A High Protein Calorie Restriction Diet Alters the Gut Microbiome in Obesity
Abstract
1. Introduction
2. Materials and Methods
2.1. Patient Recruitment
2.2. Enrollment and Randomization
2.3. Dietary Intervention
2.4. Nutritional Assessments
2.5. Fecal Sampling
2.6. Statistical Analysis
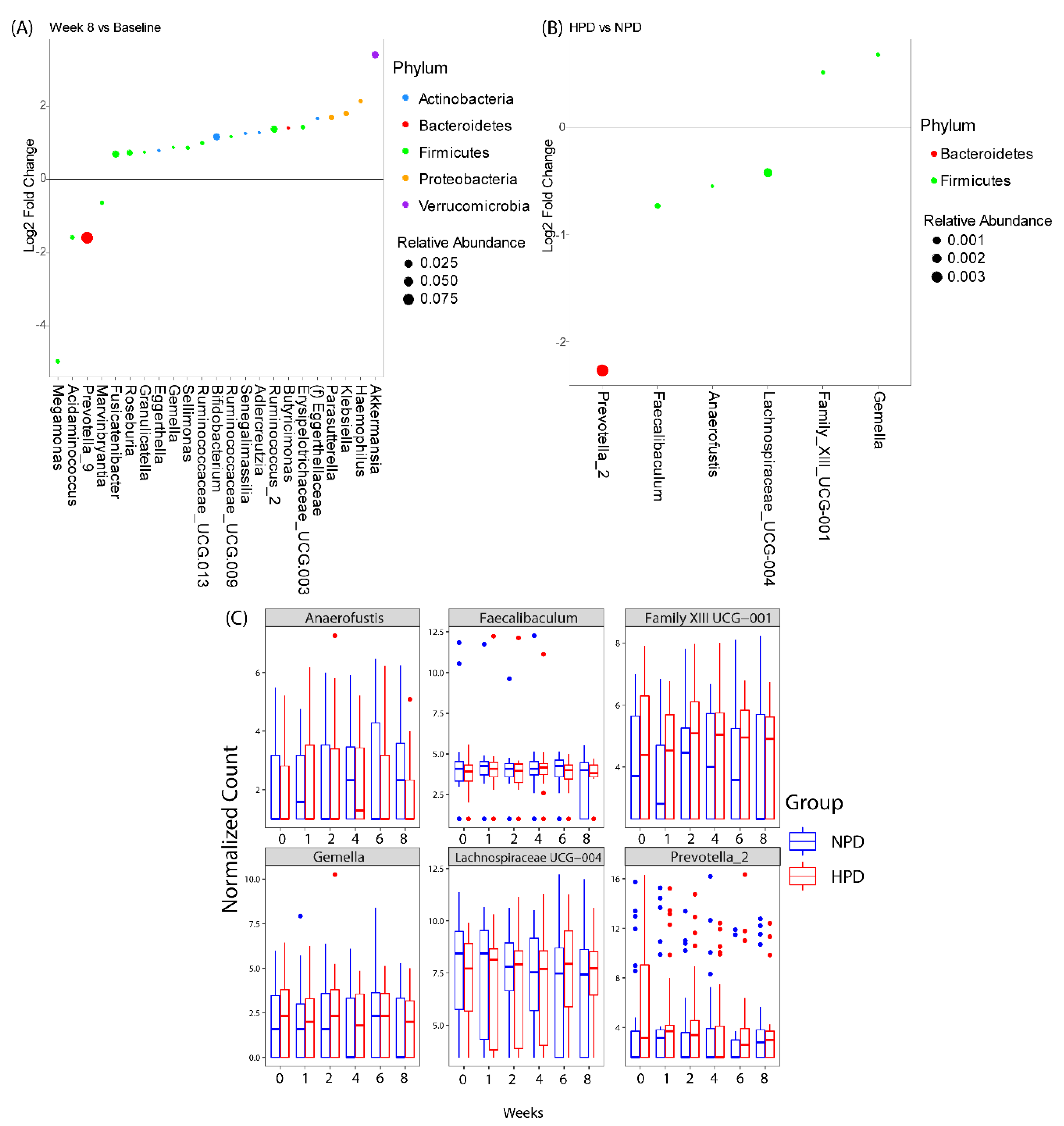
3. Results
4. Discussion
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Ogden, C.L.; Carroll, M.D.; Kit, B.K.; Flegal, K.M. Prevalence of childhood and adult obesity in the United States, 2011–2012. JAMA 2014, 311, 806–814. [Google Scholar] [CrossRef] [PubMed]
- Dietz, W.H.; Baur, L.A.; Hall, K.; Puhl, R.M.; Taveras, E.M.; Uauy, R.; Kopelman, P. Management of obesity: Improvement of health-care training and systems for prevention and care. Lancet 2015, 385, 2521–2533. [Google Scholar] [CrossRef]
- Skov, A.R.; Toubro, S.; Ronn, B.; Holm, L.; Astrup, A. Randomized trial on protein vs carbohydrate in ad libitum fat reduced diet for the treatment of obesity. Int. J. Obes. Relat. Metab. Disord. 1999, 23, 528–536. [Google Scholar] [CrossRef] [PubMed]
- Sacks, F.M.; Bray, G.A.; Carey, V.J.; Smith, S.R.; Ryan, D.H.; Anton, S.D.; McManus, K.; Champagne, C.M.; Bishop, L.M.; Laranjo, N.; et al. Comparison of weight-loss diets with different compositions of fat, protein, and carbohydrates. N. Engl. J. Med. 2009, 360, 859–873. [Google Scholar] [CrossRef]
- Larsen, T.M.; Dalskov, S.M.; van Baak, M.; Jebb, S.A.; Papadaki, A.; Pfeiffer, A.F.; Martinez, J.A.; Handjieva-Darlenska, T.; Kunesova, M.; Pihlsgard, M.; et al. Diets with high or low protein content and glycemic index for weight-loss maintenance. N. Engl. J. Med. 2010, 363, 2102–2113. [Google Scholar] [CrossRef]
- Wycherley, T.P.; Moran, L.J.; Clifton, P.M.; Noakes, M.; Brinkworth, G.D. Effects of energy-restricted high-protein, low-fat compared with standard-protein, low-fat diets: A meta-analysis of randomized controlled trials. Am. J. Clin. Nutr. 2012, 96, 1281–1298. [Google Scholar] [CrossRef]
- Weigle, D.S.; Breen, P.A.; Matthys, C.C.; Callahan, H.S.; Meeuws, K.E.; Burden, V.R.; Purnell, J.Q. A high-protein diet induces sustained reductions in appetite, ad libitum caloric intake, and body weight despite compensatory changes in diurnal plasma leptin and ghrelin concentrations. Am. J. Clin. Nutr. 2005, 82, 41–48. [Google Scholar] [CrossRef]
