Comparative Genome Analysis of Bifidobacterium longum subsp. infantis Strains Reveals Variation in Human Milk Oligosaccharide Utilization Genes among Commercial Probiotics
Abstract
1. Introduction
2. Materials and Methods
2.1. Bacterial Strains and Culture Conditions
2.2. Whole-Genome Sequencing Assembly and Annotation
2.3. Genome-Wide Comparisons, Phylogenetic Analysis and HMO Gene Analysis
2.4. Growth on HMO Standards
2.5. Purification of HMOs from Human Milk and Bacterial Growth
2.6. Glycoprofiling
2.7. Strain-Specific Primer Development
2.8. In Vitro Competitive Assays
2.9. In Vivo Strain Competition Experiments
2.10. Statistical Analysis
2.11. Data Availability
3. Results
3.1. General Genome Features of B. infantis Strains
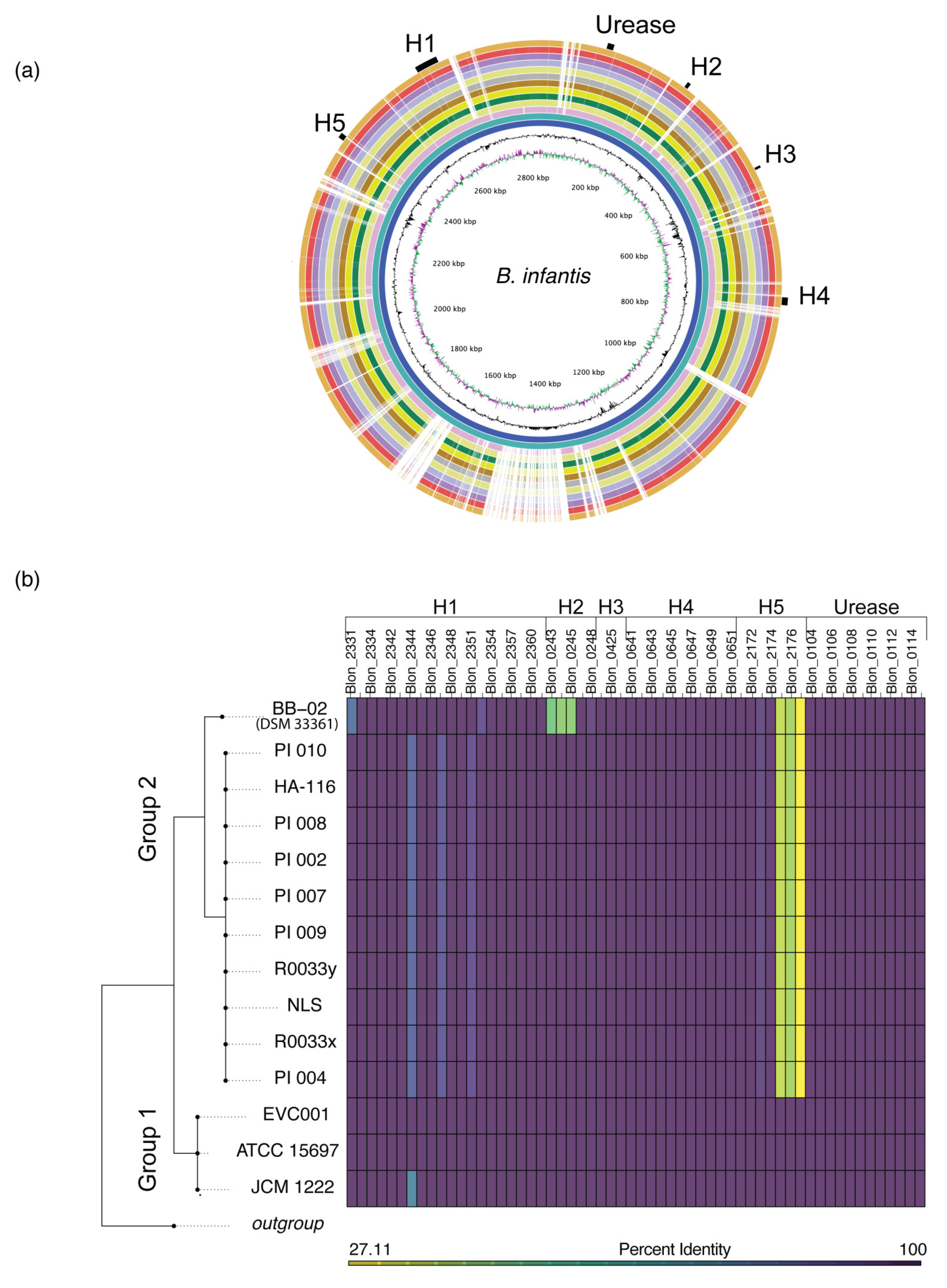
3.2. Comparative Genomic Analyses Revealed a High Degree of Genetic Conservation
3.3. Phylogenetic Grouping Corresponds with Divergence in HMO Utilization Genes
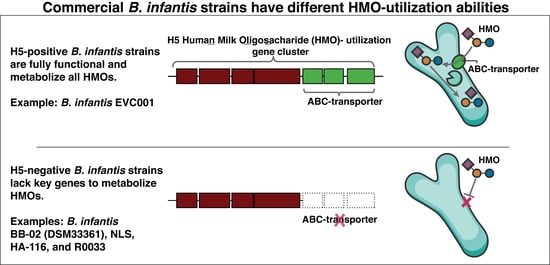
3.4. Divergence in the H5 Gene Cluster Is Associated with Poor Growth on HMOs
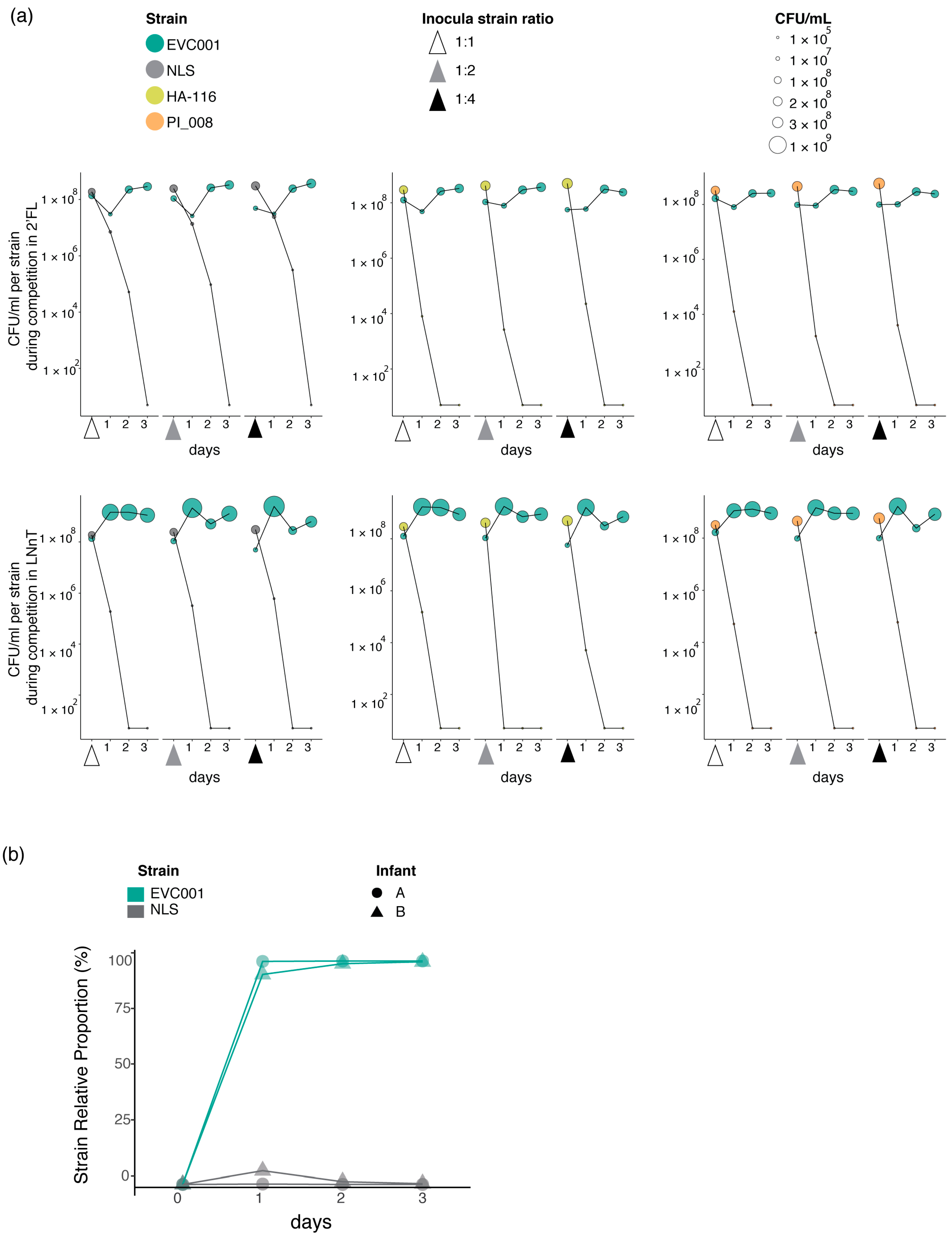
3.5. Competitive Assays
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Henderick, J.G.; Zwittink, R.D.; van Lingen, R.A.; Knol, J.; Belzer, C. The preterm gut microbiota: An inconspicuous challenge in nutritional neonatal care. Front. Cell. Infect. Microbiol. 2019, 9, 85. [Google Scholar] [CrossRef] [PubMed]
- Orivuori, L.; Mustonen, K.; De Goffau, M.; Hakala, S.; Paasela, M.; Roduit, C.; Dalphin, J.C.; Genuneit, J.; Lauener, R.; Riedler, J. High level of fecal calprotectin at age 2 months as a marker of intestinal inflammation predicts atopic dermatitis and asthma by age 6. Clin. Exp. Allergy 2015, 45, 928–939. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, M.C.; Stiemsma, L.T.; Dimitriu, P.A.; Thorson, L.; Russell, S.; Yurist-Doutsch, S.; Kuzeljevic, B.; Gold, M.J.; Britton, H.M.; Lefebvre, D.L.; et al. Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 2015, 7, 307ra152. [Google Scholar] [CrossRef] [PubMed]
- Denning, N.-L.; Prince, J.M. Neonatal intestinal dysbiosis in necrotizing enterocolitis. Mol. Med. 2018, 24, 1–10. [Google Scholar] [CrossRef]
- Underwood, M.A.; German, J.B.; Lebrilla, C.B.; Mills, D.A. Bifidobacterium longum subspecies infantis: Champion colonizer of the infant gut. Pediatr. Res. 2015, 77, 229. [Google Scholar] [CrossRef] [PubMed]
