Anti-Inflammatory Potential of Cow, Donkey and Goat Milk Extracellular Vesicles as Revealed by Metabolomic Profile
Abstract
:1. Introduction
2. Materials and Methods
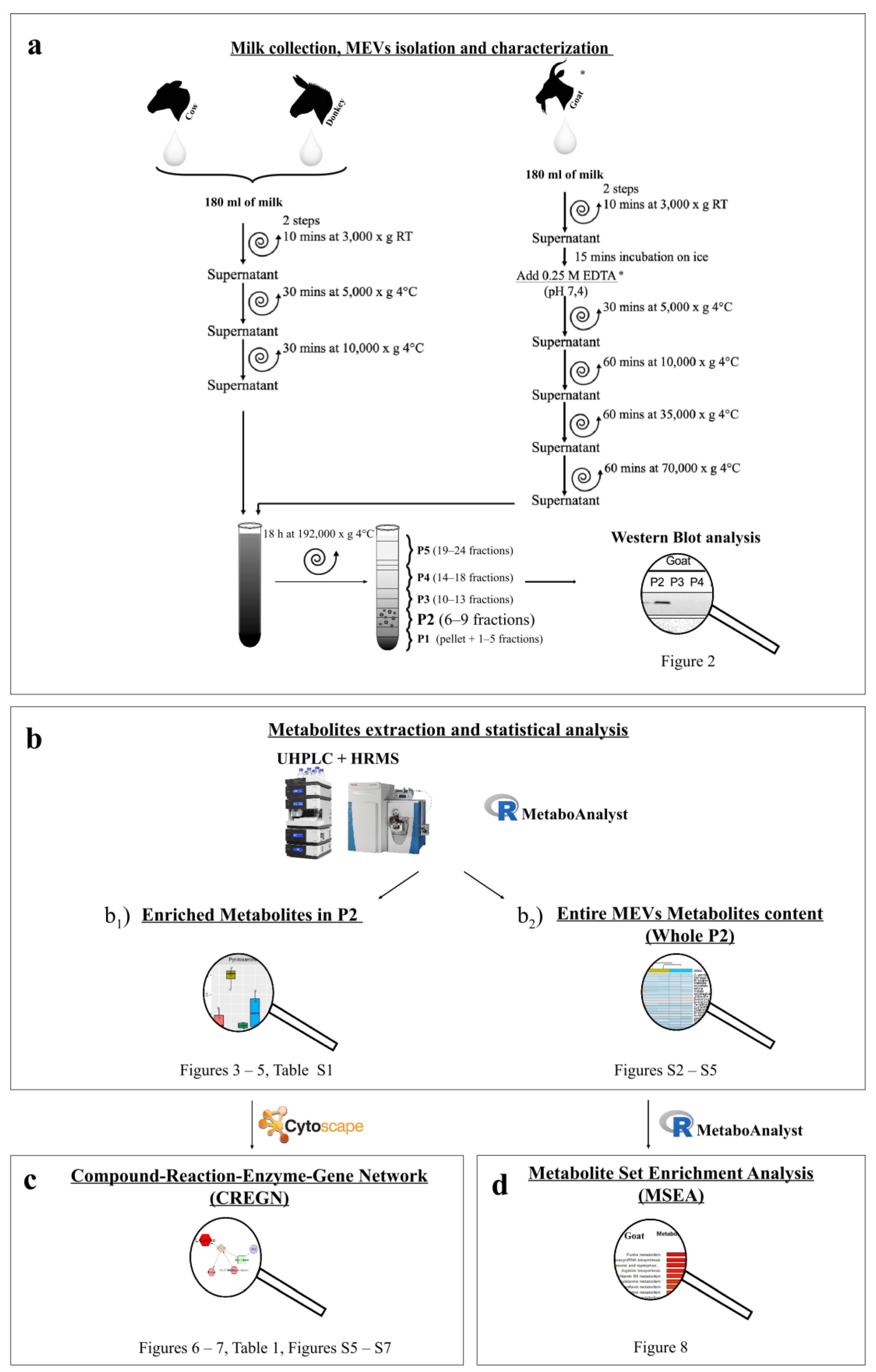
2.1. Milk Collection
2.2. EVs Isolation
2.3. MEVs Characterization
2.3.1. Western Blotting
2.3.2. Transmission Electron Microscopy (TEM)
2.4. Metabolomic Analysis
2.4.1. Metabolite Extraction
2.4.2. UHPLC-HRMS
2.5. Data Elaboration and Statistical Analysis
3. Results
3.1. MEVs Characterization
3.1.1. Western Blotting
3.1.2. Transmission Electron Microscopy
3.2. Metabolomic Analysis
3.3. Network and Pathway Enrichment Analysis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Cacho, N.T.; Lawrence, R.M. Innate Immunity and Breast Milk. Front. Immunol. 2017, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Melnik, B.C.; Schmitz, G. MicroRNAs: Milk’s epigenetic regulators. Best Pract. Res. Clin. Endocrinol. Metab. 2017, 31, 427–442. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, M.I.; van Herwijnen, M.J.C.; Fernandez-Gutierrez, M.M.; Giovanazzi, A.; de Groot, A.M.; Kleinjan, M.; van Capel, T.M.M.; Sijts, A.J.A.M.; Taams, L.S.; Garssen, J.; et al. Human milk extracellular vesicles target nodes in interconnected signalling pathways that enhance oral epithelial barrier function and dampen immune responses. Immunology 2020. under review. [Google Scholar]
- Théry, C.; Witwer, K.W.; Aikawa, E.; Alcaraz, M.J.; Anderson, J.D.; Andriantsitohaina, R.; Antoniou, A.; Arab, T.; Archer, F.; Atkin-Smith, G.K.; et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles 2018, 7, 1535750. [Google Scholar] [CrossRef] [Green Version]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Théry, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Théry, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef]
- Aryani, A.; Denecke, B. Exosomes as a Nanodelivery System: A Key to the Future of Neuromedicine? Mol. Neurobiol. 2016, 53, 818–834. [Google Scholar] [CrossRef] [Green Version]
- Capomaccio, S.; Cappelli, K.; Bazzucchi, C.; Coletti, M.; Gialletti, R.; Moriconi, F.; Passamonti, F.; Pepe, M.; Petrini, S.; Mecocci, S.; et al. Equine Adipose-Derived Mesenchymal Stromal Cells Release Extracellular Vesicles Enclosing Different Subsets of Small RNAs. Stem Cells Int. 2019, 2019, 4957806. [Google Scholar] [CrossRef] [Green Version]
- Ma, Z.; Wang, Y.; Li, H. Applications of extracellular vesicles in tissue regeneration. Biomicrofluidics 2020, 14, 011501. [Google Scholar] [CrossRef]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Lucchetti, D.; Ricciardi Tenore, C.; Colella, F.; Sgambato, A. Extracellular Vesicles and Cancer: A Focus on Metabolism, Cytokines, and Immunity. Cancers 2020, 12, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Buzas, E.I.; György, B.; Nagy, G.; Falus, A.; Gay, S. Emerging role of extracellular vesicles in inflammatory diseases. Nat. Rev. Rheumatol. 2014, 10, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Greening, D.W.; Gopal, S.K.; Xu, R.; Simpson, R.J.; Chen, W. Exosomes and their roles in immune regulation and cancer. Semin. Cell Dev. Biol. 2015, 40, 72–81. [Google Scholar] [CrossRef] [PubMed]
- Alcaraz, M.J.; Compañ, A.; Guillén, M.I. Extracellular Vesicles from Mesenchymal Stem Cells as Novel Treatments for Musculoskeletal Diseases. Cells 2020, 9, 98. [Google Scholar] [CrossRef] [Green Version]
- Boere, J.; Malda, J.; van de Lest, C.H.A.; van Weeren, P.R.; Wauben, M.H.M. Extracellular Vesicles in Joint Disease and Therapy. Front. Immunol. 2018, 9. [Google Scholar] [CrossRef] [Green Version]