- Belza, A.; Ritz, C.; Sorensen, M.Q.; Holst, J.J.; Rehfeld, J.F.; Astrup, A. Contribution of gastroenteropancreatic appetite hormones to protein-induced satiety. Am. J. Clin. Nutr. 2013, 97, 980–989. [Google Scholar] [CrossRef]
- Journel, M.; Chaumontet, C.; Darcel, N.; Fromentin, G.; Tome, D. Brain responses to high-protein diets. Adv. Nutr. 2012, 3, 322–329. [Google Scholar] [CrossRef]
- Martens, E.A.; Lemmens, S.G.; Westerterp-Plantenga, M.S. Protein leverage affects energy intake of high-protein diets in humans. Am. J. Clin. Nutr. 2013, 97, 86–93. [Google Scholar] [CrossRef]
- Johnston, C.S.; Day, C.S.; Swan, P.D. Postprandial thermogenesis is increased 100% on a high-protein, low-fat diet versus a high-carbohydrate, low-fat diet in healthy, young women. J. Am. Coll. Nutr. 2002, 21, 55–61. [Google Scholar] [CrossRef] [PubMed]
- Tentolouris, N.; Pavlatos, S.; Kokkinos, A.; Perrea, D.; Pagoni, S.; Katsilambros, N. Diet-induced thermogenesis and substrate oxidation are not different between lean and obese women after two different isocaloric meals, one rich in protein and one rich in fat. Metabolism 2008, 57, 313–320. [Google Scholar] [CrossRef]
- Ebbeling, C.B.; Swain, J.F.; Feldman, H.A.; Wong, W.W.; Hachey, D.L.; Garcia-Lago, E.; Ludwig, D.S. Effects of dietary composition on energy expenditure during weight-loss maintenance. JAMA 2012, 307, 2627–2634. [Google Scholar] [CrossRef]
- Backhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [PubMed]
- Backhed, F.; Manchester, J.K.; Semenkovich, C.F.; Gordon, J.I. Mechanisms underlying the resistance to diet-induced obesity in germ-free mice. Proc. Natl. Acad. Sci. USA 2007, 104, 979–984. [Google Scholar] [CrossRef] [PubMed]
- Ley, R.E.; Backhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Turnbaugh, P.J.; Backhed, F.; Fulton, L.; Gordon, J.I. Diet-induced obesity is linked to marked but reversible alterations in the mouse distal gut microbiome. Cell Host Microbe 2008, 3, 213–223. [Google Scholar] [CrossRef]
- Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Cheng, J.; Duncan, A.E.; Kau, A.L.; Griffin, N.W.; Lombard, V.; Henrissat, B.; Bain, J.R.; et al. Gut Microbiota from Twins Discordant for Obesity Modulate Metabolism in Mice. Science 2013, 341, 1241214. [Google Scholar] [CrossRef]
- Vijay-Kumar, M.; Aitken, J.D.; Carvalho, F.A.; Cullender, T.C.; Mwangi, S.; Srinivasan, S.; Sitaraman, S.V.; Knight, R.; Ley, R.E.; Gewirtz, A.T. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 2010, 328, 228–231. [Google Scholar] [CrossRef]
