Characterizing the Composition of the Pediatric Gut Microbiome: A Systematic Review
Abstract
1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Data Extraction
2.3. Statistical Analyses
3. Results
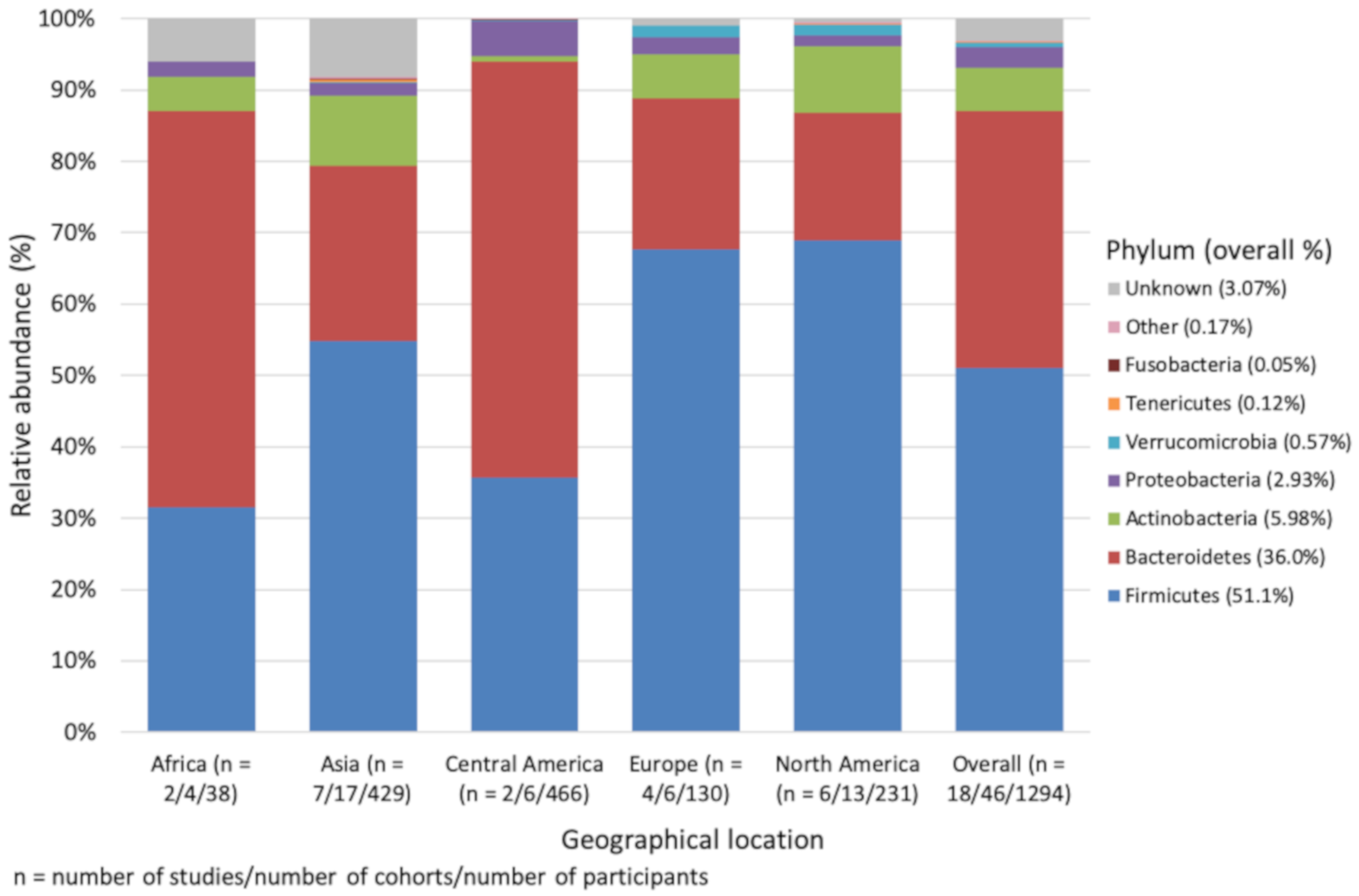
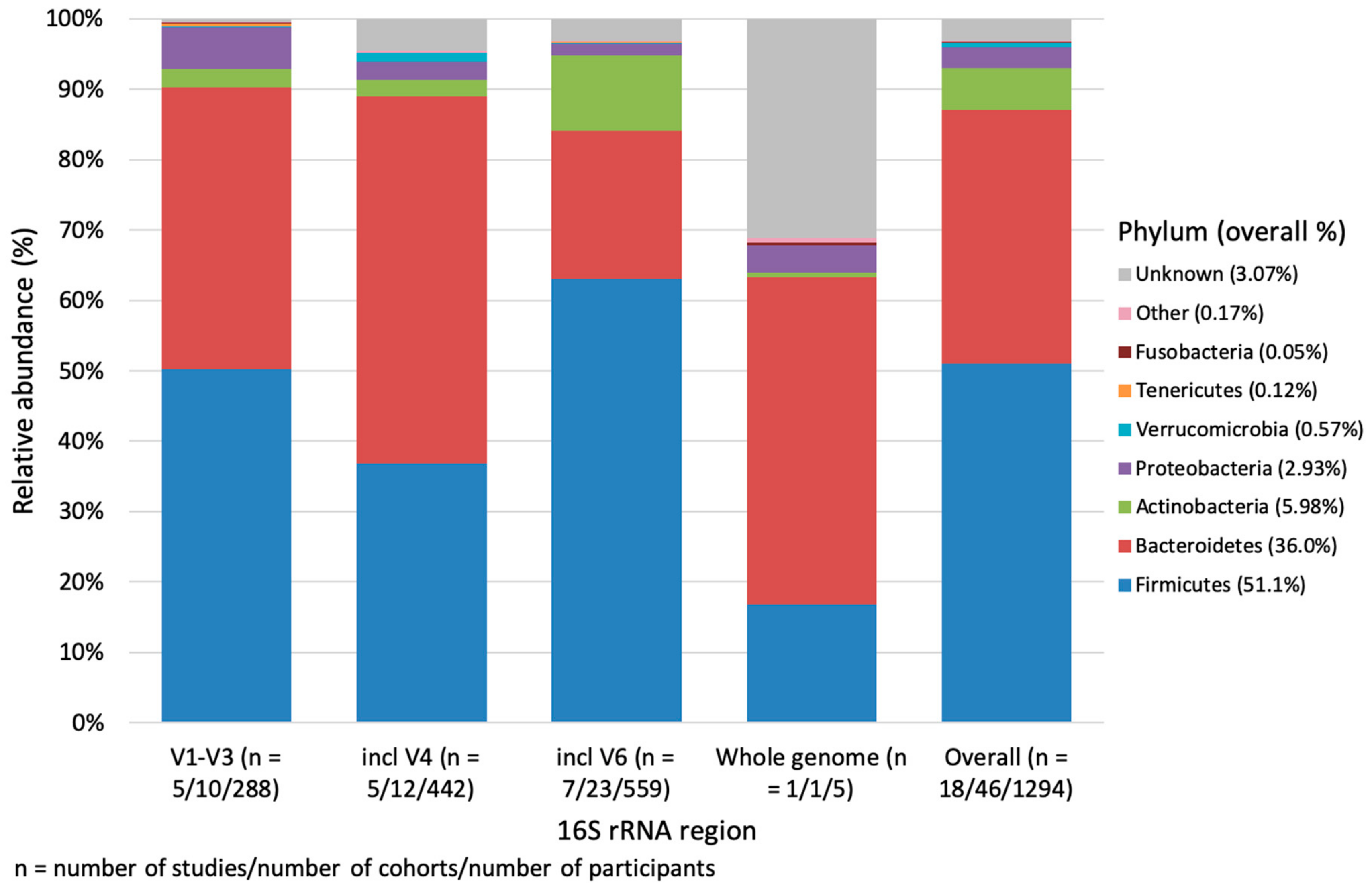
3.1. Phylum Level Impact of Geographical Location, Age and 16S rRNA Region
3.2. Family Level Impact of Geographical Location, Age and 16S rRNA Region
3.3. Genus Level Impact of Geographical Location, Age and 16S rRNA Region
3.4. α-Diversity as Reported by the Included Studies
3.5. Comparison of SCFA Concentrations
3.6. Dietary Analysis
4. Discussion
4.1. Overall Findings
4.2. 16S rRNA Sequencing Region and the Microbiome
4.3. Diet, Geographical Location, SCFA, and the Microbiome
4.4. Association Between Age and the Microbiome
4.5. Limitations of the Current Research
4.6. Strengths and Limitations of This Review
4.7. Consideration for Future Research
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Ley, R.E.; Peterson, D.A.; Gordon, J.I. Ecological and evolutionary forces shaping microbial diversity in the human intestine. Cell 2006, 124, 837–848. [Google Scholar] [CrossRef]
- Thursby, E.; Juge, N. Introduction to the human gut microbiota. Biochem. J. 2017, 474, 1823–1836. [Google Scholar] [CrossRef] [PubMed]
- Whitman, W.B.; Coleman, D.C.; Wiebe, W.J. Prokaryotes: The unseen majority. Proc. Natl. Acad. Sci. USA 1998, 95, 6578–6583. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.V.; Frassetto, A.; Kowalik, E.J., Jr.; Nawrocki, A.R.; Lu, M.M.; Kosinski, J.R.; Hubert, J.A.; Szeto, D.; Yao, X.; Forrest, G.; et al. Butyrate and propionate protect against diet-induced obesity and regulate gut hormones via free fatty acid receptor 3-independent mechanisms. PLoS ONE 2012, 7, e35240. [Google Scholar] [CrossRef] [PubMed]
- Bentley, R.; Meganathan, R. Biosynthesis of vitamin K (menaquinone) in bacteria. Microbiol. Rev. 1982, 46, 241. [Google Scholar] [PubMed]
- Dogra, S.; Sakwinska, O.; Soh, S.-E.; Ngom-Bru, C.; Brück, W.M.; Berger, B.; Brüssow, H.; Karnani, N.; Lee, Y.S.; Yap, F.; et al. Rate of establishing the gut microbiota in infancy has consequences for future health. Gut Microbes 2015, 6, 321–325. [Google Scholar] [CrossRef]
- Tannock, G.W.; Lee, P.S.; Wong, K.H.; Lawley, B. Why Don’t All Infants Have Bifidobacteria in Their Stool? Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Young, V.B. The role of the microbiome in human health and disease: An introduction for clinicians. BMJ 2017, 356. [Google Scholar] [CrossRef]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Nagpal, R.; Tsuji, H.; Takahashi, T.; Kawashima, K.; Nagata, S.; Nomoto, K.; Yamashiro, Y. Sensitive Quantitative Analysis of the Meconium Bacterial Microbiota in Healthy Term Infants Born Vaginally or by Cesarean Section. Front. Microbiol. 2016, 7. [Google Scholar] [CrossRef]
- Shi, Y.-C.; Guo, H.; Chen, J.; Sun, G.; Ren, R.-R.; Guo, M.-Z.; Peng, L.-H.; Yang, Y.-S. Initial meconium microbiome in Chinese neonates delivered naturally or by cesarean section. Sci. Rep. 2018, 8, 3255. [Google Scholar] [CrossRef] [PubMed]
- Stinson, L.F.; Boyce, M.C.; Payne, M.S.; Keelan, J.A. The Not-so-Sterile Womb: Evidence That the Human Fetus Is Exposed to Bacteria Prior to Birth. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Yassour, M.; Vatanen, T.; Siljander, H.; Hämäläinen, A.M.; Härkönen, T.; Ryhänen, S.J.; Franzosa, E.A.; Vlamakis, H.; Huttenhower, C.; Gevers, D.; et al. Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 2016, 8. [Google Scholar] [CrossRef] [PubMed]
- Robertson, R.C.; Manges, A.R.; Finlay, B.B.; Prendergast, A.J. The Human Microbiome and Child Growth—First 1000 Days and Beyond. Trends Microbiol. 2019, 27, 131–147. [Google Scholar] [CrossRef] [PubMed]