- Vella, L.J.; Hill, A.F.; Cheng, L. Focus on Extracellular Vesicles: Exosomes and Their Role in Protein Trafficking and Biomarker Potential in Alzheimer’s and Parkinson’s Disease. Int. J. Mol. Sci. 2016, 17, 173. [Google Scholar] [CrossRef]
- Braicu, C.; Tomuleasa, C.; Monroig, P.; Cucuianu, A.; Berindan-Neagoe, I.; Calin, G.A. Exosomes as divine messengers: Are they the Hermes of modern molecular oncology? Cell Death Differ. 2015, 22, 34–45. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Mittelbrunn, M.; Sánchez-Madrid, F. Intercellular communication: Diverse structures for exchange of genetic information. Nat. Rev. Mol. Cell Biol. 2012, 13. [Google Scholar] [CrossRef]
- Ogawa, Y.; Miura, Y.; Harazono, A.; Kanai-Azuma, M.; Akimoto, Y.; Kawakami, H.; Yamaguchi, T.; Toda, T.; Endo, T.; Tsubuki, M.; et al. Proteomic Analysis of Two Types of Exosomes in Human Whole Saliva. Biol. Pharm. Bull. 2011, 34, 13–23. [Google Scholar] [CrossRef] [Green Version]
- Masyuk, A.I.; Huang, B.Q.; Ward, C.J.; Gradilone, S.A.; Banales, J.M.; Masyuk, T.V.; Radtke, B.; Splinter, P.L.; LaRusso, N.F. Biliary exosomes influence cholangiocyte regulatory mechanisms and proliferation through interaction with primary cilia. Am. J. Physiol. Gastrointest. Liver Physiol. 2010, 299, G990–G999. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Admyre, C.; Grunewald, J.; Thyberg, J.; Gripenbäck, S.; Tornling, G.; Eklund, A.; Scheynius, A.; Gabrielsson, S. Exosomes with major histocompatibility complex class II and co-stimulatory molecules are present in human BAL fluid. Eur. Respir. J. 2003, 22, 578–583. [Google Scholar] [CrossRef] [PubMed]
- Dear, J.W.; Street, J.M.; Bailey, M.A. Urinary exosomes: A reservoir for biomarker discovery and potential mediators of intrarenal signalling. Proteomics 2013, 13, 1572–1580. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Qazi, K.R.; Filen, J.-J.; Lahesmaa, R.; Norman, M.; Neve, E.P.A.; Scheynius, A.; Gabrielsson, S. Exosomes with Immune Modulatory Features Are Present in Human Breast Milk. J. Immunol. 2007, 179, 1969–1978. [Google Scholar] [CrossRef] [PubMed]
- Zempleni, J. Milk exosomes: Beyond dietary microRNAs. Genes Nutr. 2017, 12. [Google Scholar] [CrossRef]
- Benmoussa, A.; Lee, C.H.C.; Laffont, B.; Savard, P.; Laugier, J.; Boilard, E.; Gilbert, C.; Fliss, I.; Provost, P. Commercial Dairy Cow Milk microRNAs Resist Digestion under Simulated Gastrointestinal Tract Conditions. J. Nutr. 2016, 146, 2206–2215. [Google Scholar] [CrossRef]
- Kusuma, R.J.; Manca, S.; Friemel, T.; Sukreet, S.; Nguyen, C.; Zempleni, J. Human vascular endothelial cells transport foreign exosomes from cow’s milk by endocytosis. Am. J. Physiol. Cell Physiol. 2016, 310, C800–C807. [Google Scholar] [CrossRef] [Green Version]
- Liao, Y.; Du, X.; Li, J.; Lönnerdal, B. Human milk exosomes and their microRNAs survive digestion in vitro and are taken up by human intestinal cells. Mol. Nutr. Food Res. 2017, 61, 1700082. [Google Scholar] [CrossRef]
- Izumi, H.; Tsuda, M.; Sato, Y.; Kosaka, N.; Ochiya, T.; Iwamoto, H.; Namba, K.; Takeda, Y. Bovine milk exosomes contain microRNA and mRNA and are taken up by human macrophages. J. Dairy Sci. 2015, 98, 2920–2933. [Google Scholar] [CrossRef] [Green Version]
- Golan-Gerstl, R.; Elbaum Shiff, Y.; Moshayoff, V.; Schecter, D.; Leshkowitz, D.; Reif, S. Characterization and biological function of milk-derived miRNAs. Mol. Nutr. Food Res. 2017, 61, 1700009. [Google Scholar] [CrossRef]