- Henao-Mejia, J.; Elinav, E.; Jin, C.; Hao, L.; Mehal, W.Z.; Strowig, T.; Thaiss, C.A.; Kau, A.L.; Eisenbarth, S.C.; Jurczak, M.J.; et al. Inflammasome-mediated dysbiosis regulates progression of NAFLD and obesity. Nature 2012, 482, 179–185. [Google Scholar] [CrossRef] [PubMed]
- Thaiss, C.A.; Itav, S.; Rothschild, D.; Meijer, M.; Levy, M.; Moresi, C.; Dohnalova, L.; Braverman, S.; Rozin, S.; Malitsky, S.; et al. Persistent microbiome alterations modulate the rate of post-dieting weight regain. Nature 2016. [Google Scholar] [CrossRef] [PubMed]
- Albenberg, L.G.; Wu, G.D. Diet and the intestinal microbiome: Associations, functions, and implications for health and disease. Gastroenterology 2014, 146, 1564–1572. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.D.; Chen, J.; Hoffmann, C.; Bittinger, K.; Chen, Y.Y.; Keilbaugh, S.A.; Bewtra, M.; Knights, D.; Walters, W.A.; Knight, R.; et al. Linking long-term dietary patterns with gut microbial enterotypes. Science 2011, 334, 105–108. [Google Scholar] [CrossRef] [PubMed]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef] [PubMed]
- McAllan, L.; Skuse, P.; Cotter, P.D.; O’Connor, P.; Cryan, J.F.; Ross, R.P.; Fitzgerald, G.; Roche, H.M.; Nilaweera, K.N. Protein quality and the protein to carbohydrate ratio within a high fat diet influences energy balance and the gut microbiota in C57BL/6J mice. PLoS ONE 2014, 9, e88904. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Blouin, J.M.; Santacruz, A.; Lan, A.; Andriamihaja, M.; Wilkanowicz, S.; Benetti, P.H.; Tome, D.; Sanz, Y.; Blachier, F.; et al. High-protein diet modifies colonic microbiota and luminal environment but not colonocyte metabolism in the rat model: The increased luminal bulk connection. Am. J. Physiol. Gastrointest. Liver Physiol. 2014, 307, G459–G470. [Google Scholar] [CrossRef]
- Mu, C.; Yang, Y.; Luo, Z.; Guan, L.; Zhu, W. The Colonic Microbiome and Epithelial Transcriptome Are Altered in Rats Fed a High-Protein Diet Compared with a Normal-Protein Diet. J. Nutr. 2016, 146, 474–483. [Google Scholar] [CrossRef]
- Kiilerich, P.; Myrmel, L.S.; Fjaere, E.; Hao, Q.; Hugenholtz, F.; Sonne, S.B.; Derrien, M.; Pedersen, L.M.; Petersen, R.K.; Mortensen, A.; et al. Effect of a long-term high-protein diet on survival, obesity development, and gut microbiota in mice. Am. J. Physiol. Endocrinol. Metab. 2016, 310, E886–E899. [Google Scholar] [CrossRef]