- Pannaraj, P.S.; Li, F.; Cerini, C.; Bender, J.M.; Yang, S.; Rollie, A.; Adisetiyo, H.; Zabih, S.; Lincez, P.J.; Bittinger, K.; et al. Association Between Breast Milk Bacterial Communities and Establishment and Development of the Infant Gut Microbiome. JAMA Pediatrics 2017, 171, 647–654. [Google Scholar] [CrossRef] [PubMed]
- Levin, A.M.; Sitarik, A.R.; Havstad, S.L.; Fujimura, K.E.; Wegienka, G.; Cassidy-Bushrow, A.E.; Kim, H.; Zoratti, E.M.; Lukacs, N.W.; Boushey, H.A.; et al. Joint effects of pregnancy, sociocultural, and environmental factors on early life gut microbiome structure and diversity. Sci. Rep. 2016, 6, 31775. [Google Scholar] [CrossRef] [PubMed]
- Biasucci, G.; Rubini, M.; Riboni, S.; Morelli, L.; Bessi, E.; Retetangos, C. Mode of delivery affects the bacterial community in the newborn gut. Early Hum. Dev. 2010, 86, 13–15. [Google Scholar] [CrossRef]
- Dominguez-Bello, M.G.; Costello, E.K.; Contreras, M.; Magris, M.; Hidalgo, G.; Fierer, N.; Knight, R. Delivery mode shapes the acquisition and structure of the initial microbiota across multiple body habitats in newborns. Proc. Natl. Acad. Sci. USA 2010, 107, 11971–11975. [Google Scholar] [CrossRef]
- Jakobsson, H.E.; Abrahamsson, T.R.; Jenmalm, M.C.; Harris, K.; Quince, C.; Jernberg, C.; Björkstén, B.; Engstrand, L.; Andersson, A.F. Decreased gut microbiota diversity, delayed Bacteroidetes colonisation and reduced Th1 responses in infants delivered by caesarean section. Gut 2014, 63, 559–566. [Google Scholar] [CrossRef]
- Backhed, F.; Roswall, J.; Peng, Y.; Feng, Q.; Jia, H.; Kovatcheva-Datchary, P.; Li, Y.; Xia, Y.; Xie, H.; Zhong, H.; et al. Dynamics and Stabilization of the Human Gut Microbiome during the First Year of Life. Cell Host Microbe 2015, 17, 690–703. [Google Scholar] [CrossRef]
- Bergstrom, A.; Skov, T.H.; Bahl, M.I.; Roager, H.M.; Christensen, L.B.; Ejlerskov, K.T.; Molgaard, C.; Michaelsen, K.F.; Licht, T.R. Establishment of intestinal microbiota during early life: A longitudinal, explorative study of a large cohort of Danish infants. Appl. Environ. Microbiol. 2014, 80, 2889–2900. [Google Scholar] [CrossRef] [PubMed]
- Guittar, J.; Shade, A.; Litchman, E. Trait-based community assembly and succession of the infant gut microbiome. Nat. Commun. 2019, 10, 512. [Google Scholar] [CrossRef] [PubMed]
- Laursen, M.F.; Andersen, L.B.; Michaelsen, K.F.; Molgaard, C.; Trolle, E.; Bahl, M.I.; Licht, T.R. Infant Gut Microbiota Development Is Driven by Transition to Family Foods Independent of Maternal Obesity. mSphere 2016, 1, e00069-00015. [Google Scholar] [CrossRef] [PubMed]
- Lim, E.S.; Zhou, Y.; Zhao, G.; Bauer, I.K.; Droit, L.; Ndao, I.M.; Warner, B.B.; Tarr, P.I.; Wang, D.; Holtz, L.R. Early life dynamics of the human gut virome and bacterial microbiome in infants. Nat. Med. 2015, 21, 1228–1234. [Google Scholar] [CrossRef] [PubMed]
- Koenig, J.E.; Spor, A.; Scalfone, N.; Fricker, A.D.; Stombaugh, J.; Knight, R.; Angenent, L.T.; Ley, R.E. Succession of microbial consortia in the developing infant gut microbiome. Proc. Natl. Acad. Sci. USA 2011, 108, 4578–4585. [Google Scholar] [CrossRef]
- Palmer, C.; Bik, E.M.; DiGiulio, D.B.; Relman, D.A.; Brown, P.O. Development of the Human Infant Intestinal Microbiota. PLoS Biol. 2007, 5, e177. [Google Scholar] [CrossRef]
- Agans, R.; Rigsbee, L.; Kenche, H.; Michail, S.; Khamis, H.J.; Paliy, O. Distal gut microbiota of adolescent children is different from that of adults. FEMS Microbiol. Ecol. 2011, 77, 404–412. [Google Scholar] [CrossRef]
- Cheng, J.; Ringel-Kulka, T.; Heikamp-de Jong, I.; Ringel, Y.; Carroll, I.; de Vos, W.M.; Salojärvi, J.; Satokari, R. Discordant temporal development of bacterial phyla and the emergence of core in the fecal microbiota of young children. ISME J. 2016, 10, 1002–1014. [Google Scholar] [CrossRef]