- Chen, T.; Xie, M.-Y.; Sun, J.-J.; Ye, R.-S.; Cheng, X.; Sun, R.-P.; Wei, L.-M.; Li, M.; Lin, D.-L.; Jiang, Q.-Y.; et al. Porcine milk-derived exosomes promote proliferation of intestinal epithelial cells. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Zempleni, J.; Aguilar-Lozano, A.; Sadri, M.; Sukreet, S.; Manca, S.; Wu, D.; Zhou, F.; Mutai, E. Biological Activities of Extracellular Vesicles and Their Cargos from Bovine and Human Milk in Humans and Implications for Infants. J. Nutr. 2017, 147, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Manca, S.; Upadhyaya, B.; Mutai, E.; Desaulniers, A.T.; Cederberg, R.A.; White, B.R.; Zempleni, J. Milk exosomes are bioavailable and distinct microRNA cargos have unique tissue distribution patterns. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trinchese, G.; Cavaliere, G.; De Filippo, C.; Aceto, S.; Prisco, M.; Chun, J.T.; Penna, E.; Negri, R.; Muredda, L.; Demurtas, A.; et al. Human Milk and Donkey Milk, Compared to Cow Milk, Reduce Inflammatory Mediators and Modulate Glucose and Lipid Metabolism, Acting on Mitochondrial Function and Oleylethanolamide Levels in Rat Skeletal Muscle. Front. Physiol. 2018, 9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Quintanilha, B.; Reis, B.; Duarte, G.; Cozzolino, S.; Rogero, M. Nutrimiromics: Role of microRNAs and Nutrition in Modulating Inflammation and Chronic Diseases. Nutrients 2017, 9, 1168. [Google Scholar] [CrossRef]
- Chiofalo, B.; Dugo, P.; Bonaccorsi, I.L.; Mondello, L. Comparison of major lipid components in human and donkey milk: New perspectives for a hypoallergenic diet in humans. Immunopharmacol. Immunotoxicol. 2011, 33, 633–644. [Google Scholar] [CrossRef]
- Huang, X.; Yuan, T.; Tschannen, M.; Sun, Z.; Jacob, H.; Du, M.; Liang, M.; Dittmar, R.L.; Liu, Y.; Liang, M.; et al. Characterization of human plasma-derived exosomal RNAs by deep sequencing. BMC Genom. 2013, 14, 319. [Google Scholar] [CrossRef] [Green Version]
- Abels, E.R.; Breakefield, X.O. Introduction to Extracellular Vesicles: Biogenesis, RNA Cargo Selection, Content, Release, and Uptake. Cell. Mol. Neurobiol. 2016, 36, 301–312. [Google Scholar] [CrossRef]
- Freedman, J.E.; Gerstein, M.; Mick, E.; Rozowsky, J.; Levy, D.; Kitchen, R.; Das, S.; Shah, R.; Danielson, K.; Beaulieu, L.; et al. Diverse human extracellular RNAs are widely detected in human plasma. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef]
- Squadrito, M.L.; Baer, C.; Burdet, F.; Maderna, C.; Gilfillan, G.D.; Lyle, R.; Ibberson, M.; De Palma, M. Endogenous RNAs Modulate MicroRNA Sorting to Exosomes and Transfer to Acceptor Cells. Cell Rep. 2014, 8, 1432–1446. [Google Scholar] [CrossRef] [Green Version]
- Stevanato, L.; Thanabalasundaram, L.; Vysokov, N.; Sinden, J.D. Investigation of Content, Stoichiometry and Transfer of miRNA from Human Neural Stem Cell Line Derived Exosomes. PLoS ONE 2016, 11, e0146353. [Google Scholar] [CrossRef]
- Hadacek, F. Low-molecular-weight metabolite systems chemistry. Front. Environ. Sci. 2015, 3. [Google Scholar] [CrossRef]
- Kosmides, A.K.; Kamisoglu, K.; Calvano, S.E.; Corbett, S.A.; Androulakis, I.P. Metabolomic Fingerprinting: Challenges and Opportunities. Crit. Rev. Biomed. Eng. 2013, 41, 205–221. [Google Scholar] [CrossRef] [PubMed]
- Luo, X.; An, M.; Cuneo, K.C.; Lubman, D.M.; Li, L. High-Performance Chemical Isotope Labeling Liquid Chromatography Mass Spectrometry for Exosome Metabolomics. Anal. Chem. 2018, 90, 8314–8319. [Google Scholar] [CrossRef] [PubMed]
- Puhka, M.; Takatalo, M.; Nordberg, M.-E.; Valkonen, S.; Nandania, J.; Aatonen, M.; Yliperttula, M.; Laitinen, S.; Velagapudi, V.; Mirtti, T.; et al. Metabolomic Profiling of Extracellular Vesicles and Alternative Normalization Methods Reveal Enriched Metabolites and Strategies to Study Prostate Cancer-Related Changes. Theranostics 2017, 7, 3824–3841. [Google Scholar] [CrossRef] [PubMed]
