C.T.; Cowley, S.M.; Young, R.A. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 2012, 482, 221–225. [Google Scholar] [CrossRef] [Green Version]
- Zhou, H.Y.; Katsman, Y.; Dhaliwal, N.K.; Davidson, S.; Macpherson, N.N.; Sakthidevi, M.; Collura, F.; Mitchell, J.A. A Sox2 distal enhancer cluster regulates embryonic stem cell differentiation potential. Genes Dev. 2014, 28, 2699–2711. [Google Scholar] [CrossRef]
- Papapetrou, E.P.; Tomishima, M.J.; Chambers, S.M.; Mica, Y.; Reed, E.; Menon, J.; Tabar, V.; Mo, Q.; Studer, L.; Sadelain, M. Stoichiometric and temporal requirements of Oct4, Sox2, Klf4, and c-Myc expression for efficient human iPSC induction and differentiation. Proc. Natl. Acad. Sci. USA 2009, 106, 12759–12764. [Google Scholar] [CrossRef] [Green Version]
- Tomioka, M.; Nishimoto, M.; Miyagi, S.; Katayanagi, T.; Fukui, N.; Niwa, H.; Muramatsu, M.; Okuda, A. Identification of Sox-2 regulatory region which is under the control of Oct-3/4-Sox-2 complex. Nucleic Acids Res. 2002, 30, 3202–3213. [Google Scholar] [CrossRef]
- Kondo, T.; Raff, M. Chromatin remodeling and histone modification in the conversion of oligodendrocyte precursors to neural stem cells. Genes Dev. 2004, 18, 2963–2972. [Google Scholar] [CrossRef] [Green Version]
- You, A.; Tong, J.K.; Grozinger, C.M.; Schreiber, S.L. CoREST is an integral component of the CoREST—Human histone deacetylase complex. Proc. Natl. Acad. Sci. USA 2001, 98, 1454–1458. [Google Scholar] [CrossRef]
- Baron, R.; Vellore, N.A. LSD1/CoREST is an allosteric nanoscale clamp regulated by H3-histone-tail molecular recognition. Proc. Natl. Acad. Sci. USA 2012, 109, 12509–12514. [Google Scholar] [CrossRef] [Green Version]
- Wu, Y.; Wang, Y.; Yang, X.H.; Kang, T.; Zhao, Y.; Wang, C.; Evers, B.M.; Zhou, B.P. The deubiquitinase USP28 stabilizes LSD1 and confers stem-cell-like traits to breast cancer cells. Cell Rep. 2013, 5, 224–236. [Google Scholar] [CrossRef]
- Boulding, T.; McCuaig, R.D.; Tan, A.; Hardy, K.; Wu, F.; Dunn, J.; Kalimutho, M.; Sutton, C.R.; Forwood, J.K.; Bert, A.G.; et al. LSD1 activation promotes inducible EMT programs and modulates the tumour microenvironment in breast cancer. Sci. Rep. 2018, 8, 73. [Google Scholar] [CrossRef] [PubMed]
- Hino, S.; Sakamoto, A.; Nagaoka, K.; Anan, K.; Wang, Y.; Mimasu, S.; Umehara, T.; Yokoyama, S.; Kosai, K.; Nakao, M. FAD-dependent lysine-specific demethylase-1 regulates cellular energy expenditure. Nat. Commun. 2012, 3, 758. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ambrosio, S.; Majello, B. Targeting Histone Demethylase LSD1/KDM1a in Neurodegenerative Diseases. J. Exp. Neurosci. 2018, 12, 1179069518765743. [Google Scholar] [CrossRef]




| PDB | Ligand | Chain | Docking Energy (kcal/mol) a | MM/GBSA (kcal/mol) a |
| 2IW5 | FAD | A–B | −7.2/−7.5 b | −39.9541 |
| PDB | Chain | Docking Energy (kcal/mol) c | MM/GBSA (kcal/mol) c | |
| 2IW5 | A–B | −6.1/−6.1 b | −8.7173 | |
| Cluster Number | ∆G, (kcal/mol) | Dissoc. Constant, (µM) | Members | MM/PBSA, Solvation Binding Energy, (kcal/mol) | Residues of the Receptor That Contact Oleacein |
|---|---|---|---|---|---|