- McVey Neufeld, K.-A.; Luczynski, P.; Dinan, T.G.; Cryan, J.F. Reframing the Teenage Wasteland: Adolescent Microbiota-Gut-Brain Axis. Can. J. Psychiatry. Rev. Can. De Psychiatr. 2016, 61, 214–221. [Google Scholar] [CrossRef]
- Yatsunenko, T.; Rey, F.E.; Manary, M.J.; Trehan, I.; Dominguez-Bello, M.G.; Contreras, M.; Magris, M.; Hidalgo, G.; Baldassano, R.N.; Anokhin, A.P.; et al. Human gut microbiome viewed across age and geography. Nature 2012, 486, 222–227. [Google Scholar] [CrossRef]
- De Filippo, C.; Cavalieri, D.; Di Paola, M.; Ramazzotti, M.; Poullet, J.B.; Massart, S.; Collini, S.; Pieraccini, G.; Lionetti, P. Impact of diet in shaping gut microbiota revealed by a comparative study in children from Europe and rural Africa. Proc. Natl. Acad. Sci. USA 2010, 107, 14691–14696. [Google Scholar] [CrossRef] [PubMed]
- Kisuse, J.; La-ongkham, O.; Nakphaichit, M.; Therdtatha, P.; Momoda, R.; Tanaka, M.; Fukuda, S.; Popluechai, S.; Kespechara, K.; Sonomoto, K.; et al. Urban diets linked to gut microbiome and metabolome alterations in children: A comparative cross-sectional study in Thailand. Front. Microbiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- Fragiadakis, G.K.; Smits, S.A.; Sonnenburg, E.D.; Van Treuren, W.; Reid, G.; Knight, R.; Manjurano, A.; Changalucha, J.; Dominguez-Bello, M.G.; Leach, J.; et al. Links between environment, diet, and the hunter-gatherer microbiome. Gut Microbes 2018, 10, 216–227. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef]
- Louis, P.; Hold, G.L.; Flint, H.J. The gut microbiota, bacterial metabolites and colorectal cancer. Nat. Rev. Microbiol. 2014, 12, 661–672. [Google Scholar] [CrossRef]
- Cummings, J.H.; Pomare, E.W.; Branch, W.J.; Naylor, C.P.; Macfarlane, G.T. Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 1987, 28, 1221–1227. [Google Scholar] [CrossRef]
- Ríos-Covián, D.; Ruas-Madiedo, P.; Margolles, A.; Gueimonde, M.; de Los Reyes-Gavilán, C.G.; Salazar, N. Intestinal Short Chain Fatty Acids and their Link with Diet and Human Health. Front. Microbiol. 2016, 7, 185. [Google Scholar] [CrossRef]
- Van de Peer, Y.; Chapelle, S.; De Wachter, R. A Quantitative Map of Nucleotide Substitution Rates in Bacterial rRNA. Nucleic Acids Res. 1996, 24, 3381–3391. [Google Scholar] [CrossRef]
- Yang, B.; Wang, Y.; Qian, P.-Y. Sensitivity and correlation of hypervariable regions in 16S rRNA genes in phylogenetic analysis. BMC Bioinform. 2016, 17, 135. [Google Scholar] [CrossRef] [PubMed]
- Tremblay, J.; Singh, K.; Fern, A.; Kirton, E.; He, S.; Woyke, T.; Lee, J.; Chen, F.; Dangl, J.; Tringe, S. Primer and platform effects on 16S rRNA tag sequencing. Front. Microbiol. 2015, 6, 771. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.; Morrison, M.; Yu, Z. Evaluation of different partial 16S rRNA gene sequence regions for phylogenetic analysis of microbiomes. J. Microbiol. Methods 2011, 84, 81–87. [Google Scholar] [CrossRef] [PubMed]
- Soergel, D.A.W.; Dey, N.; Knight, R.; Brenner, S.E. Selection of primers for optimal taxonomic classification of environmental 16S rRNA gene sequences. ISME J. 2012, 6, 1440. [Google Scholar] [CrossRef]
- Derrien, M.; Alvarez, A.S.; de Vos, W.M. The Gut Microbiota in the First Decade of Life. Trends Microbiol. 2019, 27, 997–1010. [Google Scholar] [CrossRef]
- Moher, D.; Liberati, A.; Tetzlaff, J.; Altman, D.G.; The, P.G. Preferred Reporting Items for Systematic Reviews and Meta-Analyses: The PRISMA Statement. PLoS Med. 2009, 6, e1000097. [Google Scholar] [CrossRef]
- World Health Organization (WHO). Health Topics: Obesity. Available online: https://www.who.int/topics/obesity/en/ (accessed on 20 November 2019).