- Zonneveld, M.I.; Brisson, A.R.; van Herwijnen, M.J.C.; Tan, S.; van de Lest, C.H.A.; Redegeld, F.A.; Garssen, J.; Wauben, M.H.M.; Nolte-’t Hoen, E.N.M. Recovery of extracellular vesicles from human breast milk is influenced by sample collection and vesicle isolation procedures. J. Extracell. Vesicles 2014, 3. [Google Scholar] [CrossRef] [Green Version]
- Vaswani, K.; Mitchell, M.D.; Holland, O.J.; Qin Koh, Y.; Hill, R.J.; Harb, T.; Davies, P.S.W.; Peiris, H. A Method for the Isolation of Exosomes from Human and Bovine Milk. J. Nutr. Metab. 2019, 2019. [Google Scholar] [CrossRef]
- Cavinato, L.; Genise, E.; Luly, F.R.; Di Domenico, E.G.; Del Porto, P.; Ascenzioni, F. Escaping the Phagocytic Oxidative Burst: The Role of SODB in the Survival of Pseudomonas aeruginosa within Macrophages. Front. Microbiol. 2020, 11. [Google Scholar] [CrossRef] [PubMed]
- Chong, J.; Xia, J. MetaboAnalystR: An R package for flexible and reproducible analysis of metabolomics data. Bioinformatics 2018, 34, 4313–4314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karnovsky, A.; Weymouth, T.; Hull, T.; Tarcea, V.G.; Scardoni, G.; Laudanna, C.; Sartor, M.A.; Stringer, K.A.; Jagadish, H.V.; Burant, C.; et al. Metscape 2 bioinformatics tool for the analysis and visualization of metabolomics and gene expression data. Bioinformatics 2012, 28, 373–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Pang, Z.; Chong, J.; Li, S.; Xia, J. MetaboAnalystR 3.0: Toward an Optimized Workflow for Global Metabolomics. Metabolites 2020, 10, 186. [Google Scholar] [CrossRef] [PubMed]
- Blans, K.; Hansen, M.S.; Sørensen, L.V.; Hvam, M.L.; Howard, K.A.; Möller, A.; Wiking, L.; Larsen, L.B.; Rasmussen, J.T. Pellet-free isolation of human and bovine milk extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles 2017, 6, 1294340. [Google Scholar] [CrossRef] [PubMed]
- Pascucci, L.; Coccè, V.; Bonomi, A.; Ami, D.; Ceccarelli, P.; Ciusani, E.; Viganò, L.; Locatelli, A.; Sisto, F.; Doglia, S.M.; et al. Paclitaxel is incorporated by mesenchymal stromal cells and released in exosomes that inhibit in vitro tumor growth: A new approach for drug delivery. J. Control. Release 2014, 192, 262–270. [Google Scholar] [CrossRef] [PubMed]
- Mentkowski, K.I.; Snitzer, J.D.; Rusnak, S.; Lang, J.K. Therapeutic Potential of Engineered Extracellular Vesicles. AAPS J. 2018, 20. [Google Scholar] [CrossRef] [Green Version]
- Burrello, J.; Monticone, S.; Gai, C.; Gomez, Y.; Kholia, S.; Camussi, G. Stem Cell-Derived Extracellular Vesicles and Immune-Modulation. Front. Cell Dev. Biol. 2016, 4. [Google Scholar] [CrossRef] [Green Version]
- Benmoussa, A.; Ly, S.; Shan, S.T.; Laugier, J.; Boilard, E.; Gilbert, C.; Provost, P. A subset of extracellular vesicles carries the bulk of microRNAs in commercial dairy cow’s milk. J. Extracell. Vesicles 2017, 6, 1401897. [Google Scholar] [CrossRef] [Green Version]
- Pearce, E.L.; Pearce, E.J. Metabolic pathways in immune cell activation and quiescence. Immunity 2013, 38, 633–643. [Google Scholar] [CrossRef] [Green Version]
- Hachimura, S.; Totsuka, M.; Hosono, A. Immunomodulation by food: Impact on gut immunity and immune cell function. Biosci. Biotechnol. Biochem. 2018, 82, 584–599. [Google Scholar] [CrossRef]
- Kunisawa, J.; Hashimoto, E.; Ishikawa, I.; Kiyono, H. A Pivotal Role of Vitamin B9 in the Maintenance of Regulatory T Cells In Vitro and In Vivo. PLoS ONE 2012, 7, e32094. [Google Scholar] [CrossRef] [Green Version]