- Nijman, R.M.; Liu, Y.; Bunyatratchata, A.; Smilowitz, J.T.; Stahl, B.; Barile, D. Characterization and quantification of oligosaccharides in human milk and infant formula. J. Agric. Food Chem. 2018, 66, 6851–6859. [Google Scholar] [CrossRef]
- McGuire, M.K.; Meehan, C.L.; McGuire, M.A.; Williams, J.E.; Foster, J.; Sellen, D.W.; Kamau-Mbuthia, E.W.; Kamundia, E.W.; Mbugua, S.; Moore, S.E. What’s normal? Oligosaccharide concentrations and profiles in milk produced by healthy women vary geographically. Am. J. Clin. Nutr. 2017, 105, 1086–1100. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; Lebrilla, C.B.; Mills, D.A.; German, J.B.; Freeman, S.L. Breast milk oligosaccharides: Structure-function relationships in the neonate. Annu. Rev. Nutr. 2014, 34, 143–169. [Google Scholar] [CrossRef]
- Tao, N.; Wu, S.; Kim, J.; An, H.J.; Hinde, K.; Power, M.L.; Gagneux, P.; German, J.B.; Lebrilla, C.B. Evolutionary glycomics: Characterization of milk oligosaccharides in primates. J. Proteome Res. 2011, 10, 1548–1557. [Google Scholar] [CrossRef]
- Ninonuevo, M.R.; Park, Y.; Yin, H.; Zhang, J.; Ward, R.E.; Clowers, B.H.; German, J.B.; Freeman, S.L.; Killeen, K.; Grimm, R.; et al. A strategy for annotating the human milk glycome. J. Agric. Food Chem. 2006, 54, 7471–7480. [Google Scholar] [CrossRef]
- Urashima, T.; Asakuma, S.; Leo, F.; Fukuda, K.; Messer, M.; Oftedal, O.T. The Predominance of Type I Oligosaccharides Is a Feature Specific to Human Breast Milk. Adv. Nutr. 2012, 3, 473S–482S. [Google Scholar] [CrossRef]
- Sakanaka, M.; Gotoh, A.; Yoshida, K.; Odamaki, T.; Koguchi, H.; Xiao, J.-Z.; Kitaoka, M.; Katayama, T. Varied Pathways of Infant Gut-Associated Bifidobacterium to Assimilate Human Milk Oligosaccharides: Prevalence of the Gene Set and Its Correlation with Bifidobacteria-Rich Microbiota Formation. Nutrients 2020, 12, 71. [Google Scholar] [CrossRef] [PubMed]
- Garrido, D.; Barile, D.; Mills, D.A. A molecular basis for bifidobacterial enrichment in the infant gastrointestinal tract. Adv. Nutr. 2012, 3, 415S–421S. [Google Scholar] [CrossRef] [PubMed]
- Sela, D.A.; Chapman, J.; Adeuya, A.; Kim, J.H.; Chen, F.; Whitehead, T.R.; Lapidus, A.; Rokhsar, D.S.; Lebrilla, C.B.; German, J.B.; et al. The genome sequence of Bifidobacterium longum subsp. infantis reveals adaptations for milk utilization within the infant microbiome. Proc. Natl. Acad. Sci. USA 2008, 105, 18964–18969. [Google Scholar] [CrossRef] [PubMed]
