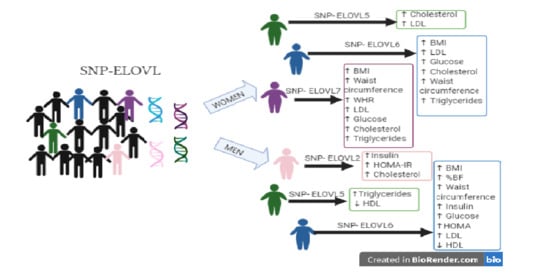
Influence of Single Nucleotide Polymorphisms of ELOVL on Biomarkers of Metabolic Alterations in the Mexican Population
Abstract
1. Introduction
2. Materials and Methods
2.1. Evaluation of Nutritional Status and Body Composition
2.2. Evaluation of Biochemical Markers
2.3. DNA Purification and Genotyping
2.4. Genetic and Statistical Analyses
3. Results
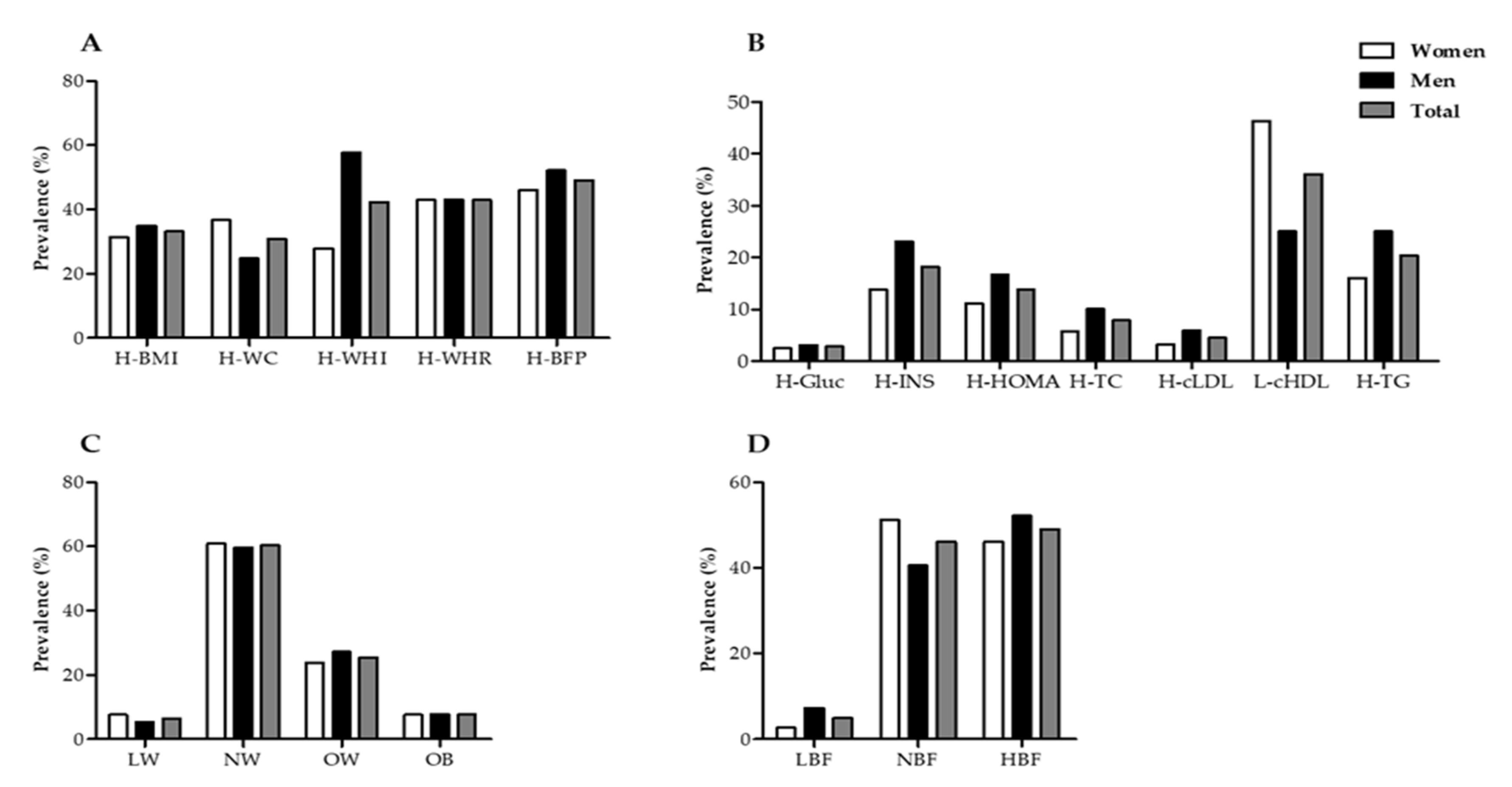
3.1. Characteristics of the Subjects
3.2. Nutritional Alterations of Subjects
3.3. Allelic Frequencies, HWE Analysis, and Private Alleles
3.4. Association of SNPs with Clinical Markers of Risk to Chronic Non-Communicable Diseases
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Green, C.D.; Ozguden-Akkoc, C.G.; Wang, Y.; Jump, D.B.; Olson, L.K. Role of fatty acid elongases in determination of de novo synthesized monounsaturated fatty acid species. J. Lipid Res. 2010, 51, 1871–1877. [Google Scholar] [CrossRef] [PubMed]
- Jakobsson, A.; Westerberg, R.; Jacobsson, A. Fatty acid elongases in mammals: Their regulation and roles in metabolism. Prog. Lipid Res. 2006, 45, 237–249. [Google Scholar] [CrossRef] [PubMed]
- Cruciani-Guglielmacci, C.; Bellini, L.; Denom, J.; Oshima, M.; Fernandez, N.; Normandie-Levi, P.; Berney, X.P.; Kassis, N.; Rouch, C.; Dairou, J.; et al. Molecular phenotyping of multiple mouse strains under metabolic challenge uncovers a role for Elovl2 in glucose-induced insulin secretion. Mol. Metab. 2017, 6, 340–351. [Google Scholar] [CrossRef] [PubMed]
- Zadravec, D.; Brolinson, A.; Fisher, R.M.; Carneheim, C.; Csikasz, R.I.; Bertrand-Michel, J.; Borén, J.; Guillou, H.; Rudling, M.; Jacobsson, A. Ablation of the very-long-chain fatty acid elongase ELOVL3 in mice leads to constrained lipid storage and resistance to diet-induced obesity. FASEB J. 2010, 24, 4366–4377. [Google Scholar]
- Conn, J.R.; Catchpoole, E.M.; Runnegar, N.; Mapp, S.J.; Markey, K.A. Low rates of antibiotic resistance and infectious mortality in a cohort of high-risk hematology patients: A single center, retrospective analysis of blood stream infection. PLoS ONE 2017, 12, e0178059. [Google Scholar] [CrossRef]
- Vasireddy, V.; Sharon, M.; Salem, N.; Ayyagari, R. Role of ELOVL4 in Fatty Acid Metabolism. In Advances in Experimental Medicine and Biology; Anderson, R.E., LaVail, M.M., Hollyfield, J.G., Eds.; Springer: New York, NY, USA, 2008; Volume 613, pp. 283–290. ISBN 9780387749020. [Google Scholar]
- Tripathy, S.; Jump, D.B. Elovl5 regulates the mTORC2-Akt-FOXO1 pathway by controlling hepatic cis -vaccenic acid synthesis in diet-induced obese mice. J. Lipid Res. 2013, 54, 71–84. [Google Scholar] [CrossRef]
- Kumadaki, S.; Matsuzaka, T.; Kato, T.; Yahagi, N.; Yamamoto, T.; Okada, S.; Kobayashi, K.; Takahashi, A.; Yatoh, S.; Suzuki, H.; et al. Mouse Elovl-6 promoter is an SREBP target. Biochem. Biophys. Res. Commun. 2008, 368, 261–266. [Google Scholar] [CrossRef]