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Bonferroni, C. Teoria statistica delle classi e calcolo delle probabilita. Pubbl. Del R Ist. Super. Di Sci. Econ. E Commericiali Di Firenze 1936, 8, 3–62. [Google Scholar]
- Anderson, M. Permutational Multivariate Analysis of Variance (PERMANOVA). Wiley StatsRef Stat. Ref. Online 2017. [Google Scholar] [CrossRef]
- Berni Canani, R.; De Filippis, F.; Nocerino, R.; Laiola, M.; Paparo, L.; Calignano, A.; De Caro, C.; Coretti, L.; Chiariotti, L.; Gilbert, J.A.; et al. Specific Signatures of the Gut Microbiota and Increased Levels of Butyrate in Children Treated with Fermented Cow‘s Milk Containing Heat-Killed Lactobacillus paracasei CBA L74. Appl. Environ. Microbiol. 2017, 83, e01206-17. [Google Scholar] [CrossRef]
- Dicksved, J.; Flöistrup, H.; Bergström, A.; Rosenquist, M.; Pershagen, G.; Scheynius, A.; Roos, S.; Alm, J.S.; Engstrand, L.; Braun-Fahrländer, C.; et al. Molecular fingerprinting of the fecal microbiota of children raised according to different lifestyles. Appl. Environ. Microbiol. 2007, 73, 2284–2289. [Google Scholar] [CrossRef] [PubMed]
- Ignacio, A.; Fernandes, M.R.; Rodrigues, V.A.A.; Groppo, F.C.; Cardoso, A.L.; Avila-Campos, M.J.; Nakano, V. Correlation between body mass index and faecal microbiota from children. Clin. Microbiol. Infect. 2016, 22, 258-e1. [Google Scholar] [CrossRef] [PubMed]
- Karlsson, C.L.J.; Onnerfält, J.; Xu, J.; Molin, G.; Ahrné, S.; Thorngren-Jerneck, K. The microbiota of the gut in preschool children with normal and excessive body weight. Obesity 2012, 20, 2257–2261. [Google Scholar] [CrossRef] [PubMed]
- La-ongkham, O.; Nakphaichit, M.; Leelavatcharamas, V.; Keawsompong, S.; Nitisinprasert, S. Distinct gut microbiota of healthy children from two different geographic regions of Thailand. Arch. Microbiol. 2015, 197, 561–573. [Google Scholar] [CrossRef] [PubMed]
- Mello, C.S.; Carmo-Rodrigues, M.S.; Filho, H.B.A.; Melli, L.C.F.L.; Tahan, S.; Pignatari, A.C.C.; De Morais, M.B. Gut microbiota differences in children from distinct socioeconomic levels living in the same urban area in Brazil. J. Pediatric Gastroenterol. Nutr. 2016, 63, 460–465. [Google Scholar] [CrossRef]
- Mousavi, S.H.; Mehrara, S.; Barzegari, A.; Ostadrahimi, A. Correlation of gut microbiota profile with body mass index among school age children. Iran. Red Crescent Med. J. 2018, 20, e58049. [Google Scholar] [CrossRef]
- Salminen, S.; Gibson, G.R.; McCartney, A.L.; Isolauri, E. Influence of mode of delivery on gut microbiota composition in seven year old children. Gut 2004, 53, 1388–1389. [Google Scholar] [CrossRef]
- Xu, P.; Li, M.; Zhang, J.; Zhang, T. Correlation of intestinal microbiota with overweight and obesity in Kazakh school children. BMC Microbiol. 2012, 12, 283. [Google Scholar] [CrossRef]
- Zhang, C.; Yin, A.; Li, H.; Wang, R.; Wu, G.; Shen, J.; Zhang, M.; Wang, L.; Hou, Y.; Ouyang, H.; et al. Dietary Modulation of Gut Microbiota Contributes to Alleviation of Both Genetic and Simple Obesity in Children. EBioMedicine 2015, 2, 968–984. [Google Scholar] [CrossRef]
- Zimmermann, M.B.; Chassard, C.; Rohner, F.; N‘Goran, E.K.; Nindjin, C.; Dostal, A.; Utzinger, J.; Ghattas, H.; Lacroix, C.; Hurrell, R.F. The effects of iron fortification on the gut microbiota in African children: A randomized controlled trial in Côte d’Ivoire. Am. J. Clin. Nutr. 2010, 92, 1406–1415. [Google Scholar] [CrossRef]
- Payne, A.N.; Chassard, C.; Zimmermann, M.; Muller, P.; Stinca, S.; Lacroix, C. The metabolic activity of gut microbiota in obese children is increased compared with normal-weight children and exhibits more exhaustive substrate utilization. Nutr. Diabetes 2011, 1, e12. [Google Scholar] [CrossRef] [PubMed]
- Avershina, E.; Storrø, O.; Øien, T.; Johnsen, R.; Pope, P.; Rudi, K. Major faecal microbiota shifts in composition and diversity with age in a geographically restricted cohort of mothers and their children. FEMS Microbiol. Ecol. 2014, 87, 280–290. [Google Scholar] [CrossRef] [PubMed]
- Berding, K.; Holscher, H.D.; Arthur, A.E.; Donovan, S.M. Fecal microbiome composition and stability in 4- to 8-year old children is associated with dietary patterns and nutrient intake. J. Nutr. Biochem. 2018, 56, 165–174. [Google Scholar] [CrossRef] [PubMed]
- Bisanz, J.E.; Enos, M.K.; Mwanga, J.R.; Changalucha, J.; Burton, J.P.; Gloor, G.B.; Reid, G. Randomized Open-Label pilot study of the influence of probiotics and the gut microbiome on toxic metal levels in Tanzanian pregnant women and school children. MBio 2014, 5. [Google Scholar] [CrossRef]
- Ringel-Kulka, T.; Cheng, J.; Ringel, Y.; Salojarvi, J.; Carroll, I.; Palva, A.; de Vos, W.M.; Satokari, R. Intestinal microbiota in healthy U.S. young children and adults--a high throughput microarray analysis. PLoS ONE 2013, 8, e64315. [Google Scholar] [CrossRef]
- De Filippo, C.; Di Paola, M.; Ramazzotti, M.; Albanese, D.; Pieraccini, G.; Banci, E.; Miglietta, F.; Cavalieri, D.; Lionetti, P. Diet, environments, and gut microbiota. A preliminary investigation in children living in rural and Urban Burkina Faso and Italy. Front. Microbiol. 2017, 8, 1979. [Google Scholar] [CrossRef]
- Schloss, P.D.; Iverson, K.D.; Petrosino, J.F.; Schloss, S.J. The dynamics of a family’s gut microbiota reveal variations on a theme. Microbiome 2014, 2. [Google Scholar] [CrossRef]
- Ghosh, T.S.; Gupta, S.S.; Bhattacharya, T.; Yadav, D.; Barik, A.; Chowdhury, A.; Das, B.; Mande, S.S.; Nair, G.B. Gut microbiomes of Indian children of varying nutritional status. PLoS ONE 2014, 9, e95547. [Google Scholar] [CrossRef]