| 1 | −9.63 | 0.08779 | 13% | −66.844 | Leu-625, Val-629, Leu-630, Lys-631, Gln-632, Gln-633, Pro-635, Ala-636, Val-637, Gln-638, Phe-639, Pro-642, Leu-643, Thr-648, Val-651 (chain A) |
| 2 | −9.58 | 0.09509 | 14% | −155.861 | Gly-287, Val-288, Gly-314, Gly-315, Arg-316, Val-317, Leu-329, Gly-330, Ala-331, Phe-538, Leu-659, Asn-660, Lys-661, Trp-751, Ser-760, Tyr-761, Glu-801, Ala-809, Thr-810, Val-811, Ala-814 (chain A) |
| 3 | −9.33 | 0.14574 | 5% | −41.285 | Thr-335, Ala-539, Asn-540, Leu-547, Trp-552, Asp-553, Asp-555, Asp-556, Phe-558, Glu-559, Phe-560, His-564, Ser-762, Tyr-763, Val-764, Tyr-773, Asn-806, Tyr-807, Pro-808, Ala-809, Thr-810, His-812 (chain A) |
| 4 | −8.30 | 0.82642 | 17% | −10.762 | Pro-171, Glu-175, Ala-178, Phe-179, Arg-182, Leu-183, Pro-184, His-185, Asp-186, Arg-187, Gly-338, Gly-339, Asp-557, Phe-558, Glu-559, Phe-560, Thr-561, Tyr-807 (chain A) |
| 5 | −7.85 | 1.75000 | 6% | −6.730 | Ala-541, Thr-542, Pro-543, Thr-546, Leu-627, Gly-628, Lys-631, Gly-655, Phe-656, Gly-657, Asn-658, Gly-709, Ala-712, Gly-713, Arg-752, Arg-758, Gly-759, Tyr-763 (chain A) |
| 6 | −7.62 | 2.61000 | 5% | −49.536 | Arg-182, Gln-191, His-259, Leu-261, Tyr-773, Asp-774, Ala-777, Thr-803, Ile-804, Arg-805, Asn-806, Leu-816, Ser-817, Leu-819, Arg-820, Arg-824 (chain A) |
| 7 | −7.28 | 7.18000 | 25% | −40.682 | Lys-481, His-484, Arg-485, Thr-488 (chain A) and Leu-372, Pro-373, Glu-374, Val-375, Ile- 376, Gln-377, Asp-407, Val-408, Gly-410 (chain B) |
| 8 | −6.65 | 13.32000 | 10% | −30.051 | Lys-374, Met-377, Val-378, Glu-381, Arg-384, Gln-419, His-422, Val-423, Ser-517, Val-519, Tyr-520, Leu-521, Ser-522, Asp-525 (chain A) and Gly-313, Met-314, Phe-315 (chain B) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cuyàs, E.; Gumuzio, J.; Lozano-Sánchez, J.; Carreras, D.; Verdura, S.; Llorach-Parés, L.; Sanchez-Martinez, M.; Selga, E.; Pérez, G.J.; Scornik, F.S.; et al. Extra Virgin Olive Oil Contains a Phenolic Inhibitor of the Histone Demethylase LSD1/KDM1A. Nutrients 2019, 11, 1656. https://doi.org/10.3390/nu11071656
Cuyàs E, Gumuzio J, Lozano-Sánchez J, Carreras D, Verdura S, Llorach-Parés L, Sanchez-Martinez M, Selga E, Pérez GJ, Scornik FS, et al. Extra Virgin Olive Oil Contains a Phenolic Inhibitor of the Histone Demethylase LSD1/KDM1A. Nutrients. 2019; 11(7):1656. https://doi.org/10.3390/nu11071656
Chicago/Turabian StyleCuyàs, Elisabet, Juan Gumuzio, Jesús Lozano-Sánchez, David Carreras, Sara Verdura, Laura Llorach-Parés, Melchor Sanchez-Martinez, Elisabet Selga, Guillermo J. Pérez, Fabiana S. Scornik, and et al. 2019. "Extra Virgin Olive Oil Contains a Phenolic Inhibitor of the Histone Demethylase LSD1/KDM1A" Nutrients 11, no. 7: 1656. https://doi.org/10.3390/nu11071656
APA StyleCuyàs, E., Gumuzio, J., Lozano-Sánchez, J., Carreras, D., Verdura, S., Llorach-Parés, L., Sanchez-Martinez, M., Selga, E., Pérez, G. J., Scornik, F. S., Brugada, R., Bosch-Barrera, J., Segura-Carretero, A., Martin, Á. G., Encinar, J. A., & Menendez, J. A. (2019). Extra Virgin Olive Oil Contains a Phenolic Inhibitor of the Histone Demethylase LSD1/KDM1A. Nutrients, 11(7), 1656. https://doi.org/10.3390/nu11071656