- Wang, L.; Jacobs, J.P.; Lagishetty, V.; Yuan, P.Q.; Wu, S.V.; Million, M.; Reeve, J.R., Jr.; Pisegna, J.R.; Tache, Y. High-protein diet improves sensitivity to cholecystokinin and shifts the cecal microbiome without altering brain inflammation in diet-induced obesity in rats. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2017, 313, R473–R486. [Google Scholar] [CrossRef]
- Zhang, H.; DiBaise, J.K.; Zuccolo, A.; Kudrna, D.; Braidotti, M.; Yu, Y.; Parameswaran, P.; Crowell, M.D.; Wing, R.; Rittmann, B.E.; et al. Human gut microbiota in obesity and after gastric bypass. Proc. Natl. Acad. Sci. USA 2009, 106, 2365–2370. [Google Scholar] [CrossRef]
- Liou, A.P.; Paziuk, M.; Luevano, J.M., Jr.; Machineni, S.; Turnbaugh, P.J.; Kaplan, L.M. Conserved shifts in the gut microbiota due to gastric bypass reduce host weight and adiposity. Sci. Transl. Med. 2013, 5, 178ra141. [Google Scholar] [CrossRef]
- Everard, A.; Belzer, C.; Geurts, L.; Ouwerkerk, J.P.; Druart, C.; Bindels, L.B.; Guiot, Y.; Derrien, M.; Muccioli, G.G.; Delzenne, N.M.; et al. Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. USA 2013, 110, 9066–9071. [Google Scholar] [CrossRef] [PubMed]
- Duncan, S.H.; Belenguer, A.; Holtrop, G.; Johnstone, A.M.; Flint, H.J.; Lobley, G.E. Reduced dietary intake of carbohydrates by obese subjects results in decreased concentrations of butyrate and butyrate-producing bacteria in feces. Appl. Environ. Microbiol. 2007, 73, 1073–1078. [Google Scholar] [CrossRef]
- Beaumont, M.; Portune, K.J.; Steuer, N.; Lan, A.; Cerrudo, V.; Audebert, M.; Dumont, F.; Mancano, G.; Khodorova, N.; Andriamihaja, M.; et al. Quantity and source of dietary protein influence metabolite production by gut microbiota and rectal mucosa gene expression: A randomized, parallel, double-blind trial in overweight humans. Am. J. Clin. Nutr. 2017, 106, 1005–1019. [Google Scholar] [CrossRef]
- Grundy, S.M.; Cleeman, J.I.; Daniels, S.R.; Donato, K.A.; Eckel, R.H.; Franklin, B.A.; Gordon, D.J.; Krauss, R.M.; Savage, P.J.; Smith, S.C., Jr.; et al. Diagnosis and management of the metabolic syndrome: An American Heart Association/National Heart, Lung, and Blood Institute Scientific Statement. Circulation 2005, 112, 2735–2752. [Google Scholar] [CrossRef]
- Diet Health Questionnaire III. Available online: https://epi.grants.cancer.gov/dhq3/ (accessed on 20 October 2020).