- Matsuzaka, T.; Shimano, H. Elovl6: A new player in fatty acid metabolism and insulin sensitivity. J. Mol. Med. 2009, 87, 379–384. [Google Scholar] [CrossRef] [PubMed]
- Li, G.; Cui, S.; Du, J.; Liu, J.; Zhang, P.; Fu, Y.; He, Y.; Zhou, H.; Ma, J.; Chen, S. Association of GALC, ZNF184, IL1R2 and ELOVL7 With Parkinson’s Disease in Southern Chinese. Front. Aging Neurosci. 2018, 10, 1–6. [Google Scholar] [CrossRef]
- Morales, E.; Bustamante, M.; Gonzalez, J.R.; Guxens, M.; Torrent, M.; Mendez, M.; Garcia-Esteban, R.; Julvez, J.; Forns, J.; Vrijheid, M.; et al. Genetic variants of the FADS gene cluster and ELOVL gene family, colostrums LC-PUFA levels, breastfeeding, and child cognition. PLoS ONE 2011, 6, e17181. [Google Scholar] [CrossRef] [PubMed]
- Aslibekyan, S.; Jensen, M.K.; Campos, H.; Linkletter, C.D.; Loucks, E.B.; Ordovas, J.M.; Deka, R.; Rimm, E.B.; Baylin, A. Genetic variation in fatty acid elongases is not associated with intermediate cardiovascular phenotypes or myocardial infarction. Eur. J. Clin. Nutr. 2012, 66, 353–359. [Google Scholar] [CrossRef]
- World Medical Association. World Medical Association Declaration of Helsinki. Ethical principles for medical research involving human subjects. Bull. World Health Organ. 2001, 79, 373. [Google Scholar]
- WHO. WHO World Health Organization. Available online: http://www.scopus.com/inward/record.url?eid=2-s2.0-84894477459&partnerID=tZOtx3y1 (accessed on 25 March 2020).
- Parra, I.; Jonguitud, V. La fórmula de Friedewald no debe ser utilizada para el cálculo de colesterol de baja densidad en pacientes con triglicéridos elevados. Rev Mex Patol Clin 2007, 54, 112–115. [Google Scholar]
- Alberti, K.G.M.M.; Eckel, R.H.; Grundy, S.M.; Zimmet, P.Z.; Cleeman, J.I.; Donato, K.A.; Fruchart, J.-C.; James, W.P.T.; Loria, C.M.; Smith, S.C. Harmonizing the Metabolic Syndrome. Circulation 2009, 120, 1640–1645. [Google Scholar] [CrossRef]
- Roccella, E.J. Update on the 1987 Task Force Report on High Blood Pressure in Children and Adolescents: A working group report from the National High Blood Pressure Education Program. National High Blood Pressure Education Program Working Group on Hypertension Control i. Pediatrics 1996, 98, 649–658. [Google Scholar]
- Murguía-Romero, M.; Jiménez-Flores, J.R.; Sigrist-Flores, S.C.; Espinoza-Camacho, M.A.; Jiménez-Morales, M.; Piña, E.; Méndez-Cruz, A.R.; Villalobos-Molina, R.; Reaven, G.M. Plasma triglyceride/HDL-cholesterol ratio, insulin resistance, and cardiometabolic risk in young adults. J. Lipid Res. 2013, 54, 2795–2799. [Google Scholar] [CrossRef]
- Ilumina Inc Infinium HTS Assay: Reference Guide; Ilumina Inc.: San Diego, CA, USA, 2019; Volume 2, pp. 1–83. ISBN 1000000074604.
- IBM Corp IBM SPSS Statistics for Windows, Armonk, NY, USA. 2015. Available online: https://www.ibm.com/support/home/ (accessed on 1 April 2020).