- Hollister, E.B.; Riehle, K.; Luna, R.A.; Weidler, E.M.; Rubio-Gonzales, M.; Mistretta, T.-A.; Raza, S.; Doddapaneni, H.V.; Metcalf, G.A.; Muzny, D.M.; et al. Structure and function of the healthy pre-adolescent pediatric gut microbiome. Microbiome 2015, 3, 36. [Google Scholar] [CrossRef]
- Chong, C.W.; Ahmad, A.F.; Lim, Y.A.L.; Teh, C.S.J.; Yap, I.K.S.; Lee, S.C.; Chin, Y.T.; Loke, P.n.; Chua, K.H. Effect of ethnicity and socioeconomic variation to the gut microbiota composition among pre-adolescent in Malaysia. Sci. Rep. 2015, 5, 13338. [Google Scholar] [CrossRef]
- Lin, A.; Bik, E.M.; Costello, E.K.; Dethlefsen, L.; Haque, R.; Relman, D.A.; Singh, U. Distinct Distal Gut Microbiome Diversity and Composition in Healthy Children from Bangladesh and the United States. PLoS ONE 2013, 8, e53838. [Google Scholar] [CrossRef] [PubMed]
- López-Contreras, B.E.; Morán-Ramos, S.; Villarruel-Vázquez, R.; Macías-Kauffer, L.; Villamil-Ramírez, H.; León-Mimila, P.; Vega-Badillo, J.; Sánchez-Muñoz, F.; Llanos-Moreno, L.E.; Canizalez-Román, A.; et al. Composition of gut microbiota in obese and normal-weight Mexican school-age children and its association with metabolic traits. Pediatric Obes. 2018, 13, 381–388. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Watanabe, K.; Jiang, J.; Matsuda, K.; Chao, S.-H.; Haryono, P.; La-ongkham, O.; Sarwoko, M.-A.; Sujaya, I.N.; Zhao, L.; et al. Diversity in gut bacterial community of school-age children in Asia. Sci. Rep. 2015, 5, 8397. [Google Scholar] [CrossRef] [PubMed]
- Nakayama, J.; Yamamoto, A.; Palermo-Conde, L.A.; Higashi, K.; Sonomoto, K.; Tan, J.; Lee, Y.K. Impact of westernized diet on gut microbiota in children on Leyte island. Front. Microbiol. 2017, 8, 197. [Google Scholar] [CrossRef] [PubMed]
- Nicolucci, A.C.; Hume, M.P.; Martinez, I.; Mayengbam, S.; Walter, J.; Reimer, R.A. Prebiotics Reduce Body Fat and Alter Intestinal Microbiota in Children Who Are Overweight or With Obesity. Gastroenterology 2017, 153, 711–722. [Google Scholar] [CrossRef] [PubMed]
- Riva, A.; Borgo, F.; Lassandro, C.; Verduci, E.; Morace, G.; Borghi, E.; Berry, D. Pediatric obesity is associated with an altered gut microbiota and discordant shifts in Firmicutes populations. Environ. Microbiol. 2017, 19, 95–105. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2009, 457, 480–484. [Google Scholar] [CrossRef]
- Ley, R.; Turnbaugh, P.; Klein, S.; Gordon, J. Microbial ecology: Human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Smith-Brown, P.; Morrison, M.; Krause, L.; Davies, P.S.W. Dairy and plant based food intakes are associated with altered faecal microbiota in 2 to 3 year old Australian children. Sci. Rep. 2016, 6, 32385. [Google Scholar] [CrossRef]
- Smith-Brown, P.; Morrison, M.; Krause, L.; Davies, P.S.W. Male-specific association between fat-free mass index and fecal microbiota in 2- to 3-year-old Australian children. J. Pediatric Gastroenterol. Nutr. 2018, 66, 147–151. [Google Scholar] [CrossRef]
- Hollister, E.B.; Foster, B.A.; Dahdouli, M.; Ramirez, J.; Lai, Z. Characterization of the Stool Microbiome in Hispanic Preschool Children by Weight Status and Time. Child. Obes. Print 2018, 14, 122–130. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Xu, R.; He, F.; Zhou, J.; Wang, Y.; Zhou, J.; Wang, M.; Zhou, W. Diversity of gut microbiota metabolic pathways in 10 pairs of Chinese infant twins. PLoS ONE 2016, 11, e0161627. [Google Scholar] [CrossRef] [PubMed]
- Murugesan, S.; Ulloa-Martinez, M.; Martinez-Rojano, H.; Galvan-Rodriguez, F.M.; Miranda-Brito, C.; Romano, M.C.; Pina-Escobedo, A.; Pizano-Zarate, M.L.; Hoyo-Vadillo, C.; Garcia-Mena, J. Study of the diversity and short-chain fatty acids production by the bacterial community in overweight and obese Mexican children. Eur. J. Clin. Microbiol. Infect. Dis. 2015, 34, 1337–1346. [Google Scholar] [CrossRef] [PubMed]
- Monira, S.; Nakamura, S.; Gotoh, K.; Izutsu, K.; Watanabe, H.; Alam, N.; Endtz, H.; Cravioto, A.; Ali, S.; Nakaya, T.; et al. Gut Microbiota of Healthy and Malnourished Children in Bangladesh. Front. Microbiol. 2011, 2, 228. [Google Scholar] [CrossRef]
- Shin, J.-H.; Sim, M.; Lee, J.-Y.; Shin, D.-M. Lifestyle and geographic insights into the distinct gut microbiota in elderly women from two different geographic locations. J. Physiol. Anthropol. 2016, 35, 31. [Google Scholar] [CrossRef]
- Kelly, M. The Nutrition Transition in Developing Asia: Dietary Change, Drivers and Health Impacts. In Eating, Drinking: Surviving: The International Year of Global Understanding—IYGU; Jackson, P., Spiess, W.E.L., Sultana, F., Eds.; Springer: Cham, Switzerland, 2016; pp. 83–90. [Google Scholar] [CrossRef]
- Koliada, A.; Syzenko, G.; Moseiko, V.; Budovska, L.; Puchkov, K.; Perederiy, V.; Gavalko, Y.; Dorofeyev, A.; Romanenko, M.; Tkach, S.; et al. Association between body mass index and Firmicutes/Bacteroidetes ratio in an adult Ukrainian population. BMC Microbiol. 2017, 17, 120. [Google Scholar] [CrossRef]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Indiani, C.; Rizzardi, K.F.; Castelo, P.M.; Ferraz, L.F.C.; Darrieux, M.; Parisotto, T.M. Childhood Obesity and Firmicutes/Bacteroidetes Ratio in the Gut Microbiota: A Systematic Review. Child. Obes. Print 2018, 14, 501–509. [Google Scholar] [CrossRef]
- Walters, W.A.; Xu, Z.; Knight, R. Meta-analyses of human gut microbes associated with obesity and IBD. FEBS Lett. 2014, 588, 4223–4233. [Google Scholar] [CrossRef]