- Subar, A.F.; Thompson, F.E.; Kipnis, V.; Midthune, D.; Hurwitz, P.; McNutt, S.; McIntosh, A.; Rosenfeld, S. Comparative validation of the Block, Willett, and National Cancer Institute food frequency questionnaires: The Eating at America’s Table Study. Am. J. Epidemiol. 2001, 154, 1089–1099. [Google Scholar] [CrossRef] [PubMed]
- Song, S.J.; Amir, A.; Metcalf, J.L.; Amato, K.R.; Xu, Z.Z.; Humphrey, G.; Knight, R. Preservation Methods Differ in Fecal Microbiome Stability, Affecting Suitability for Field Studies. mSystems 2016, 1. [Google Scholar] [CrossRef] [PubMed]
- Tong, M.; Jacobs, J.P.; McHardy, I.H.; Braun, J. Sampling of intestinal microbiota and targeted amplification of bacterial 16S rRNA genes for microbial ecologic analysis. Curr. Protoc. Immunol. 2014, 107, 7.41.1–7.41.11. [Google Scholar] [CrossRef]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Martino, C.; Morton, J.T.; Marotz, C.A.; Thompson, L.R.; Tripathi, A.; Knight, R.; Zengler, K. A Novel Sparse Compositional Technique Reveals Microbial Perturbations. MSystems 2019, 4. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yi, N. Fast zero-inflated negative binomial mixed modeling approach for analyzing longitudinal metagenomics data. Bioinformatics 2020, 36, 2345–2351. [Google Scholar] [CrossRef] [PubMed]
- Kaplan, R.C.; Wang, Z.; Usyk, M.; Sotres-Alvarez, D.; Daviglus, M.L.; Schneiderman, N.; Talavera, G.A.; Gellman, M.D.; Thyagarajan, B.; Moon, J.Y.; et al. Gut microbiome composition in the Hispanic Community Health Study/Study of Latinos is shaped by geographic relocation, environmental factors, and obesity. Genome Biol. 2019, 20, 219. [Google Scholar] [CrossRef]
- Gilbert, J.A.; Quinn, R.A.; Debelius, J.; Xu, Z.Z.; Morton, J.; Garg, N.; Jansson, J.K.; Dorrestein, P.C.; Knight, R. Microbiome-wide association studies link dynamic microbial consortia to disease. Nature 2016, 535, 94–103. [Google Scholar] [CrossRef]
- Sze, M.A.; Schloss, P.D. Erratum for Sze and Schloss, “Looking for a Signal in the Noise: Revisiting Obesity and the Microbiome”. mBio 2017, 8. [Google Scholar] [CrossRef]
- Le Chatelier, E.; Nielsen, T.; Qin, J.; Prifti, E.; Hildebrand, F.; Falony, G.; Almeida, M.; Arumugam, M.; Batto, J.M.; Kennedy, S.; et al. Richness of human gut microbiome correlates with metabolic markers. Nature 2013, 500, 541–546. [Google Scholar] [CrossRef]
- Cotillard, A.; Kennedy, S.P.; Kong, L.C.; Prifti, E.; Pons, N.; Le Chatelier, E.; Almeida, M.; Quinquis, B.; Levenez, F.; Galleron, N.; et al. Dietary intervention impact on gut microbial gene richness. Nature 2013, 500, 585–588. [Google Scholar] [CrossRef]
- Wallen, Z.D.; Appah, M.; Dean, M.N.; Sesler, C.L.; Factor, S.A.; Molho, E.; Zabetian, C.P.; Standaert, D.G.; Payami, H. Characterizing dysbiosis of gut microbiome in PD: Evidence for overabundance of opportunistic pathogens. NPJ Parkinson’s Dis. 2020, 6, 11. [Google Scholar] [CrossRef]
- Kelly, T.N.; Bazzano, L.A.; Ajami, N.J.; He, H.; Zhao, J.; Petrosino, J.F.; Correa, A.; He, J. Gut Microbiome Associates With Lifetime Cardiovascular Disease Risk Profile Among Bogalusa Heart Study Participants. Circ. Res. 2016, 119, 956–964. [Google Scholar] [CrossRef] [PubMed]