- Albert, K.; Rani, A.; Sela, D.A. Comparative Pangenomics of the Mammalian Gut Commensal Bifidobacterium longum. Microorganisms 2020, 8, 7. [Google Scholar] [CrossRef] [PubMed]
- LoCascio, R.G.; Desai, P.; Sela, D.A.; Weimer, B.; Mills, D.A. Broad conservation of milk utilization genes in Bifidobacterium longum subsp. infantis as revealed by comparative genomic hybridization. Appl. Environ. Microbiol. 2010, 76, 7373–7381. [Google Scholar] [CrossRef] [PubMed]
- Asakuma, S.; Hatakeyama, E.; Urashima, T.; Yoshida, E.; Katayama, T.; Yamamoto, K.; Kumagai, H.; Ashida, H.; Hirose, J.; Kitaoka, M. Physiology of consumption of human milk oligosaccharides by infant gut-associated bifidobacteria. J. Biol. Chem. 2011, 286, 34583–34592. [Google Scholar] [CrossRef]
- Pokusaeva, K.; Fitzgerald, G.F.; van Sinderen, D. Carbohydrate metabolism in Bifidobacteria. Genes Nutr. 2011, 6, 285–306. [Google Scholar] [CrossRef]
- Thomson, P.; Medina, D.A.; Garrido, D. Human milk oligosaccharides and infant gut bifidobacteria: Molecular strategies for their utilization. Food Microbiol. 2018, 75, 37–46. [Google Scholar] [CrossRef]
- Fukuda, S.; Toh, H.; Hase, K.; Oshima, K.; Nakanishi, Y.; Yoshimura, K.; Tobe, T.; Clarke, J.M.; Topping, D.L.; Suzuki, T.; et al. Bifidobacteria can protect from enteropathogenic infection through production of acetate. Nature 2011, 469, 543–547. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Hutton, A.A.; Contreras, L.N.; Shaw, C.A.; Palumbo, M.C.; Casaburi, G.; Xu, G.; Davis, J.C.C.; Lebrilla, C.B.; Henrick, B.M.; et al. Persistence of Supplemented Bifidobacterium longum subsp. infantis EVC001 in Breastfed Infants. mSphere 2017, 2. [Google Scholar] [CrossRef] [PubMed]
- Henrick, B.M.; Hutton, A.A.; Palumbo, M.C.; Casaburi, G.; Mitchell, R.D.; Underwood, M.A.; Smilowitz, J.T.; Frese, S.A. Elevated Fecal pH Indicates a Profound Change in the Breastfed Infant Gut Microbiome Due to Reduction of Bifidobacterium over the Past Century. mSphere 2018, 3. [Google Scholar] [CrossRef] [PubMed]
- Duar, R.M.; Kyle, D.; Casaburi, G. Colonization Resistance in the Infant Gut: The Role of B. Infantis in Reducing pH and Preventing Pathogen Growth. High-Throughput 2020, 9, 7. [Google Scholar] [CrossRef] [PubMed]
- Lewis, Z.T.; Shani, G.; Masarweh, C.F.; Popovic, M.; Frese, S.A.; Sela, D.A.; Underwood, M.A.; Mills, D.A. Validating bifidobacterial species and subspecies identity in commercial probiotic products. Pediatr. Res. 2016, 79, 445–452. [Google Scholar] [CrossRef]