- Gallagher, D.; Heymsfield, S.B.; Heo, M.; Jebb, S.A.; Murgatroyd, P.R.; Sakamoto, Y. Healthy percentage body fat ranges: An approach for developing guidelines based on body mass index. Am. J. Clin. Nutr. 2000, 72, 694–701. [Google Scholar] [CrossRef]
- Shamah, T.; Cuevas, L.; Gaona, E.B.; Gómez, L.M.; Morales, M.C.; Hernández, M.; Rivera, J.Á. Sobrepeso y obesidad en niños y adolescentes en México, actualización de la Encuesta Nacional de Salud y Nutrición de Medio Camino 2016. Salud Publica Mex. 2018, 60, 244–253. [Google Scholar] [CrossRef]
- Shamah-Levy, T.; Romero-Martínez, M.; Cuevas-Nasu, L.; Gómez-Humaran, I.M.; Avila-Arcos, M.A.; Rivera-Dommarco, J.A. The Mexican national health and nutrition survey as a basis for public policy planning: Overweight and obesity. Nutrients 2019, 11, 1727. [Google Scholar] [CrossRef]
- González Sandoval, C.E.; Díaz Burke, Y.; Mendizabal-Ruiz, A.P.; Medina Díaz, E.; Alejandro Morales, J. Prevalencia de obesidad y perfil lipídico alterado en jóvenes universitarios. Nutr. Hosp. 2014, 29, 315–321. [Google Scholar]
- Secretaría de Salud (SSA) Sobrepeso y Obesidad, Factores de Riesgos Para Desarrollar Diabetes|Secretaría de Salud|Gobierno|Gob.mx. Available online: https://www.gob.mx/salud/articulos/sobrepeso-y-obesidad-factores-de-riesgos-para-desarrollar-diabetes (accessed on 17 April 2020).
- González Jiménez, E.; Aguilar Cordero, M.J.; Padilla López, C.A.; García García, I. Obesidad monogénica humana: Papel del sistema leptina-melanocortina en la regulación de la ingesta de alimentos y el peso corporal en humanos. An. Sist. Sanit. Navar. 2012, 35, 285–293. [Google Scholar] [CrossRef]
- Zhou, Y.-T.; Grayburn, P.; Karim, A.; Shimabukuro, M.; Higa, M.; Baetens, D.; Orci, L.; Unger, R.H. Lipotoxic heart disease in obese rats: Implications for human obesity. Proc. Natl. Acad. Sci. USA 2000, 97, 1784–1789. [Google Scholar] [CrossRef]
- Izquierdo, A.; Medina-Gómez, G. Papel de la lipotoxicidad en el desarrollo de la lesión renal en el síndrome metabólico y el envejecimiento. Diálisis y Traspl. 2012, 33, 89–96. [Google Scholar] [CrossRef]
- Cazanave, S.C.; Sanyal, A.J. Molecular mechanisms of lipotoxicity in nonalcoholic fatty liver disease. Hepatic Novo Lipogenes. Regul. Metab. 2015, 65, 101–129. [Google Scholar]
- Murguia-Romero, M.; Jimenez-Flores, J.R.; Sigrist-Flores, S.C.; Tapia-Pancardo, D.C.; Jimenez-Ramos, A.; Mendez-Cruz, A.R.; Villalobos-Molina, R. Prevalence of Metabolic Syndrome in Young Mexicans: A Sensitivity Analysis on Its Components. Nutr. Hosp. 2015, 32, 189–195. [Google Scholar]
- Sugawara, E.; Nikaido, H. Properties of AdeABC and AdeIJK Efflux Systems of Acinetobacter baumannii Compared with Those of the AcrAB-TolC System of Escherichia coli. Antimicrob. Agents Chemother. 2014, 58, 7250–7257. [Google Scholar] [CrossRef] [PubMed]
- Campos-Nonato, I.; Hernández-Barrera, L.; Rojas-Martínez, R. Hipertensión arterial: Prevalencia, diagnóstico oportuno, control y tendencias en adultos mexicanos. Salud Publica Mex. 2013, 55, 144. [Google Scholar] [CrossRef]
- Christine, A. Andrews Natural Selection, Genetic Drift, and Gene Flow Do Not Act in Isolation in Natural Populations. Nat. Educ. Knowl. 2010, 3, 5. [Google Scholar]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research--an update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Hwang, J.-Y.; Lee, H.J.; Go, M.J.; Jang, H.B.; Choi, N.-H.; Bae, J.B.; Castillo-Fernandez, J.E.; Bell, J.T.; Spector, T.D.; Lee, H.-J.; et al. Genome-wide methylation analysis identifies ELOVL5 as an epigenetic biomarker for the risk of type 2 diabetes mellitus. Sci. Rep. 2018, 8, 14862. [Google Scholar] [CrossRef]
- Morcillo, S.; Martín-Núñez, G.M.; Rojo-Martínez, G.; Almaraz, M.C.; García-Escobar, E.; Mansego, M.L.; de Marco, G.; Chaves, F.J.; Soriguer, F. ELOVL6 Genetic Variation Is Related to Insulin Sensitivity: A New Candidate Gene in Energy Metabolism. PLoS ONE 2011, 6, e21198. [Google Scholar] [CrossRef] [PubMed]
- Matsuzaka, T.; Shimano, H.; Yahagi, N.; Kato, T.; Atsumi, A.; Yamamoto, T.; Inoue, N.; Ishikawa, M.; Okada, S.; Ishigaki, N.; et al. Crucial role of a long-chain fatty acid elongase, Elovl6, in obesity-induced insulin resistance. Nat. Med. 2007, 13, 1193–1202. [Google Scholar] [CrossRef]