- Vasapolli, R.; Schutte, K.; Schulz, C.; Vital, M.; Schomburg, D.; Pieper, D.H.; Vilchez-Vargas, R.; Malfertheiner, P. Analysis of Transcriptionally Active Bacteria Throughout the Gastrointestinal Tract of Healthy Individuals. Gastroenterology 2019, 157, 1081–1092.e3. [Google Scholar] [CrossRef] [PubMed]
- Holmes, A.J.; Chew, Y.V.; Colakoglu, F.; Cliff, J.B.; Klaassens, E.; Read, M.N.; Solon-Biet, S.M.; McMahon, A.C.; Cogger, V.C.; Ruohonen, K.; et al. Diet-Microbiome Interactions in Health Are Controlled by Intestinal Nitrogen Source Constraints. Cell Metab. 2017, 25, 140–151. [Google Scholar] [CrossRef]
- Gratz, S.W.; Hazim, S.; Richardson, A.J.; Scobbie, L.; Johnstone, A.M.; Fyfe, C.; Holtrop, G.; Lobley, G.E.; Russell, W.R. Dietary carbohydrate rather than protein intake drives colonic microbial fermentation during weight loss. Eur. J. Nutr. 2019, 58, 1147–1158. [Google Scholar] [CrossRef]
- Prokopidis, K.; Cervo, M.M.; Gandham, A.; Scott, D. Impact of Protein Intake in Older Adults with Sarcopenia and Obesity: A Gut Microbiota Perspective. Nutrients 2020, 12, 2285. [Google Scholar] [CrossRef]
- Butteiger, D.N.; Hibberd, A.A.; McGraw, N.J.; Napawan, N.; Hall-Porter, J.M.; Krul, E.S. Soy Protein Compared with Milk Protein in a Western Diet Increases Gut Microbial Diversity and Reduces Serum Lipids in Golden Syrian Hamsters. J. Nutr. 2016, 146, 697–705. [Google Scholar] [CrossRef]
- Zou, H.; Wang, D.; Ren, H.; Cai, K.; Chen, P.; Fang, C.; Shi, Z.; Zhang, P.; Wang, J.; Yang, H.; et al. Effect of Caloric Restriction on BMI, Gut Microbiota, and Blood Amino Acid Levels in Non-Obese Adults. Nutrients 2020, 12, 631. [Google Scholar] [CrossRef]
- Ott, B.; Skurk, T.; Hastreiter, L.; Lagkouvardos, I.; Fischer, S.; Buttner, J.; Kellerer, T.; Clavel, T.; Rychlik, M.; Haller, D.; et al. Effect of caloric restriction on gut permeability, inflammation markers, and fecal microbiota in obese women. Sci. Rep. 2017, 7, 11955. [Google Scholar] [CrossRef]
- Cani, P.D.; Neyrinck, A.M.; Fava, F.; Knauf, C.; Burcelin, R.G.; Tuohy, K.M.; Gibson, G.R.; Delzenne, N.M. Selective increases of bifidobacteria in gut microflora improve high-fat-diet-induced diabetes in mice through a mechanism associated with endotoxaemia. Diabetologia 2007, 50, 2374–2383. [Google Scholar] [CrossRef] [PubMed]
- Amar, J.; Chabo, C.; Waget, A.; Klopp, P.; Vachoux, C.; Bermudez-Humaran, L.G.; Smirnova, N.; Berge, M.; Sulpice, T.; Lahtinen, S.; et al. Intestinal mucosal adherence and translocation of commensal bacteria at the early onset of type 2 diabetes: Molecular mechanisms and probiotic treatment. EMBO Mol. Med. 2011, 3, 559–572. [Google Scholar] [CrossRef]
- Heianza, Y.; Sun, D.; Ma, W.; Zheng, Y.; Champagne, C.M.; Bray, G.A.; Sacks, F.M.; Qi, L. Gut-microbiome-related LCT genotype and 2-year changes in body composition and fat distribution: The POUNDS Lost Trial. Int. J. Obes. 2018, 42, 1565–1573. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, H.K.; Gudmundsdottir, V.; Nielsen, H.B.; Hyotylainen, T.; Nielsen, T.; Jensen, B.A.; Forslund, K.; Hildebrand, F.; Prifti, E.; Falony, G.; et al. Human gut microbes impact host serum metabolome and insulin sensitivity. Nature 2016, 535, 376–381. [Google Scholar] [CrossRef]