- Milani, C.; Lugli, G.A.; Turroni, F.; Mancabelli, L.; Duranti, S.; Viappiani, A.; Mangifesta, M.; Segata, N.; van Sinderen, D.; Ventura, M. Evaluation of bifidobacterial community composition in the human gut by means of a targeted amplicon sequencing (ITS) protocol. FEMS Microbiol. Ecol. 2014, 90, 493–503. [Google Scholar] [CrossRef]
- Lawley, B.; Munro, K.; Hughes, A.; Hodgkinson, A.J.; Prosser, C.G.; Lowry, D.; Zhou, S.J.; Makrides, M.; Gibson, R.A.; Lay, C.; et al. Differentiation of Bifidobacterium longum subspecies longum and infantis by quantitative PCR using functional gene targets. PeerJ 2017, 5, e3375. [Google Scholar] [CrossRef]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. J. Comput. Mol. Cell Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef]
- Antipov, D.; Korobeynikov, A.; McLean, J.S.; Pevzner, P.A. hybridSPAdes: An algorithm for hybrid assembly of short and long reads. Bioinformatics 2016, 32, 1009–1015. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid prokaryotic genome annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Alikhan, N.F.; Petty, N.K.; Ben Zakour, N.L.; Beatson, S.A. BLAST Ring Image Generator (BRIG): Simple prokaryote genome comparisons. BMC Genom. 2011, 12, 402. [Google Scholar] [CrossRef]
- Sullivan, M.J.; Petty, N.K.; Beatson, S.A. Easyfig: A genome comparison visualizer. Bioinformatics 2011, 27, 1009–1010. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Glover, R.H.; Humphris, S.; Elphinstone, J.G.; Toth, I.K. Genomics and taxonomy in diagnostics for food security: Soft-rotting enterobacterial plant pathogens. Anal. Methods 2016, 8, 12–24. [Google Scholar] [CrossRef]
- Page, A.J.; Cummins, C.A.; Hunt, M.; Wong, V.K.; Reuter, S.; Holden, M.T.; Fookes, M.; Falush, D.; Keane, J.A.; Parkhill, J. Roary: Rapid large-scale prokaryote pan genome analysis. Bioinformatics 2015, 31, 3691–3693. [Google Scholar] [CrossRef] [PubMed]
- Milani, C.; Duranti, S.; Lugli, G.A.; Bottacini, F.; Strati, F.; Arioli, S.; Foroni, E.; Turroni, F.; van Sinderen, D.; Ventura, M. Comparative genomics of Bifidobacterium animalis subsp. lactis reveals a strict monophyletic bifidobacterial taxon. Appl. Environ. Microbiol. 2013, 79, 4304–4315. [Google Scholar] [CrossRef]
- Eren, A.M.; Esen, Ö.C.; Quince, C.; Vineis, J.H.; Morrison, H.G.; Sogin, M.L.; Delmont, T.O. Anvi’o: An advanced analysis and visualization platform for ‘omics data. PeerJ 2015, 3, e1319. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.D. GToTree: A user-friendly workflow for phylogenomics. Bioinformatics 2019, 35, 4162–4164. [Google Scholar] [CrossRef]