- Tamura, K.; Makino, A.; Hullin-Matsuda, F.; Kobayashi, T.; Furihata, M.; Chung, S.; Ashida, S.; Miki, T.; Fujioka, T.; Shuin, T.; et al. Novel Lipogenic Enzyme ELOVL7 Is Involved in Prostate Cancer Growth through Saturated Long-Chain Fatty Acid Metabolism. Cancer Res. 2009, 69, 8133–8140. [Google Scholar] [CrossRef]
- Mansego, M.; Milagro, F.; Zulet, M.; Moreno-Aliaga, M.; Martínez, J. Differential DNA Methylation in Relation to Age and Health Risks of Obesity. Int. J. Mol. Sci. 2015, 16, 16816–16832. [Google Scholar] [CrossRef]
- Bellini, L.; Campana, M.; Rouch, C.; Chacinska, M.; Bugliani, M.; Meneyrol, K.; Hainault, I.; Lenoir, V.; Denom, J.; Véret, J.; et al. Protective role of the ELOVL2/docosahexaenoic acid axis in glucolipotoxicity-induced apoptosis in rodent beta cells and human islets. Diabetologia 2018, 61, 1780–1793. [Google Scholar] [CrossRef]
- Khamlaoui, W.; Mehri, S.; Hammami, S.; Hammouda, S.; Chraeif, I.; Elosua, R.; Hammami, M. Association Between Genetic Variants in FADS1-FADS2 and ELOVL2 and Obesity, Lipid Traits, and Fatty Acids in Tunisian Population. Clin. Appl. Thromb. 2020, 26, 107602962091528. [Google Scholar] [CrossRef]
- Maguolo, A.; Zusi, C.; Giontella, A.; Miraglia Del Giudice, E.; Tagetti, A.; Fava, C.; Morandi, A.; Maffeis, C. Influence of genetic variants in FADS2 and ELOVL2 genes on BMI and PUFAs homeostasis in children and adolescents with obesity. Int. J. Obes. 2020, 613, 283–290. [Google Scholar] [CrossRef]
| Gene | Genetic Variant | Alleles | Functional Consequence |
|---|---|---|---|
| ELOVL2 | rs8523 | (A/G) | genic downstream transcript variant, 3′ UTR variant. |
| ELOVL2 | rs3734396 | (A/G) | genic downstream transcript variant, 3′ UTR variant. |
| ELOVL2 | rs17606561 | (A/G) | genic downstream transcript variant, 3′ UTR variant. |
| ELOVL2 | rs3734398 | (T/C) | genic downstream transcript variant, 3′ UTR variant. |
| ELOVL2 | rs2281591 | (A/G) | intron variant, genic downstream transcript variant. |
| ELOVL2 | rs2236212 | (G/C) | intron variant, genic downstream transcript variant. |
| ELOVL2 | rs3798713 | (G/C) | intron variant. |
| ELOVL2 | rs7765206 | (A/C) | intron variant, genic upstream transcript variant. |
| ELOVL2 | rs116279801 | (T/C) | intron variant, genic upstream transcript variant. |
| ELOVL2 | rs9295757 | (T/G) | intron variant, genic upstream transcript variant. |
| ELOVL2 | rs3798721 | (A/C) | intron variant, genic upstream transcript variant. |
| ELOVL2 | rs16870899 | (A/G) | intron variant, genic upstream transcript variant. |
| ELOVL2 | rs3798722 | (A/G) | intron variant, genic upstream transcript variant. |
| ELOVL2 | rs9393903 | (A/G) | intron variant, upstream transcript variant, genic upstream transcript variant. |
| ELOVL2 | rs4532436 | (G/C) | genic downstream transcript variant, 3′ UTR variant. |
| ELOVL2 | rs12195587 | (A/G) | downstream gene transcription variant, synonym variant, coding sequence variant. |
| ELOVL3 | rs10748816 | (A/G) | intron variant. |
| ELOVL3 | rs36103207 | (A/G) | nonsense variant, coding sequence variant. |
| ELOVL4 | rs3812153 | (T/C) | coding sequence variant, nonsense variant. |
| ELOVL4 | 6:80628844 | (A/G) | |
| ELOVL4 | rs117891930 | (T/C) | intron variant. |
| ELOVL4 | rs144198896 | (A/G) | intron variant. |
| ELOVL4 | rs80246554 | (T/C) | intron variant. |
| ELOVL4 | rs12196014 | (A/G) | intron variant. |
| ELOVL4 | rs9448863 | (A/G) | intron variant. |
| ELOVL4 | rs16891339 | (A/G) | intron variant. |
| ELOVL5 | rs41273878 | (A/C) | coding sequence variant, synonym variant, downstream gene transcription variant. |
| ELOVL5 | rs41273880 | (T/C) | coding sequence variant, nonsense variant, synonym variant, downstream gene transcription variant. |
| ELOVL5 | rs72938776 | (A/G) | intron variant, genic downstream transcript variant. |
| ELOVL5 | rs182937551 | (T/C) | downstream gene transcription variant, intron variant |
| ELOVL5 | rs36054518 | (A/G) | intron variant. |
| ELOVL5 | rs209487 | (T/G) | intron variant. |