- Levy, S.E.; Myers, R.M. Advancements in Next-Generation Sequencing. Annu. Rev. Genom. Hum. Genet. 2016, 17, 95–115. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Q.; Zhao, J.; Gong, L.; Zhang, Y.; Wang, X.; Yuan, Z. Altered diversity and composition of the gut microbiome in patients with cervical cancer. AMB Express 2019, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Long, W.; Zhang, C.; Liu, S.; Zhao, L.; Hamaker, B.R. Fiber-utilizing capacity varies in Prevotella- versus Bacteroides-dominated gut microbiota. Sci. Rep. 2017, 7, 2594. [Google Scholar] [CrossRef] [PubMed]
- Primec, M.; Mičetić-Turk, D.; Langerholc, T. Analysis of short-chain fatty acids in human feces: A scoping review. Anal. Biochem. 2017, 526, 9–21. [Google Scholar] [CrossRef] [PubMed]
- Baxter, N.T.; Schmidt, A.W.; Venkataraman, A.; Kim, K.S.; Waldron, C.; Schmidt, T.M. Dynamics of Human Gut Microbiota and Short-Chain Fatty Acids in Response to Dietary Interventions with Three Fermentable Fibers. MBio 2019, 10, e02566-02518. [Google Scholar] [CrossRef] [PubMed]
- den Besten, G.; van Eunen, K.; Groen, A.K.; Venema, K.; Reijngoud, D.-J.; Bakker, B.M. The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 2013, 54, 2325–2340. [Google Scholar] [CrossRef] [PubMed]
- Leahy, S.C.; Higgins, D.G.; Fitzgerald, G.F.; van Sinderen, D. Getting better with bifidobacteria. J. Appl. Microbiol. 2005, 98, 1303–1315. [Google Scholar] [CrossRef]
- O‘Callaghan, A.; van Sinderen, D. Bifidobacteria and Their Role as Members of the Human Gut Microbiota. Front. Microbiol. 2016, 7, 925. [Google Scholar] [CrossRef]
- Pool-Zobel, B.L.; Neudecker, C.; Domizlaff, I.; Ji, S.; Schillinger, U.; Rumney, C.; Moretti, M.; Vilarini, I.; Scassellati-Sforzolini, R.; Rowland, I. Lactobacillus- and bifidobacterium-mediated antigenotoxicity in the colon of rats. Nutr. Cancer 1996, 26, 365–380. [Google Scholar] [CrossRef]
- Le Leu, R.K.; Hu, Y.; Brown, I.L.; Woodman, R.J.; Young, G.P. Synbiotic intervention of Bifidobacterium lactis and resistant starch protects against colorectal cancer development in rats. Carcinogenesis 2010, 31, 246–251. [Google Scholar] [CrossRef]
- Correa, N.B.; Peret Filho, L.A.; Penna, F.J.; Lima, F.M.; Nicoli, J.R. A randomized formula controlled trial of Bifidobacterium lactis and Streptococcus thermophilus for prevention of antibiotic-associated diarrhea in infants. J. Clin. Gastroenterol. 2005, 39, 385–389. [Google Scholar] [CrossRef]
- Chenoll, E.; Rivero, M.; Codoner, F.M.; Martinez-Blanch, J.F.; Ramon, D.; Genoves, S.; Moreno Munoz, J.A. Complete Genome Sequence of Bifidobacterium longum subsp. infantis Strain CECT 7210, a Probiotic Strain Active against Rotavirus Infections. Genome Announc. 2015, 3, e00105-15. [Google Scholar] [CrossRef] [PubMed]
- Rizzatti, G.; Lopetuso, L.R.; Gibiino, G.; Binda, C.; Gasbarrini, A. Proteobacteria: A Common Factor in Human Diseases. BioMed Res. Int. 2017, 2017, 9351507. [Google Scholar] [CrossRef] [PubMed]
- Hiippala, K.; Kainulainen, V.; Kalliomäki, M.; Arkkila, P.; Satokari, R. Mucosal Prevalence and Interactions with the Epithelium Indicate Commensalism of Sutterella spp. Front. Microbiol. 2016, 7, 1706. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Christophersen, C.T.; Sorich, M.J.; Gerber, J.P.; Angley, M.T.; Conlon, M.A. Increased abundance of Sutterella spp. and Ruminococcus torques in feces of children with autism spectrum disorder. Mol. Autism 2013, 4, 42. [Google Scholar] [CrossRef] [PubMed]
- Mills, S.; Stanton, C.; Lane, J.A.; Smith, G.J.; Ross, R.P. Precision Nutrition and the Microbiome, Part I: Current State of the Science. Nutrients 2019, 11, 923. [Google Scholar] [CrossRef] [PubMed]
- Allali, I.; Arnold, J.W.; Roach, J.; Cadenas, M.B.; Butz, N.; Hassan, H.M.; Koci, M.; Ballou, A.; Mendoza, M.; Ali, R.; et al. A comparison of sequencing platforms and bioinformatics pipelines for compositional analysis of the gut microbiome. BMC Microbiol. 2017, 17, 194. [Google Scholar] [CrossRef] [PubMed]
- López-García, A.; Pineda-Quiroga, C.; Atxaerandio, R.; Pérez, A.; Hernández, I.; García-Rodríguez, A.; González-Recio, O. Comparison of Mothur and QIIME for the Analysis of Rumen Microbiota Composition Based on 16S rRNA Amplicon Sequences. Front. Microbiol. 2018, 9, 3010. [Google Scholar] [CrossRef]
- Plummer, E.; Twin, J. A Comparison of Three Bioinformatics Pipelines for the Analysis of Preterm Gut Microbiota using 16S rRNA Gene Sequencing Data. J. Proteom. Bioinform. 2015, 8, 283–291. [Google Scholar] [CrossRef]
- Galisteo, M.; Duarte, J.; Zarzuelo, A. Effects of dietary fibers on disturbances clustered in the metabolic syndrome. J. Nutr. Biochem. 2008, 19, 71–84. [Google Scholar] [CrossRef]
- Knight, R.; Vrbanac, A.; Taylor, B.C.; Aksenov, A.; Callewaert, C.; Debelius, J.; Gonzalez, A.; Kosciolek, T.; McCall, L.I.; McDonald, D.; et al. Best practices for analysing microbiomes. Nat. Rev. Microbiol. 2018. [Google Scholar] [CrossRef]
- Pollock, J.; Glendinning, L.; Wisedchanwet, T.; Watson, M. The Madness of Microbiome: Attempting To Find Consensus “Best Practice” for 16S Microbiome Studies. Appl. Environ. Microbiol. 2018, 84, e02627-02617. [Google Scholar] [CrossRef] [PubMed]
- Thompson, L.R.; Sanders, J.G.; McDonald, D.; Amir, A.; Ladau, J.; Locey, K.J.; Prill, R.J.; Tripathi, A.; Gibbons, S.M.; Ackermann, G.; et al. A communal catalogue reveals Earth’s multiscale microbial diversity. Nature 2017, 551, 457–463. [Google Scholar] [CrossRef] [PubMed]