- Edgar, R.C. MUSCLE: A multiple sequence alignment method with reduced time and space complexity. BMC Bioinform. 2004, 5, 113. [Google Scholar] [CrossRef]
- Price, M.N.; Dehal, P.S.; Arkin, A.P. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS ONE 2010, 5, e9490. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Sprouffske, K.; Wagner, A. Growthcurver: An R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 2016, 17, 172. [Google Scholar] [CrossRef]
- De Moura Bell, J.M.; Cohen, J.L.; de Aquino, L.F.; Lee, H.; de Melo Silva, V.L.; Liu, Y.; Domizio, P.; Barile, D. An integrated bioprocess to recover bovine milk oligosaccharides from colostrum whey permeate. J. Food Eng. 2018, 216, 27–35. [Google Scholar] [CrossRef]
- Bhattacharya, M.; Salcedo, J.; Robinson, R.C.; Henrick, B.M.; Barile, D. Peptidomic and glycomic profiling of commercial dairy products: Identification, quantification and potential bioactivities. NPJ Sci. Food 2019, 3, 1–13. [Google Scholar] [CrossRef]
- Lee, H.; de MeloSilva, V.L.; Liu, Y.; Barile, D. Short communication: Quantification of carbohydrates in whey permeate products using high-performance anion-exchange chromatography with pulsed amperometric detection. J. Dairy Sci. 2015, 98, 7644–7649. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Darling, A.E.; Mau, B.; Perna, N.T. progressiveMauve: Multiple genome alignment with gene gain, loss and rearrangement. PLoS ONE 2010, 5, e11147. [Google Scholar] [CrossRef] [PubMed]
- Arndt, D.; Grant, J.R.; Marcu, A.; Sajed, T.; Pon, A.; Liang, Y.; Wishart, D.S. PHASTER: A better, faster version of the PHAST phage search tool. Nucleic Acids Res. 2016, 44, W16–W21. [Google Scholar] [CrossRef] [PubMed]
- Locascio, R.G.; Ninonuevo, M.R.; Kronewitter, S.R.; Freeman, S.L.; German, J.B.; Lebrilla, C.B.; Mills, D.A. A versatile and scalable strategy for glycoprofiling bifidobacterial consumption of human milk oligosaccharides. Microb. Biotechnol. 2009, 2, 333–342. [Google Scholar] [CrossRef]
- Katayama, T. Host-derived glycans serve as selected nutrients for the gut microbe: Human milk oligosaccharides and bifidobacteria. Biosci. Biotechnol. Biochem. 2016, 80, 621–632. [Google Scholar] [CrossRef]