| ELOVL5 | rs115397424 | (A/G) | intron variant. |
| ELOVL5 | rs13208390 | (T/G) | intron variant. |
| ELOVL5 | rs72940713 | (T/C) | intron variant. |
| ELOVL5 | rs114271869 | (T/G) | intron variant. |
| ELOVL5 | rs2073040 | (A/G) | intron variant, genic downstream transcript variant. |
| ELOVL5 | rs9370194 | (C/T) | intron variant. |
| ELOVL6 | rs11098065 | (A/G) | intron variant. |
| ELOVL6 | rs17041284 | (T/C) | intron variant. |
| ELOVL6 | rs7662161 | (T/C) | intron variant. |
| ELOVL6 | rs77958351 | (A/G) | intron variant. |
| ELOVL6 | rs77808755 | (A/G) | intron variant. |
| ELOVL6 | rs59634436 | (A/C) | intron variant. |
| ELOVL6 | rs78160528 | (T/C) | intron variant. |
| ELOVL6 | rs16997129 | (T/C) | intron variant. |
| ELOVL6 | rs3813827 | (A/G) | intron variant. |
| ELOVL6 | rs11737840 | (T/C) | intron variant. |
| ELOVL6 | rs10033691 | (T/C) | intron variant. |
| ELOVL6 | rs2005701 | (T/C) | intron variant. |
| ELOVL6 | rs76145164 | (T/C) | intron variant. |
| ELOVL6 | rs76338299 | (T/G) | intron variant. |
| ELOVL6 | rs6533491 | (T/C) | intron variant. |
| ELOVL6 | rs72679222 | (A/G) | intron variant. |
| ELOVL6 | rs11937052 | (A/G) | intron variant. |
| ELOVL6 | rs11098070 | (A/G) | intron variant. |
| ELOVL6 | rs80343897 | (T/C) | intron variant. |
| ELOVL6 | rs373773495 | (T/C) | intron variant. |
| ELOVL6 | rs114422025 | (T/C) | intron variant. |
| ELOVL6 | rs6533495 | (A/G) | intron variant. |
| ELOVL6 | rs28722886 | (T/C) | intron variant. |
| ELOVL6 | rs6533497 | (T/C) | intron variant. |
| ELOVL6 | rs77504516 | (A/G) | intron variant. |
| ELOVL6 | rs6815102 | (T/C) | intron variant. |
| ELOVL6 | rs4326075 | (A/C) | intron variant. |
| ELOVL6 | rs116418972 | (A/G) | intron variant. |
| ELOVL6 | rs11729740 | (T/C) | intron variant. |
| ELOVL6 | rs2035415 | (T/C) | intron variant. |
| ELOVL6 | rs17041402 | (A/C) | intron variant. |
| ELOVL6 | rs59111930 | (A/G) | intron variant. |
| ELOVL6 | rs74874270 | (A/G) | intron variant. |
| ELOVL6 | rs1384331 | (T/G) | intron variant. |
| ELOVL6 | rs72679246 | (A/C) | intron variant. |
| ELOVL6 | rs78563565 | (T/C) | intron variant. |
| ELOVL6 | rs6533498 | (A/C) | intron variant. |
| ELOVL6 | rs9997926 | (C/T) | intron variant. |
| ELOVL6 | rs6824447 | (A/G) | upstream transcription variant. |
| ELOVL6 | rs17041272 | (C/G) | 3′ UTR region variant. |
| ELOVL7 | rs75621404 | (A/G) | intron variant, genic downstream transcript variant. |
| ELOVL7 | rs143990657 | (A/G) | downstream gene transcription variant, intron variant. |
| ELOVL7 | rs115862620 | (T/C) | intron variant, genic downstream transcript variant. |
| ELOVL7 | rs1563517 | (T/G) | intron variant, genic downstream transcript variant. |
| ELOVL7 | rs12188996 | (A/C) | downstream gene transcription variant, intron variant. |
| ELOVL7 | rs60258111 | (T/C) | intron variant, genic downstream transcript variant. |
| ELOVL7 | rs16878426 | (T/C) | downstream gene transcription variant, intron variant. |
| ELOVL7 | rs6872863 | (A/G) | intron variant, genic downstream transcript variant. |
| ELOVL7 | rs76641655 | (T/G) | intron variant, genic downstream transcript variant, coding sequence variant, synonym variant. |
| ELOVL7 | rs145299240 | (T/C) | downstream gene transcription variant, intron variant, genic upstream transcript variant. |
| ELOVL7 | rs114011218 | (T/C) | upstream gene transcription variant, intron variant, genic downstream transcript variant. |
| ELOVL7 | rs115159664 | (T/C) | upstream gene transcription variant, intron variant, genic downstream transcript variant. |
| ELOVL7 | rs4700398 | (A/G) | upstream gene transcription variant, intron variant. |
| Biomarkers | Total (N = 599) | Women (N = 311) | Men (N = 288) | ||||
|---|---|---|---|---|---|---|---|
| Mean | S.D. | Mean | S.D. | Mean | S.D. | p-Value | |
| Age (years) | 19.18 | 1.95 | 19.07 | 1.80 | 19.29 | 2.09 | 0.162 |
| Weight (kg) | 65.07 | 13.70 | 59.88 | 12.06 | 70.65 | 13.18 | 0.000 |
| BMI (kg/m2) | 23.76 | 4.30 | 23.50 | 4.44 | 20.04 | 4.14 | 0.127 |
| Waist circumference (cm) | 80.98 | 11.83 | 78.24 | 11.62 | 83.92 | 11.36 | 0.000 |
| Waist–Hip Ratio | 0.83 | 0.07 | 0.81 | 0.07 | 0.86 | 0.06 | 0.000 |