- Kinoshita, M.; Kayama, H.; Kusu, T.; Yamaguchi, T.; Kunisawa, J.; Kiyono, H.; Sakaguchi, S.; Takeda, K. Dietary Folic Acid Promotes Survival of Foxp3+ Regulatory T Cells in the Colon. J. Immunol. 2012, 189, 2869–2878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hegyi, J.; Schwartz, R.A.; Hegyi, V. Pellagra: Dermatitis, dementia, and diarrhea. Int. J. Dermatol. 2004, 43, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Mandrika, I.; Tilgase, A.; Petrovska, R.; Klovins, J. Hydroxycarboxylic Acid Receptor Ligands Modulate Proinflammatory Cytokine Expression in Human Macrophages and Adipocytes without Affecting Adipose Differentiation. Biol. Pharm. Bull. 2018, 41, 1574–1580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gambhir, D.; Ananth, S.; Veeranan-Karmegam, R.; Elangovan, S.; Hester, S.; Jennings, E.; Offermanns, S.; Nussbaum, J.J.; Smith, S.B.; Thangaraju, M.; et al. GPR109A as an Anti-Inflammatory Receptor in Retinal Pigment Epithelial Cells and Its Relevance to Diabetic Retinopathy. Investig. Opthalmol. Vis. Sci. 2012, 53, 2208. [Google Scholar] [CrossRef]
- Thangaraju, M.; Cresci, G.A.; Liu, K.; Ananth, S.; Gnanaprakasam, J.P.; Browning, D.D.; Mellinger, J.D.; Smith, S.B.; Digby, G.J.; Lambert, N.A.; et al. GPR109A Is a G-protein-Coupled Receptor for the Bacterial Fermentation Product Butyrate and Functions as a Tumor Suppressor in Colon. Cancer Res. 2009, 69, 2826–2832. [Google Scholar] [CrossRef] [Green Version]
- Singh, N.; Gurav, A.; Sivaprakasam, S.; Brady, E.; Padia, R.; Shi, H.; Thangaraju, M.; Prasad, P.D.; Manicassamy, S.; Munn, D.H.; et al. Activation of the receptor (Gpr109a) for niacin and the commensal metabolite butyrate suppresses colonic inflammation and carcinogenesis. Immunity 2014, 40, 128–139. [Google Scholar] [CrossRef] [Green Version]
- Bhatt, B.; Zeng, P.; Zhu, H.; Sivaprakasam, S.; Li, S.; Xiao, H.; Dong, L.; Shiao, P.; Kolhe, R.; Patel, N.; et al. Gpr109a limits microbiota-induced IL-23 production to constrain ILC3-mediated colonic inflammation. J. Immunol. 2018, 200, 2905–2914. [Google Scholar] [CrossRef] [Green Version]
- Farnier, M.; Chen, E.; Johnson-Levonas, A.O.; McCrary Sisk, C.; Mitchel, Y.B. Effects of extended-release niacin/laropiprant, simvastatin, and the combination on correlations between apolipoprotein B, LDL cholesterol, and non-HDL cholesterol in patients with dyslipidemia. Vasc. Health Risk Manag. 2014, 10, 279–290. [Google Scholar] [CrossRef] [Green Version]
- Meyer-Ficca, M.; Kirkland, J.B. Niacin12. Adv. Nutr. 2016, 7, 556–558. [Google Scholar] [CrossRef] [Green Version]
- Lukasova, M.; Malaval, C.; Gille, A.; Kero, J.; Offermanns, S. Nicotinic acid inhibits progression of atherosclerosis in mice through its receptor GPR109A expressed by immune cells. J. Clin. Investig. 2011, 121, 1163–1173. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Bin, P.; Wu, W.; Chang, Y.; Zhu, G. Tryptophan Metabolism, Regulatory T Cells, and Inflammatory Bowel Disease: A Mini Review. Mediat. Inflamm. 2020, 2020, 1–10. [Google Scholar] [CrossRef]
- Agus, A.; Planchais, J.; Sokol, H. Gut Microbiota Regulation of Tryptophan Metabolism in Health and Disease. Cell Host Microbe 2018, 23, 716–724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taleb, S. Tryptophan Dietary Impacts Gut Barrier and Metabolic Diseases. Front. Immunol. 2019, 10. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Wang, X.; Hu, C.-A. Therapeutic Potential of Amino Acids in Inflammatory Bowel Disease. Nutrients 2017, 9, 920. [Google Scholar] [CrossRef] [Green Version]
- Andrade, M.E.R.; Araújo, R.S.; de Barros, P.A.V.; Soares, A.D.N.; Abrantes, F.A.; de Vasconcelos Generoso, S.; Fernandes, S.O.A.; Cardoso, V.N. The role of immunomodulators on intestinal barrier homeostasis in experimental models. Clin. Nutr. 2015, 34, 1080–1087. [Google Scholar] [CrossRef]
- Lamas, B.; Vergnaud-Gauduchon, J.; Goncalves-Mendes, N.; Perche, O.; Rossary, A.; Vasson, M.-P.; Farges, M.-C. Altered functions of natural killer cells in response to L-Arginine availability. Cell. Immunol. 2012, 280, 182–190. [Google Scholar] [CrossRef]