- Dong, T.S.; Katzka, W.; Lagishetty, V.; Luu, K.; Hauer, M.; Pisegna, J.; Jacobs, J.P. A Microbial Signature Identifies Advanced Fibrosis in Patients with Chronic Liver Disease Mainly Due to NAFLD. Sci. Rep. 2020, 10, 2771. [Google Scholar] [CrossRef] [PubMed]



| NPD | HPD | |
|---|---|---|
| Meal Plans (% of Total Calories/Day) Including Supplement | ||
| Grains/Starch | 20 | 10 |
| Vegetables | 15 | 15 |
| Protein | 15 | 30 |
| Fruit | 20 | 15 |
| Fats and Oils | 30 | 30 |
| Supplement Characteristics per Serving | ||
| Calories | 100 | 100 |
| Total Fat | 0 g | 1 g |
| Total Carbohydrate | 22 g | 1 g |
| Protein | 0 g | 20 g |
| Sodium | 160 mg | 290 mg |
| Potassium | 45 mg | 35 mg |
| NPD (n = 37) | HPD (n = 43) | p-Value | |
|---|---|---|---|
| Gender (% Males) | 78.4 | 76.7 | 0.86 |
| Age (years) (SD) | 55.7 (11.4) | 55.9 (10.1) | 0.91 |
| BMI | 34.6 (5.1) | 34.9 (4.5) | 0.8 |
| Weight (kg) (SD) | 104.3 (18.1) | 104.7 (18.4) | 0.93 |
| Presence of DM (%) | 27.0 | 32.6 | 0.59 |
| Presence of Metabolic Syndrome (%) | 32.4 | 41.9 | 0.39 |
| Race (%) | |||
| White (n = 34) | 56.8 | 30.2 | 0.15 |
| African American (n = 26) | 27.0 | 37.2 | |
| Hispanic (n = 17) | 13.5 | 27.9 | |
| Asian (n = 2) | 2.7 | 2.3 | |
| Other (n = 1) | 0.0 | 2.3 | |
| Average Weight Change at Week 8 (kg) (SD) | −2.83 (2.25) | −3.46 (2.67) | 0.34 |
| Baseline Diet | |||
| Total Calories (kcal) (SD) | 2561.1 (1580.7) | 2003.7 (1629.9) | 0.13 |
| Protein (% of total kcal/day) (SD) | 16.4 (3.9) | 17.7 (4.3) | 0.16 |
| Fat (% of total kcal/day) (SD) | 37.3 (6.7) | 37.5 (6.9) | 0.93 |
| Carbohydrate (% of total kcal/day) (SD) | 47.4 (7.5) | 45.8 (8.9) | 0.39 |
| Fiber (g) (SD) | 24.7 (14.7) | 19.2 (17.2) | 0.14 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dong, T.S.; Luu, K.; Lagishetty, V.; Sedighian, F.; Woo, S.-L.; Dreskin, B.W.; Katzka, W.; Chang, C.; Zhou, Y.; Arias-Jayo, N.; et al. A High Protein Calorie Restriction Diet Alters the Gut Microbiome in Obesity. Nutrients 2020, 12, 3221. https://doi.org/10.3390/nu12103221
Dong TS, Luu K, Lagishetty V, Sedighian F, Woo S-L, Dreskin BW, Katzka W, Chang C, Zhou Y, Arias-Jayo N, et al. A High Protein Calorie Restriction Diet Alters the Gut Microbiome in Obesity. Nutrients. 2020; 12(10):3221. https://doi.org/10.3390/nu12103221
Chicago/Turabian StyleDong, Tien S., Kayti Luu, Venu Lagishetty, Farzaneh Sedighian, Shih-Lung Woo, Benjamin W. Dreskin, William Katzka, Candace Chang, Yi Zhou, Nerea Arias-Jayo, and et al. 2020. "A High Protein Calorie Restriction Diet Alters the Gut Microbiome in Obesity" Nutrients 12, no. 10: 3221. https://doi.org/10.3390/nu12103221
APA StyleDong, T. S., Luu, K., Lagishetty, V., Sedighian, F., Woo, S.-L., Dreskin, B. W., Katzka, W., Chang, C., Zhou, Y., Arias-Jayo, N., Yang, J., Ahdoot, A., Li, Z., Pisegna, J. R., & Jacobs, J. P. (2020). A High Protein Calorie Restriction Diet Alters the Gut Microbiome in Obesity. Nutrients, 12(10), 3221. https://doi.org/10.3390/nu12103221