| Waist–height Ratio | 0.49 | 0.07 | 0.49 | 0.07 | 0.49 | 0.07 | 0.970 |
| Body Fat (%) | 26.41 | 0.56 | 31.39 | 7.34 | 21.08 | 8.06 | 0.000 |
| Glucose (mg/dL) | 83.53 | 9.04 | 82.25 | 9.03 | 84.91 | 8.86 | 0.000 |
| Insulin (µg/mL) | 7.87 | 5.65 | 7.99 | 0.07 | 7.73 | 5.14 | 0.581 |
| HOMA-IR Index | 1.63 | 1.20 | 1.63 | 1.24 | 1.63 | 1.15 | 0.999 |
| Triglycerides (mg/dL) | 105.18 | 64.11 | 95.57 | 53.58 | 114.45 | 72.76 | 0.001 |
| Total Cholesterol (mg/dL) | 157.43 | 30.19 | 157.44 | 27.71 | 157.41 | 32.71 | 0.989 |
| HDL (mg/dL) | 50.72 | 12.55 | 53.25 | 13.33 | 48.02 | 11.07 | 0.000 |
| LDL (mg/dL) | 85.51 | 23.71 | 84.63 | 22.50 | 85.47 | 24.95 | 0.346 |
| Sex | Gene | Biomarker | Clinical Diagnosis | Locus | Allele | Frequency |
|---|---|---|---|---|---|---|
| Males | ELOVL2 | %BF | Low body fat | rs116279801 | A | 0.025 |
| Glucose | Normal levels | rs7765206 | A | 0.028 | ||
| HOMA | No insulin resistance | rs7765206 | A | 0.031 | ||
| ELOVL4 | WHR | Without cardiovascular risk | rs16891339 | G | 0.023 | |
| LDL | Without cardiovascular risk | rs117891930 | A | 0.020 | ||
| Without cardiovascular risk | rs80246554 | G | 0.037 | |||
| Without cardiovascular risk | rs12196014 | A | 0.095 | |||
| ELOVL5 | WHR | Without cardiovascular risk | rs36054518 | G | 0.021 | |
| TG | Normal levels | rs36054518 | G | 0.021 | ||
| ELOVL6 | Glucose | Normal levels | rs76145164 | A | 0.021 | |
| Normal levels | rs76338299 | A | 0.053 | |||
| HOMA | No insulin resistance | rs77958351 | A | 0.021 | ||
| Cholesterol | Normal levels | rs76145164 | A | 0.021 | ||
| Normal levels | rs74874270 | G | 0.031 | |||
| LDL | Without cardiovascular risk | rs76145164 | A | 0.021 | ||
| Without cardiovascular risk | rs74874270 | G | 0.030 | |||
| ELOVL7 | HOMA | No insulin resistance | rs115159664 | G | 0.024 | |
| TG | Normal levels | rs115159664 | G | 0.023 | ||
| Females | ELOVL2 | Cholesterol | Normal levels | rs7765206 | A | 0.031 |
| Normal levels | rs16870899 | G | 0.022 | |||
| LDL | Without cardiovascular risk | 587/ELOVL2 | A | 0.071 | ||
| ELOVL5 | WHR | Without cardiovascular risk | rs72938776 | A | 0.019 | |
| %BF | High levels | rs72938776 | A | 0.018 | ||
| High levels | rs72940713 | G | 0.019 | |||
| ELOVL6 | Glucose | Normal levels | rs76145164 | A | 0.020 | |
| Normal levels | rs74874270 | G | 0.029 | |||
| HOMA | No insulin resistance | rs17041402 | C | 0.023 | ||
| Cholesterol | Normal levels | rs17041402 | C | 0.022 | ||
| Normal levels | rs74874270 | G | 0.027 | |||
| Normal levels | rs78563565 | G | 0.024 | |||
| TG | Normal levels | rs17041402 | C | 0.025 | ||
| LDL | Without cardiovascular risk | rs11729740 | A | 0.062 | ||
| Without cardiovascular risk | rs17041402 | C | 0.022 | |||
| Without cardiovascular risk | rs74874270 | G | 0.027 | |||
| Without cardiovascular risk | rs78563565 | G | 0.023 | |||
| ELOVL7 | WHI | Normal distribution | rs76641655 | C | 0.022 |
| Gene | SNP | Clinical Marker | OR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| ELOVL2 | rs8523 | H-Insulin | 2.048 | 1.238 | 3.388 | 0.005 |
| H-HOMA | 1.847 | 1.132 | 3.013 | 0.013 | ||
| H-Cholesterol | 2.628 | 1.400 | 4.935 | 0.002 | ||
| rs3734398 | H-BMI | 1.441 | 1.020 | 2.036 | 0.038 | |
| H-Insulin | 1.894 | 1.146 | 3.130 | 0.012 | ||
| H-HOMA | 1.780 | 1.088 | 2.912 | 0.020 | ||
| H-Cholesterol | 2.341 | 1.251 | 4.378 | 0.006 | ||
| rs2236212 | H-Insulin | 1.922 | 1.135 | 3.254 | 0.014 | |
| H-HOMA | 1.995 | 1.184 | 3.361 | 0.009 | ||
| H-Cholesterol | 2.432 | 1.236 | 4.785 | 0.008 | ||
| H-LDL | 2.856 | 1.136 | 7.181 | 0.020 | ||
| rs3798713 | H-HOMA | 1.765 | 1.058 | 2.944 | 0.028 | |
| H-Cholesterol | 2.485 | 1.263 | 4.888 | 0.007 | ||
| H-LDL | 2.915 | 1.159 | 7.330 | 0.018 | ||
| rs4532436 | H-Insulin | 2.248 | 1.357 | 3.722 | 0.001 | |
| H-HOMA | 2.025 | 1.237 | 3.315 | 0.004 | ||
| H-Cholesterol | 2.654 | 1.418 | 4.966 | 0.002 | ||
| ELOVL4 | rs80246554 | H-HOMA | 2.622 | 1.164 | 5.908 | 0.016 |
| ELOVL5 | rs72938776 | H-BMI | 12.447 | 1.488 | 104.127 | 0.003 |
| H-Insulin | 6.973 | 1.145 | 42.469 | 0.015 | ||
| rs72940713 | H-BMI | 7.32 | 1.506 | 35.579 | 0.004 | |