- Gilbert, J.A.; Jansson, J.K.; Knight, R. The Earth Microbiome project: Successes and aspirations. BMC Biol. 2014, 12. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Finegold, S.M.; Song, Y.; Lawson, P.A. Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int. J. Syst. Evol. Microbiol. 2008, 58, 1896–1902. [Google Scholar] [CrossRef] [PubMed]
- Sakamoto, M.; Ohkuma, M. Reclassification of Xylanibacter oryzae Ueki et al. 2006 as Prevotella oryzae comb. nov., with an emended description of the genus Prevotella. Int. J. Syst. Evol. Microbiol. 2012, 62, 2637–2642. [Google Scholar] [CrossRef] [PubMed]
- Greenblum, S.; Chiu, H.-C.; Levy, R.; Carr, R.; Borenstein, E. Towards a predictive systems-level model of the human microbiome: Progress, challenges, and opportunities. Curr. Opin. Biotechnol. 2013, 24, 810–820. [Google Scholar] [CrossRef]
- Shaffer, M.; Armstrong, A.J.S.; Phelan, V.V.; Reisdorph, N.; Lozupone, C.A. Microbiome and metabolome data integration provides insight into health and disease. Transl. Res. 2017, 189, 51–64. [Google Scholar] [CrossRef]
- Surana, N.K.; Kasper, D.L. Moving beyond microbiome-wide associations to causal microbe identification. Nature 2017, 552, 244. [Google Scholar] [CrossRef]

| First Author Year [REF] | Cohort Location | Study Population | Study Design | Age Range | Number of Participants | 16S rRNA Target Region | Sequencing Platform | Summary |
|---|---|---|---|---|---|---|---|---|
| Avershina 2014 [63] | Norway | Healthy infants (IMPACT Cohort) | Longitudinal | 2 years | 39 | V3-V4 | 454 | The butyrate producing family Lachnospiraceae dominated the microbiome of the two-year-old children (46%). Followed by unclassified Actinobacteria and Faecalibacterium. |
| Berding 2018 [64] | USA | Healthy preadolescents | Longitudinal | 4–8 years | 22 | V3-V4 | MiSeq | Baseline dietary patterns are associated with temporal stability of microbiota over a 6-month period |
| Bisanz 2014 [65] | Tanzania | Healthy children exposed to heavy metals | Longitudinal | 6–10 years | 40 | V6 | Ion torrent | Probiotic yoghurt did not significantly impact community composition. Gut was dominated by Prevotellaceae, similar to the other African study (De Filippo et al. [31]). |
| Ringel-Kulka 2013 [66] | USA | Healthy children | Cross-sectional | 1–4 years | 28 | V1 and V6 | HITChip Microarray | Firmicutes dominated the gut microbiome at a phylum rank. Clostridium cluster XIVa was the most abundant taxon. Authors suggest microbiota is not adult-like at 4 years. |
| Cheng 2016 [28] | USA | Healthy children | Longitudinal | 1–5 years | 28 | As above | As above | Bacterial diversity did not increase with age. All age groups were dominated by Firmicutes at the phylum. At genus rank, it was Ruminococcus, Clostridium and Bifidobacterium. The authors conclude that at age five the microbiome is still developing towards a stable adult-like composition. |
| De Filippo 2010 [31] | Italy (EU), Burkina Faso (BF) | Healthy children | Cross-sectional | 1–6 years | 29 | V5-V6 | 454 | Significant difference in community structure. BF gut dominated by genera Prevotella and Xylanibacter. EU gut dominated by genera Faecalibacterium and Bacteroides. |
| De Filippo 2017 [67] | As above | As above | Cross-sectional | 2–8 years | 37 | As above | As above | Burkina Faso rural children had a gut dominated by bacteria of the Prevotellaceae family (66.8%). This trended downwards as the participants became less rural and was even more extreme for the Italian children, where the relative abundance of Prevotellaceae was only 0.44%. |
| Schloss 2014 [68] | USA | Healthy children | Longitudinal | 2–10 years | 4 | V3-V5; WGS | 454 | As others have concluded, intra-individual variability is less than inter-individual microbiome variability. |
| Ghosh 2014 [69] | India | Data of healthy children only | Cross-sectional | 2.5–6 years | 5 | WGS only | 454 | Apparently healthy Indian children’s gut microbiome were dominated at the phylum rank by Bacteroidetes and Firmicutes and at the genus rank by Prevotella. This is similar to those in other developing countries, such as Burkina Faso [31] and Tanzania [65] |
| Hollister 2015 [70] | USA | Healthy preadolescents | Cross-sectional | 7–12 years | 37 | V3-V5; WGS | HiSeq 2000 | In 16S rRNA analysis the gut microbiome was dominated at the genus rank by Bacteroides, Faecalibacterium and Alistipes. In the WGS subgroup analysis, Bacteroides, Faecalibacterium, Bifidobacterium and Alistipes dominated at the genus rank. |
| Jakobsson 2014 [19] | Sweden | Healthy infants | Longitudinal | 2 years | 24 | V3-V4 | 454 | At two years old, the gut microbiome was dominated at the phylum rank by Firmicutes and at the genus rank by uncl. Lachnospiraceae, Bacteroides and Bifidobacterium. Similar to results from Avershina et al. [63]. |
| Chong 2015 [71] | Malaysia | Healthy children | Cross-section | 7–12 years | 61 | V3-V5 | 454 | Despite differences in hygiene practices and socioeconomic status, Chinese and Malays were not significantly different. Orang Asli were significantly different to other ethnicities. |
| Kisuse 2018 [32] | Thailand | Healthy children | Cross-sectional | 9–11 years | 45 | V1-V2 | MiSeq | Rural children followed a more traditional plant-based diet and had higher SCFA production and a functional gut reflecting this. City of residence significantly associated with community structure even after adjustment for age and gender. At phylum rank, the microbiome was dominated by Bacteroidetes and Firmicutes. At the genera rank, Bacteroides, Prevotella and Faecalibacterium were the most dominant. |