- Hong, M.Y.; Beidler, J.; Hooshmand, S.; Figueroa, A.; Kern, M. Watermelon and l-arginine consumption improve serum lipid profile and reduce inflammation and oxidative stress by altering gene expression in rats fed an atherogenic diet. Nutr. Res. 2018, 58, 46–54. [Google Scholar] [CrossRef]
- Khalaf, D.; Krüger, M.; Wehland, M.; Infanger, M.; Grimm, D. The Effects of Oral l-Arginine and l-Citrulline Supplementation on Blood Pressure. Nutrients 2019, 11, 1679. [Google Scholar] [CrossRef] [Green Version]
- Ren, W.; Zou, L.; Li, N.; Wang, Y.; Liu, G.; Peng, Y.; Ding, J.; Cai, L.; Yin, Y.; Wu, G. Dietary arginine supplementation enhances immune responses to inactivated Pasteurella multocida vaccination in mice. Br. J. Nutr. 2013, 109, 867–872. [Google Scholar] [CrossRef] [Green Version]
- Coburn, L.A.; Horst, S.N.; Allaman, M.M.; Brown, C.T.; Williams, C.S.; Hodges, M.E.; Druce, J.P.; Beaulieu, D.B.; Schwartz, D.A.; Wilson, K.T. L-arginine Availability and Metabolism Is Altered in Ulcerative Colitis. Inflamm. Bowel Dis. 2016, 22, 1847–1858. [Google Scholar] [CrossRef] [Green Version]
- Timmons, J. Polyamines and Gut Mucosal Homeostasis. J. Gastrointest. Dig. Syst. 2013, 3. [Google Scholar] [CrossRef]
- Tofalo, R.; Cocchi, S.; Suzzi, G. Polyamines and Gut Microbiota. Front. Nutr. 2019, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fan, J.; Meng, Q.; Guo, G.; Xie, Y.; Li, X.; Xiu, Y.; Li, T.; Ma, L. Effects of early enteral nutrition supplemented with arginine on intestinal mucosal immunity in severely burned mice. Clin. Nutr. 2010, 29, 124–130. [Google Scholar] [CrossRef] [PubMed]
- He, F.; Wu, C.; Li, P.; Li, N.; Zhang, D.; Zhu, Q.; Ren, W.; Peng, Y. Functions and Signaling Pathways of Amino Acids in Intestinal Inflammation. BioMed. Res. Int. 2018, 2018. [Google Scholar] [CrossRef] [PubMed]
- Brasse-Lagnel, C.G.; Lavoinne, A.M.; Husson, A.S. Amino acid regulation of mammalian gene expression in the intestine. Biochimie 2010, 92, 729–735. [Google Scholar] [CrossRef]
- Chen, S.; Liu, Y.; Wang, X.; Wang, H.; Li, S.; Shi, H.; Zhu, H.; Zhang, J.; Pi, D.; Hu, C.-A.A.; et al. Asparagine improves intestinal integrity, inhibits TLR4 and NOD signaling, and differently regulates p38 and ERK1/2 signaling in weanling piglets after LPS challenge. Innate Immun. 2016, 22, 577–587. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Li, S.; Pi, D.; Zhu, H.; Hou, Y.; Shi, H.; Leng, W. Asparagine attenuates intestinal injury, improves energy status and inhibits AMP-activated protein kinase signalling pathways in weaned piglets challenged with Escherichia coli lipopolysaccharide. Br. J. Nutr. 2015, 114, 553–565. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Pi, D.; Leng, W.; Wang, X.; Hu, C.-A.A.; Hou, Y.; Xiong, J.; Wang, C.; Qin, Q.; Liu, Y. Asparagine preserves intestinal barrier function from LPS-induced injury and regulates CRF/CRFR signaling pathway. Innate Immun. 2017, 23, 546–556. [Google Scholar] [CrossRef] [Green Version]
- Torres, A.; Luke, J.D.; Kullas, A.L.; Kapilashrami, K.; Botbol, Y.; Koller, A.; Tonge, P.J.; Chen, E.I.; Macian, F.; van der Velden, A.W.M. Asparagine deprivation mediated by Salmonella asparaginase causes suppression of activation-induced T cell metabolic reprogramming. J. Leukoc. Biol. 2016, 99, 387–398. [Google Scholar] [CrossRef] [Green Version]
- Lushchak, V.I. Glutathione Homeostasis and Functions: Potential Targets for Medical Interventions. J. Amino Acids 2012, 2012, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Gaucher, C.; Boudier, A.; Bonetti, J.; Clarot, I.; Leroy, P.; Parent, M. Glutathione: Antioxidant Properties Dedicated to Nanotechnologies. Antioxidants 2018, 7, 62. [Google Scholar] [CrossRef] [Green Version]