| H-%BF | 8.561 | 1.064 | 68.899 | 0.016 | ||
| H-Insulin | 6.953 | 1.142 | 42.346 | 0.015 | ||
| ELOVL6 | rs59634436 | H-BMI | 2.129 | 1.256 | 3.608 | 0.004 |
| H-%BF | 1.879 | 1.091 | 3.238 | 0.021 | ||
| H-Insulin | 2.237 | 1.168 | 4.284 | 0.013 | ||
| H-Glucose | 3.597 | 1.225 | 10.561 | 0.013 | ||
| H-HOMA | 2.023 | 1.052 | 3.888 | 0.032 | ||
| rs78160528 | H-LDL | 4.642 | 1.258 | 17.12 | 0.012 | |
| rs10033691 | H-BMI | 1.844 | 1.14 | 2.983 | 0.012 | |
| H-%BF | 2.27 | 1.371 | 3.759 | 0.001 | ||
| H-Insulin | 2.094 | 1.132 | 3.874 | 0.017 | ||
| H-Glucose | 3.687 | 1.325 | 10.265 | 0.008 | ||
| rs76145164 | H-Triglycerides | 3.073 | 1.264 | 7.471 | 0.009 | |
| rs11937052 | H-BMI | 1.797 | 1.113 | 2.901 | 0.015 | |
| H-Waist | 1.776 | 1.104 | 2.858 | 0.017 | ||
| rs114422025 | H-HOMA | 4.678 | 1.445 | 15.141 | 0.005 | |
| rs72679246 | H-Glucose | 6.144 | 1.263 | 29.902 | 0.011 | |
| rs9997926 | H-WHR | 1.975 | 1.098 | 3.553 | 0.021 | |
| rs17041272 | H-BMI | 1.569 | 1 | 2.46 | 0.049 | |
| H-%BF | 1.794 | 1.133 | 2.839 | 0.012 | ||
| H-Insulin | 2.147 | 1.198 | 3.847 | 0.009 | ||
| H-HOMA | 1.956 | 1.096 | 3.491 | 0.021 | ||
| ELOVL7 | rs1563517 | H- Waist | 1.517 | 1.049 | 2.194 | 0.027 |
| H-Insulin | 1.844 | 1.107 | 3.074 | 0.018 | ||
| rs115159664 | H-LDL | 3.847 | 1.06 | 13.959 | 0.028 | |
| rs4700398 | H-BMI | 1.57 | 1.109 | 2.221 | 0.011 | |
| H-WHI | 1.438 | 1.036 | 1.996 | 0.03 | ||
| H-%BF | 1.473 | 1.058 | 2.051 | 0.022 | ||
| Gene | SNP | Clinical Marker | OR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| ELOVL2 | rs17606561 | H-HOMA | 0.521 | 0.298 | 0.914 | 0.021 |
| H-Triglycerides | 0.625 | 0.404 | 0.968 | 0.034 | ||
| rs2281591 | H-Waist | 0.641 | 0.445 | 0.924 | 0.017 | |
| H-Insulin | 0.517 | 0.297 | 0.899 | 0.018 | ||
| H-HOMA | 0.479 | 0.276 | 0.831 | 0.008 | ||
| rs9393903 | H-HOMA | 0.560 | 0.322 | 0.972 | 0.038 | |
| ELOVL3 | rs10748816 | H-WHR | 0.578 | 0.401 | 0.832 | 0.003 |
| ELOVL5 | rs2073040 | L-HDL | 0.686 | 0.475 | 0.992 | 0.045 |
| ELOVL6 | rs11098065 | H-Insulin | 0.499 | 0.289 | 0.861 | 0.011 |
| L-HDL | 0.686 | 0.487 | 0.966 | 0.031 | ||
| rs7662161 | H-LDL | 0.331 | 0.123 | 0.89 | 0.022 | |
| rs6533491 | H-Waist | 0.676 | 0.476 | 0.958 | 0.028 | |
| H-WHR | 0.704 | 0.508 | 0.977 | 0.035 | ||
| rs80343897 | H-Glucose | 0.202 | 0.045 | 0.895 | 0.02 | |
| L-HDL | 0.654 | 0.462 | 0.924 | 0.016 | ||
| rs6815102 | H-Insulin | 0.575 | 0.349 | 0.949 | 0.029 | |
| rs4326075 | L-HDL | 0.552 | 0.312 | 0.977 | 0.042 | |
| rs6824447 | H-Cholesterol | 0.447 | 0.242 | 0.825 | 0.008 | |
| H-LDL | 0.435 | 0.196 | 0.968 | 0.036 | ||
| Gene | SNP | Clinical Marker | OR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| ELOVL5 | rs72938776 | H-LDL | 11.037 | 1.044 | 116.696 | 0.013 |
| rs9370194 | H-Cholesterol | 2.926 | 1.085 | 7.892 | 0.027 | |
| ELOVL6 | rs59634436 | H-BMI | 2.761 | 1.314 | 5.802 | 0.006 |
| rs10033691 | H-BMI | 2.102 | 1.076 | 4.106 | 0.027 | |
| H-LDL | 4.479 | 1.21 | 16.584 | 0.015 | ||
| rs78160528 | H-LDL | 3.605 | 0.412 | 31.569 | 0.218 | |
| rs2005701 | H-BMI | 1.717 | 1.004 | 2.937 | 0.047 | |
| H-Waist | 2.163 | 1.287 | 3.634 | 0.003 | ||
| rs76145164 | H-Triglycerides | 5.667 | 1.576 | 20.37 | 0.003 | |
| rs11937052 | H-BMI | 2 | 1.03 | 3.884 | 0.038 | |
| rs72679246 | H-Glucose | 19.2 | 3.085 | 119.485 | 0.001 | |
| rs17041272 | H-Cholesterol | 3.1 | 1.101 | 8.73 | 0.025 | |
| ELOVL7 | rs1563517 | H-Waist | 1.716 | 1.051 | 2.801 | 0.03 |
| H-WHR | 1.806 | 1.075 | 3.033 | 0.025 | ||
| H-Insulin | 3.126 | 1.441 | 6.778 | 0.003 | ||
| rs12188996 | H-LDL | 10.963 | 1.037 | 115.915 | 0.013 | |
| rs76641655 | H-Triglycerides | 4.452 | 1.152 | 17.204 | 0.019 | |
| rs115159664 | H-Cholesterol | 7.125 | 1.714 | 29.621 | 0.002 | |
| H-LDL | 8.111 | 1.503 | 43.76 | 0.004 | ||
| rs4700398 | H-BMI | 1.964 | 1.196 | 3.225 | 0.007 | |
| Gene | SNP | Clinical Marker | OR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| ELOVL2 | rs2281591 | H-Waist | 0.610 | 0.377 | 0.988 | 0.044 |
| ELOVL6 | rs6533491 | H-Waist | 0.526 | 0.329 | 0.841 | 0.007 |
| H-WHR | 0.559 | 0.338 | 0.925 | 0.023 | ||
| rs11098065 | L-HDL | 0.582 | 0.369 | 0.919 | 0.02 | |