- Sakanaka, M.; Hansen, M.E.; Gotoh, A.; Katoh, T.; Yoshida, K.; Odamaki, T.; Yachi, H.; Sugiyama, Y.; Kurihara, S.; Hirose, J. Evolutionary adaptation in fucosyllactose uptake systems supports bifidobacteria-infant symbiosis. Sci. Adv. 2019, 5, eaaw7696. [Google Scholar] [CrossRef]
- Garrido, D.; Kim, J.H.; German, J.B.; Raybould, H.E.; Mills, D.A. Oligosaccharide binding proteins from Bifidobacterium longum subsp. infantis reveal a preference for host glycans. PLoS ONE 2011, 6, e17315. [Google Scholar] [CrossRef]
- Ozcan, E.; Sela, D.A. Inefficient Metabolism of the Human Milk Oligosaccharides Lacto-N-tetraose and Lacto-N-neotetraose Shifts Bifidobacterium longum subsp. infantis Physiology. Front. Nutr. 2018, 5, 46. [Google Scholar] [CrossRef] [PubMed]
- Ochman, H.; Moran, N.A. Genes Lost and Genes Found: Evolution of Bacterial Pathogenesis and Symbiosis. Science 2001, 292, 1096–1099. [Google Scholar] [CrossRef] [PubMed]
- Remigi, P.; Zhu, J.; Young, J.P.W.; Masson-Boivin, C. Symbiosis within Symbiosis: Evolving Nitrogen-Fixing Legume Symbionts. Trends Microbiol. 2016, 24, 63–75. [Google Scholar] [CrossRef] [PubMed]
- Mandel, M.J.; Wollenberg, M.S.; Stabb, E.V.; Visick, K.L.; Ruby, E.G. A single regulatory gene is sufficient to alter bacterial host range. Nature 2009, 458, 215–218. [Google Scholar] [CrossRef] [PubMed]
- Frese, S.A.; Benson, A.K.; Tannock, G.W.; Loach, D.M.; Kim, J.; Zhang, M.; Oh, P.L.; Heng, N.C.; Patil, P.B.; Juge, N.; et al. The evolution of host specialization in the vertebrate gut symbiont Lactobacillus reuteri. PLoS Genet. 2011, 7, e1001314. [Google Scholar] [CrossRef]
- Douglas, G.L.; Klaenhammer, T.R. Genomic evolution of domesticated microorganisms. Annu. Rev. Food Sci. Technol. 2010, 1, 397–414. [Google Scholar] [CrossRef]
- Young, S.L.; Simon, M.A.; Baird, M.A.; Tannock, G.W.; Bibiloni, R.; Spencely, K.; Lane, J.M.; Fitzharris, P.; Crane, J.; Town, I. Bifidobacterial species differentially affect expression of cell surface markers and cytokines of dendritic cells harvested from cord blood. Clin. Diagn. Lab. Immunol. 2004, 11, 686–690. [Google Scholar] [CrossRef]
- Lewis, Z.T.; Totten, S.M.; Smilowitz, J.T.; Popovic, M.; Parker, E.; Lemay, D.G.; Van Tassell, M.L.; Miller, M.J.; Jin, Y.S.; German, J.B.; et al. Maternal fucosyltransferase 2 status affects the gut bifidobacterial communities of breastfed infants. Microbiome 2015, 3, 13. [Google Scholar] [CrossRef]