- Avila, M.A.; García-Trevijano, E.R.; Lu, S.C.; Corrales, F.J.; Mato, J.M. Methylthioadenosine. Int. J. Biochem. Cell Biol. 2004, 36, 2125–2130. [Google Scholar] [CrossRef] [PubMed]
- Hevia, H.; Varela-Rey, M.; Corrales, F.J.; Berasain, C.; Martínez-Chantar, M.L.; Latasa, M.U.; Lu, S.C.; Mato, J.M.; García-Trevijano, E.R.; Avila, M.A. 5′-methylthioadenosine modulates the inflammatory response to endotoxin in mice and in rat hepatocytes. Hepatology 2004, 39, 1088–1098. [Google Scholar] [CrossRef]
- Moreno, B.; Hevia, H.; Santamaria, M.; Sepulcre, J.; Muñoz, J.; García-Trevijano, E.R.; Berasain, C.; Corrales, F.J.; Avila, M.A.; Villoslada, P. Methylthioadenosine reverses brain autoimmune disease. Ann. Neurol. 2006, 60, 323–334. [Google Scholar] [CrossRef] [PubMed]
- Moreno, B.; Fernandez-Diez, B.; Di Penta, A.; Villoslada, P. Preclinical studies of methylthioadenosine for the treatment of multiple sclerosis. Mult. Scler. J. 2010, 16, 1102–1108. [Google Scholar] [CrossRef] [PubMed]
- Li, T.W.H.; Yang, H.; Peng, H.; Xia, M.; Mato, J.M.; Lu, S.C. Effects of S-adenosylmethionine and methylthioadenosine on inflammation-induced colon cancer in mice. Carcinogenesis 2012, 33, 427–435. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benight, N.M.; Stoll, B.; Marini, J.C.; Burrin, D.G. Preventative oral methylthioadenosine is anti-inflammatory and reduces DSS-induced colitis in mice. Am. J. Physiol. Gastrointest. Liver Physiol. 2012, 303, G71–G82. [Google Scholar] [CrossRef] [Green Version]
- Han, H.; Yin, J.; Wang, B.; Huang, X.; Yao, J.; Zheng, J.; Fan, W.; Li, T.; Yin, Y. Effects of dietary lysine restriction on inflammatory responses in piglets. Sci. Rep. 2018, 8. [Google Scholar] [CrossRef] [Green Version]
- Mine, Y.; Zhang, H. Anti-inflammatory Effects of Poly-l-lysine in Intestinal Mucosal System Mediated by Calcium-Sensing Receptor Activation. J. Agric. Food Chem. 2015, 63, 10437–10447. [Google Scholar] [CrossRef]
- Bao, X.; Feng, Z.; Yao, J.; Li, T.; Yin, Y. Roles of Dietary Amino Acids and Their Metabolites in Pathogenesis of Inflammatory Bowel Disease. Mediat. Inflamm. 2017, 2017. [Google Scholar] [CrossRef]
- Wu, C.-H.; Ko, J.-L.; Liao, J.-M.; Huang, S.-S.; Lin, M.-Y.; Lee, L.-H.; Chang, L.-Y.; Ou, C.-C. D-methionine alleviates cisplatin-induced mucositis by restoring the gut microbiota structure and improving intestinal inflammation. Adv. Med. Oncol. 2019, 11. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez, Y.; Li, X.; Liu, G.; Bin, P.; Yan, W.; Más, D.; Valdivié, M.; Hu, C.-A.A.; Ren, W.; Yin, Y. The role of methionine on metabolism, oxidative stress, and diseases. Amino Acids 2017, 49, 2091–2098. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Tan, B.; Huang, B.; Li, J.; Wang, J.; Liao, P.; Guan, G.; Ji, P.; Yin, Y. Involvement of calcium-sensing receptor activation in the alleviation of intestinal inflammation in a piglet model by dietary aromatic amino acid supplementation. Br. J. Nutr. 2018, 120, 1321–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, Y.P.; Cheng, Y.F.; Li, X.H.; Yang, W.L.; Wen, C.; Zhuang, S.; Zhou, Y.M. Effects of threonine supplementation on the growth performance, immunity, oxidative status, intestinal integrity, and barrier function of broilers at the early age. Poult. Sci. 2017, 96, 405–413. [Google Scholar] [CrossRef]
- Sugihara, K.; Kamada, N. 10 the Role of Dietary L-Serine in the Regulation of Intestinal Mucus Barrier during Inflammation. Gastroenterology 2020, 158, S70. [Google Scholar] [CrossRef]