| Koenig 2011 [25] | USA | Healthy child | Longitudinal | 1–2.5 | 1 | V1-V2 | 454 | Bacteroidetes and Firmicutes dominate the child’s microbiome at from one year onwards. This is associated with an increase in SCFAs and an enrichment of carbohydrate utilization genes. Results suggest that the 2.5-y-old human gut microbiome has many of the functional attributes of the adult microbiome. |
| Lim 2015 [24] | USA | Healthy children | Longitudinal | 2 years | 8 | V4 | MiSeq | 24-month microbiome was dominated at the family rank by Lachnospiraceae and Ruminococcaceae. Both of which contain known butyrate producers. |
| Lin 2013 [72] | Bangladesh, USA | Healthy children | Longitudinal | 9–14 years | 10 | V1-V3 | 454 | Prevotella and lactobacillus were higher in NE children, as was vegetables. Both of which were correlated. Prevotella was higher in others following a traditional plant-based diet [31,32,65]. |
| López-Contreras 2018 [73] | Mexico | Healthy children | Cross-sectional | 6–12 years | 138 | V4 | MiSeq | Both microbiomes were dominated at the phylum rank by Bacteroidetes (67.5% in normal weight children and 69.4% in obese children) and Firmicutes (27.8% in normal weight and 26% in obese children). At the genus rank, the four most abundant bacteria were Bacteroides (39.0%), Prevotella (24.0%), unclassified Lachnospiraceae (7.2%) and unclassified Ruminococcaceae (6.1%). |
| Nakayama 2015 [74] | China, Taiwan, Japan, Indonesia, Thailand | Healthy children | Cross-sectional | 7–11 years | 303 | V6-V8 | 454 | Overall, at the phylum rank the microbiome was dominated by Firmicutes (61.98%) and at the genus rank by Bifidobacterium, Faecalibacterium and Bacteroides (overall average above 10% each). Two Indonesian cities show highest α-diversity and Japanese cities reported lowest α-diversity. |
| Nakayama 2017 [75] | Philippines | Healthy preadolescents | Cross-sectional | 7–9 years | 43 | V6-V8 | 454 | Baybay children’s microbiome was dominated by the family Prevotellaceae, whose diets had significantly less fat. Ormoc children by the families Ruminococcaceae and Lachnospiraceae. |
| Nicolucci 2017 [76] | Canada | Overweight or obese children (>85th BMI percentile) | Longitudinal | 7–12 years | 42 | V3 | MiSeq | At a phylum rank, microbiomes were dominated by Firmicutes (prebiotics group = 68.6%, placebo group = 68%) and Bacteroidetes (both 14.7%). |
| Riva 2017 [77] | Italy | Healthy children | Cross-sectional | 9–16 years | 78 | V3-V4 | MiSeq | At all three taxa ranks there was a clear distinction in microbiota composition between normal weight and obese children. F:B ratio was significantly higher in obese children. This is similar to other studies examining body composition and F:B ratio ([78,79]). |
| Smith-Brown 2016 [80] | Australia | Healthy preadolescents | Cross-sectional | 2–3 years | 37 | V6-V8 | MiSeq | Correlations and UniFrac analyses indicated that intake of several food groups is associated with various genera and microbial composition. |
| Smith-Brown 2018 [81] | As above | As above | As above | As above | As above | As above | As above | Weighted UniFrac is associated with FFMI z-scores in all participants but only significant in boys (when stratified by gender). |
| Hollister 2018 [82] | USA (Hispanic children) | Obese children | Longitudinal | 2–5 years | 52 | V1-V3 | MiSeq | Despite significant weight loss in the intervention group, limited shifts in community composition were seen, including no significant changes in α-diversity. |
| Yassour 2016 [13] | Finland | Healthy infants (DIABIMMUNE study) | Longitudinal | 2–3 years | 39 | V4 | 16S: MiSeq; WGS: HiSeq | Antibiotic positive children had reported greater instability between consecutive samples. The microbiome of the whole group was dominated at the family rank at 24 to 36 months by Bacteroidaceae (42%), Ruminococcaceae (17%) and Lachnospiraceae (14%). |
| Zhou 2016 [83] | China | Healthy twins | Cross-sectional | 1–6 years | 14 | WGS | HiSeq 2500 | The genus Bacteroides dominated the gut (36.6%), followed by Eubacterium (11.1%) and Bifidobacterium (6.98%). |
| Murugesan 2015 [84] | Mexico | Healthy children | Cross-sectional | 9–11 years | 190 | V3 | Ion torrent | Normal weight and obese children had a similar relative abundance of Actinobacteria, Bacteroidetes and Firmicutes, however, there were 2.5 times more relative Proteobacteria in normal weight children. |
| Monira 2011 [85] | Bangladesh | Data of healthy children only | Cross-sectional | 2–3 years | 7 | V5-V6 | 454 | Prevotella and Bacteroides dominated the microbiome of healthy Bangladeshi children. |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Deering, K.E.; Devine, A.; O’Sullivan, T.A.; Lo, J.; Boyce, M.C.; Christophersen, C.T. Characterizing the Composition of the Pediatric Gut Microbiome: A Systematic Review. Nutrients 2020, 12, 16. https://doi.org/10.3390/nu12010016
Deering KE, Devine A, O’Sullivan TA, Lo J, Boyce MC, Christophersen CT. Characterizing the Composition of the Pediatric Gut Microbiome: A Systematic Review. Nutrients. 2020; 12(1):16. https://doi.org/10.3390/nu12010016
Chicago/Turabian StyleDeering, Kane E., Amanda Devine, Therese A. O’Sullivan, Johnny Lo, Mary C. Boyce, and Claus T. Christophersen. 2020. "Characterizing the Composition of the Pediatric Gut Microbiome: A Systematic Review" Nutrients 12, no. 1: 16. https://doi.org/10.3390/nu12010016
APA StyleDeering, K. E., Devine, A., O’Sullivan, T. A., Lo, J., Boyce, M. C., & Christophersen, C. T. (2020). Characterizing the Composition of the Pediatric Gut Microbiome: A Systematic Review. Nutrients, 12(1), 16. https://doi.org/10.3390/nu12010016