- Matsuki, T.; Yahagi, K.; Mori, H.; Matsumoto, H.; Hara, T.; Tajima, S.; Ogawa, E.; Kodama, H.; Yamamoto, K.; Yamada, T.; et al. A key genetic factor for fucosyllactose utilization affects infant gut microbiota development. Nat. Commun. 2016, 7, 11939. [Google Scholar] [CrossRef]
- Bunesova, V.; Lacroix, C.; Schwab, C. Fucosyllactose and L-fucose utilization of infant Bifidobacterium longum and Bifidobacterium kashiwanohense. BMC Microbiol. 2016, 16, 248. [Google Scholar] [CrossRef]
- Cheng, C.C.; Duar, R.M.; Lin, X.; Perez-Munoz, M.E.; Tollenaar, S.; Oh, J.-H.; van Pijkeren, J.-P.; Li, F.; van Sinderen, D.; Gänzle, M.G. Ecological importance of cross-feeding of the intermediate metabolite 1, 2-propanediol between bacterial gut symbionts. Appl. Environ. Microbiol. 2020, 86. [Google Scholar] [CrossRef] [PubMed]
- Duar, R.M.; Henrick, B.M.; Casaburi, G.; Frese, S.A. Integrating the ecosystem services framework to define dysbiosis of the breastfed infant gut: The role of B. infantis and human milk oligosaccharides. Front. Nutr. 2020, 7, 33. [Google Scholar] [CrossRef] [PubMed]
- Casaburi, G.; Duar, R.M.; Vance, D.P.; Mitchell, R.; Contreras, L.; Frese, S.A.; Smilowitz, J.T.; Underwood, M.A. Early-life gut microbiome modulation reduces the abundance of antibiotic-resistant bacteria. Antimicrob. Resist. Infect. Control 2019, 8, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Henrick, B.M.; Chew, S.; Casaburi, G.; Brown, H.K.; Frese, S.A.; Zhou, Y.; Underwood, M.A.; Smilowitz, J.T. Colonization by B. infantis EVC001 modulates enteric inflammation in exclusively breastfed infants. Pediatr. Res. 2019. [Google Scholar] [CrossRef]
- Casaburi, G.; Frese, S.A. Colonization of breastfed infants by Bifidobacterium longum subsp. infantis EVC001 reduces virulence gene abundance. Hum. Microbiome J. 2018, 9, 7–10. [Google Scholar] [CrossRef]
- Smilowitz, J.T.; Moya, J.; Breck, M.A.; Cook, C.; Fineberg, A.; Angkustsiri, K.; Underwood, M.A. Safety and tolerability of Bifidobacterium longum subspecies infantis EVC001 supplementation in healthy term breastfed infants: A phase I clinical trial. BMC Pediatr. 2017, 17, 133. [Google Scholar]



| Group | Strain | Source Reference/Source | NCBI Accession Number | Size (Kbp) | Coverage (x) | GC (%) | CDSs | tRNAs |
|---|---|---|---|---|---|---|---|---|
| 1 | ATCC 15697 | Sela (2008) sela [14] | NC_011593 | 2832 | n/a | 59.8 | 2547 | 84 |
| 1 | JCM 1222 | Fukuda (2011) Fukuda [20] | NC_017219 | 2828 | n/a | 59.8 | 2544 | 84 |
| 1 | EVC001 | probiotic product | NZ_LR655210 | 2832 | 147 | 59.8 | 2553 | 84 |