- Xu, X.; Wang, X.; Wu, H.; Zhu, H.; Liu, C.; Hou, Y.; Dai, B.; Liu, X.; Liu, Y. Glycine Relieves Intestinal Injury by Maintaining mTOR Signaling and Suppressing AMPK, TLR4, and NOD Signaling in Weaned Piglets after Lipopolysaccharide Challenge. Int. J. Mol. Sci. 2018, 19, 1980. [Google Scholar] [CrossRef] [Green Version]
- Hess, J.R.; Greenberg, N.A. The Role of Nucleotides in the Immune and Gastrointestinal Systems. Nutr. Clin. Pract. 2012, 27, 281–294. [Google Scholar] [CrossRef]



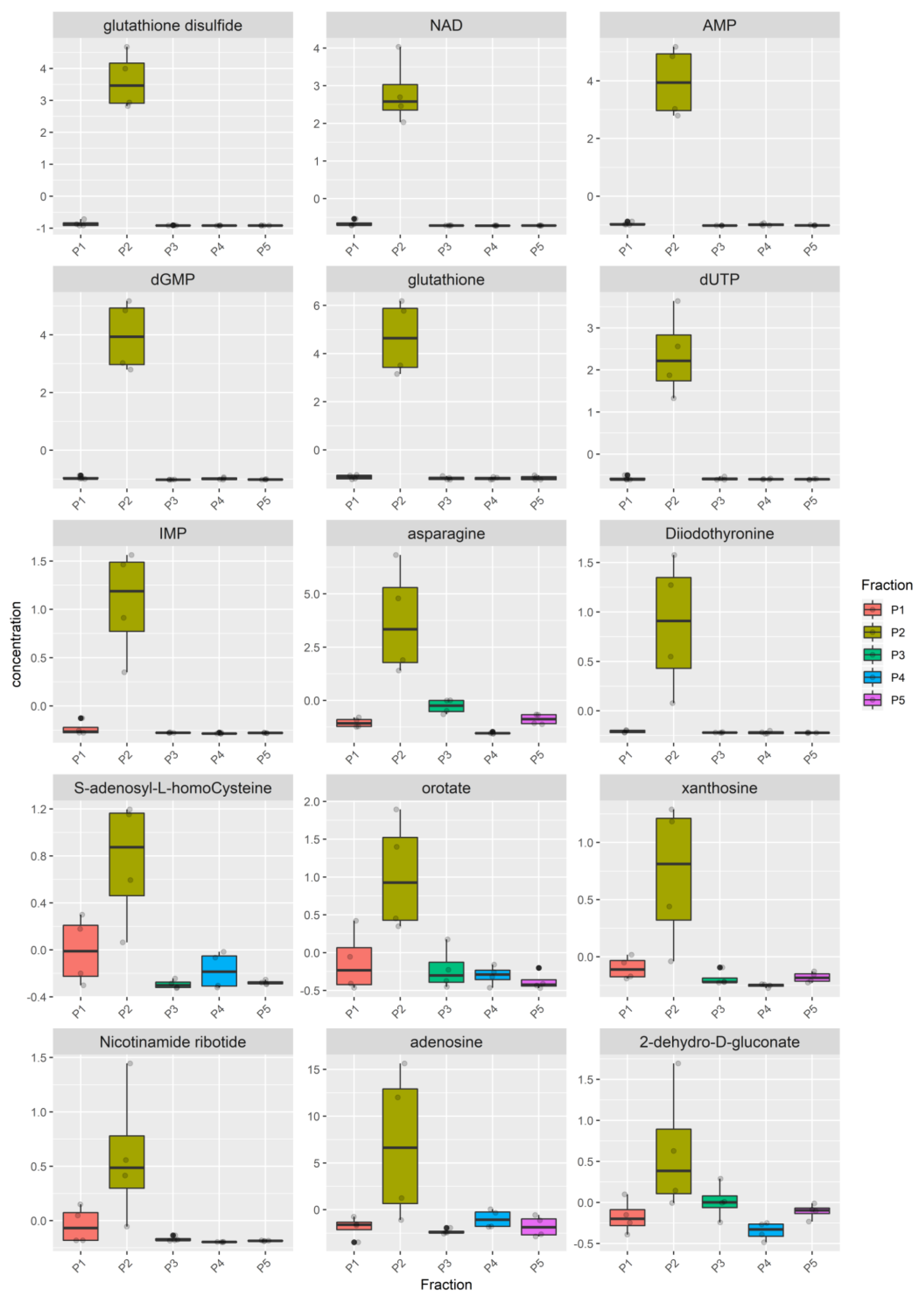



| Pathways | Pathway Recurrence among Species | Bovine Metabolites | Donkey Metabolites | Goat Metabolites |
|---|---|---|---|---|
| Purine metabolism | 3 | Adenine Adenosine; Deoxyinosine; GMP; IMP; Inosine; Guanosine | Adenosine; AMP; dGMP; IMP; Xanthosine | Adenosine; AMP; Deoxyinosine; dGMP; Guanine; Xanthosine |
| Pyrimidine metabolism | 3 | Cytidine | dUTP Orotate | dTMP; dUMP; Uridine |
| Urea cycle and metabolism of arginine, proline, glutamate, aspartate and asparagine | 3 | Lysine | Glutathione; Glutathione disulfide; Asparagine | S-methyl-5-thioadenosine; Glutathione disulfide; Arginine |
| Vitamin B3 (nicotinate and nicotinamide) metabolism | 3 | NAD; NADH; NADP; Nicotinamide ribotide | NAD; Nicotinamide ribotide | Arginine; NAD; NADH |
| Methionine and cysteine metabolism | 2 | S-Adenosyl-L-homocysteine | Methionine | |
| Tyrosine metabolism | 2 | Diiodothyronine | Diiodothyronine; Phenylalanine; Tyrosine | |
| Pentose phosphate pathway | 1 | 2-Dehydro-D-gluconate | ||
| Biopterin metabolism | 1 | Phenylalanine; Tyrosine | ||
| Galactose metabolism | 1 | Glucose-1-phosphate | ||
| Glycerophospholipid metabolism | 1 | Choline | ||
| Glycine, serine, alanine and threonine metabolism | 1 | Choline; Arginine; Methionine | ||
| Glycolysis and Gluconeogenesis | 1 | Glucose-1-phosphate | ||
| Tryptophan metabolism | 1 | Tryptophan | ||
| Vitamin B9 (folate) metabolism | 1 | 5-methyl-THF | ||
| Vitamin B2 (riboflavin) metabolism | 1 | FMN | ||
| Vitamin B6 (pyridoxine) metabolism | 1 | Pyridoxamine | ||
| Vitamin H (biotin) metabolism | 1 | Lysine | ||
| Lipoate metabolism | 1 | Lysine | ||
| Lysine metabolism | 1 | Lysine | ||
| None 1 | Allantoin; Cytosine | Acetyllysine; Cytosine; Deoxyribose-phosphate Glucose-6-phosphate Indoleacrylic acid | ||
| No KEGG ID 2 | Methionine sulfoxide; 5-deoxyribose-1-phosphate; N(alpha)-Acetyllysine |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mecocci, S.; Gevi, F.; Pietrucci, D.; Cavinato, L.; Luly, F.R.; Pascucci, L.; Petrini, S.; Ascenzioni, F.; Zolla, L.; Chillemi, G.; et al. Anti-Inflammatory Potential of Cow, Donkey and Goat Milk Extracellular Vesicles as Revealed by Metabolomic Profile. Nutrients 2020, 12, 2908. https://doi.org/10.3390/nu12102908
Mecocci S, Gevi F, Pietrucci D, Cavinato L, Luly FR, Pascucci L, Petrini S, Ascenzioni F, Zolla L, Chillemi G, et al. Anti-Inflammatory Potential of Cow, Donkey and Goat Milk Extracellular Vesicles as Revealed by Metabolomic Profile. Nutrients. 2020; 12(10):2908. https://doi.org/10.3390/nu12102908
Chicago/Turabian StyleMecocci, Samanta, Federica Gevi, Daniele Pietrucci, Luca Cavinato, Francesco R. Luly, Luisa Pascucci, Stefano Petrini, Fiorentina Ascenzioni, Lello Zolla, Giovanni Chillemi, and et al. 2020. "Anti-Inflammatory Potential of Cow, Donkey and Goat Milk Extracellular Vesicles as Revealed by Metabolomic Profile" Nutrients 12, no. 10: 2908. https://doi.org/10.3390/nu12102908
APA StyleMecocci, S., Gevi, F., Pietrucci, D., Cavinato, L., Luly, F. R., Pascucci, L., Petrini, S., Ascenzioni, F., Zolla, L., Chillemi, G., & Cappelli, K. (2020). Anti-Inflammatory Potential of Cow, Donkey and Goat Milk Extracellular Vesicles as Revealed by Metabolomic Profile. Nutrients, 12(10), 2908. https://doi.org/10.3390/nu12102908