| 2 | NLS | probiotic product | CP054528 | 2598 | 332 | 59.3 | 2226 | 59 |
| 2 | BB-02 (DSM33361) | probiotic product | CP054527 | 2758 | 486 | 59.8 | 2437 | 60 |
| 2 | PI_002 | probiotic product | CP054526 | 2604 | 185 | 59.3 | 2227 | 59 |
| 2 | HA-116 | probiotic product | CP054525 | 2612 | 88 | 59.2 | 2232 | 59 |
| 2 | PI_004 | probiotic product | CP054524 | 2604 | 172 | 59.3 | 2226 | 59 |
| 2 | R0033x | probiotic product “x” | CP054523 | 2614 | 130 | 59.2 | 2233 | 59 |
| 2 | R0033y | probiotic product “y” | CP054596 | 2615 | 171 | 59.2 | 2234 | 59 |
| 2 | PI_007 | probiotic product | CP054522 | 2604 | 220 | 59.3 | 2228 | 59 |
| 2 | PI_008 | probiotic product | CP054521 | 2612 | 213 | 59.2 | 2230 | 59 |
| 2 | PI_009 | probiotic product | CP054520 | 2604 | 162 | 59.3 | 2230 | 59 |
| 2 | PI_010 | probiotic product | CP054519 | 2609 | 117 | 59.2 | 2231 | 59 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Duar, R.M.; Casaburi, G.; Mitchell, R.D.; Scofield, L.N.C.; Ortega Ramirez, C.A.; Barile, D.; Henrick, B.M.; Frese, S.A. Comparative Genome Analysis of Bifidobacterium longum subsp. infantis Strains Reveals Variation in Human Milk Oligosaccharide Utilization Genes among Commercial Probiotics. Nutrients 2020, 12, 3247. https://doi.org/10.3390/nu12113247
Duar RM, Casaburi G, Mitchell RD, Scofield LNC, Ortega Ramirez CA, Barile D, Henrick BM, Frese SA. Comparative Genome Analysis of Bifidobacterium longum subsp. infantis Strains Reveals Variation in Human Milk Oligosaccharide Utilization Genes among Commercial Probiotics. Nutrients. 2020; 12(11):3247. https://doi.org/10.3390/nu12113247
Chicago/Turabian StyleDuar, Rebbeca M., Giorgio Casaburi, Ryan D. Mitchell, Lindsey N.C. Scofield, Camila A. Ortega Ramirez, Daniela Barile, Bethany M. Henrick, and Steven A. Frese. 2020. "Comparative Genome Analysis of Bifidobacterium longum subsp. infantis Strains Reveals Variation in Human Milk Oligosaccharide Utilization Genes among Commercial Probiotics" Nutrients 12, no. 11: 3247. https://doi.org/10.3390/nu12113247
APA StyleDuar, R. M., Casaburi, G., Mitchell, R. D., Scofield, L. N. C., Ortega Ramirez, C. A., Barile, D., Henrick, B. M., & Frese, S. A. (2020). Comparative Genome Analysis of Bifidobacterium longum subsp. infantis Strains Reveals Variation in Human Milk Oligosaccharide Utilization Genes among Commercial Probiotics. Nutrients, 12(11), 3247. https://doi.org/10.3390/nu12113247