| Gene | SNP | Clinical Marker | OR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| ELOVL2 | rs8523 | H-Insulin | 2.305 | 1.172 | 4.532 | 0.014 |
| H-HOMA | 2.099 | 1.094 | 4.027 | 0.024 | ||
| H-Cholesterol | 3.154 | 1.408 | 7.061 | 0.004 | ||
| rs3734398 | H-Insulin | 2.222 | 1.133 | 4.360 | 0.019 | |
| H-HOMA | 2.080 | 1.079 | 4.009 | 0.027 | ||
| H-Cholesterol | 3.010 | 1.320 | 6.864 | 0.006 | ||
| rs2236212 | H-Insulin | 2.198 | 1.088 | 4.441 | 0.026 | |
| H-HOMA | 2.415 | 1.200 | 4.859 | 0.012 | ||
| H-Cholesterol | 2.842 | 1.173 | 6.886 | 0.016 | ||
| rs3798713 | H-Cholesterol | 2.885 | 1.191 | 6.990 | 0.015 | |
| rs4532436 | H-Insulin | 2.924 | 1.476 | 5.792 | 0.002 | |
| H-HOMA | 2.710 | 1.395 | 5.267 | 0.003 | ||
| H-Cholesterol | 2.935 | 1.311 | 6.568 | 0.007 | ||
| ELOVL4 | rs12196014 | H-Triglycerides | 1.978 | 1.011 | 3.871 | 0.044 |
| ELOVL5 | rs41273878 | H-Triglycerides | 4.176 | 0.912 | 19.127 | 0.047 |
| rs114271869 | L-HDL | 5.274 | 1.228 | 22.651 | 0.013 | |
| ELOVL6 | rs114422025 | H-HOMA | 4.929 | 0.961 | 25.264 | 0.036 |
| rs59634436 | H-%BF | 2.333 | 1.027 | 5.299 | 0.039 | |
| H-Insulin | 2.87 | 1.212 | 6.797 | 0.013 | ||
| H-Glucose | 7.407 | 1.876 | 29.251 | 0.001 | ||
| H-HOMA | 3.635 | 1.567 | 8.433 | 0.002 | ||
| rs78160528 | L-HDL | 5.299 | 1.234 | 22.757 | 0.013 | |
| H-LDL | 5.889 | 1.095 | 31.683 | 0.02 | ||
| rs10033691 | H-%BF | 3.299 | 1.493 | 7.288 | 0.002 | |
| H-Insulin | 2.824 | 1.232 | 6.469 | 0.011 | ||
| H-Glucose | 5.765 | 1.475 | 22.524 | 0.005 | ||
| H-HOMA | 2.957 | 1.342 | 6.514 | 0.005 | ||
| rs11937052 | H-Waist | 2.441 | 1.206 | 4.94 | 0.011 | |
| H-%BF | 2.101 | 1.009 | 4.375 | 0.044 | ||
| H-Glucose | 5.577 | 1.429 | 21.766 | 0.006 | ||
| L-HDL | 2.105 | 1.035 | 4.28 | 0.037 | ||
| rs9997926 | H-Insulin | 5.714 | 2.035 | 16.048 | 0.001 | |
| H-BMI | 2.068 | 1.118 | 3.826 | 0.019 | ||
| H-Waist | 2.131 | 1.116 | 4.069 | 0.02 | ||
| rs17041272 | H-%BF | 1.933 | 1.016 | 3.68 | 0.043 | |
| Gene | SNP | Clinical marker | OR | 95% CI | p-Value | |
|---|---|---|---|---|---|---|
| ELOVL2 | rs17606561 | H-Insulin | 0.291 | 0.122 | 0.693 | 0.004 |
| H-HOMA | 0.204 | 0.077 | 0.539 | 0.001 | ||
| rs2281591 | H-Insulin | 0.240 | 0.101 | 0.572 | 0.001 | |
| H-HOMA | 0.204 | 0.083 | 0.503 | 0.001 | ||
| rs9295757 | H-HOMA | 0.403 | 0.184 | 0.881 | 0.020 | |
| rs3798721 | H-HOMA | 0.381 | 0.174 | 0.833 | 0.013 | |
| rs3798722 | H-Insulin | 0.422 | 0.199 | 0.894 | 0.022 | |
| H-HOMA | 0.357 | 0.168 | 0.759 | 0.006 | ||
| rs9393903 | H-Insulin | 0.299 | 0.125 | 0.713 | 0.004 | |
| H-HOMA | 0.204 | 0.077 | 0.539 | 0.001 | ||
| ELOVL3 | rs10748816 | H-WHR | 0.508 | 0.298 | 0.866 | 0.012 |
| ELOVL5 | rs9370194 | H-%BF | 0.537 | 0.319 | 0.907 | 0.019 |
| ELOVL6 | rs11098065 | H-Insulin | 0.436 | 0.21 | 0.906 | 0.024 |
| rs7662161 | H-Cholesterol | 0.34 | 0.134 | 0.863 | 0.018 | |
| H-LDL | 0.177 | 0.04 | 0.789 | 0.011 | ||
| rs6815102 | H-Insulin | 0.478 | 0.244 | 0.939 | 0.03 | |
| rs4326075 | H-LDL | 0.552 | 0.312 | 0.977 | 0.04 | |
| rs59111930 | H-Triglycerides | 0.5 | 0.291 | 0.859 | 0.011 | |
| L-HDL | 0.582 | 0.34 | 0.995 | 0.047 | ||
| rs6824447 | H-BFI | 0.516 | 0.313 | 0.85 | 0.009 | |
| H-Waist | 0.551 | 0.319 | 0.952 | 0.031 | ||
| H-Cholesterol | 0.268 | 0.114 | 0.628 | 0.001 | ||
| H-LDL | 0.227 | 0.072 | 0.714 | 0.006 | ||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Maycotte-Cervantes, M.L.; Aguilar-Galarza, A.; Anaya-Loyola, M.A.; Anzures-Cortes, M.d.L.; Haddad-Talancón, L.; Méndez-Rangel, A.S.; García-Gasca, T.; Rodríguez-García, V.M.; Moreno-Celis, U. Influence of Single Nucleotide Polymorphisms of ELOVL on Biomarkers of Metabolic Alterations in the Mexican Population. Nutrients 2020, 12, 3389. https://doi.org/10.3390/nu12113389
Maycotte-Cervantes ML, Aguilar-Galarza A, Anaya-Loyola MA, Anzures-Cortes MdL, Haddad-Talancón L, Méndez-Rangel AS, García-Gasca T, Rodríguez-García VM, Moreno-Celis U. Influence of Single Nucleotide Polymorphisms of ELOVL on Biomarkers of Metabolic Alterations in the Mexican Population. Nutrients. 2020; 12(11):3389. https://doi.org/10.3390/nu12113389
Chicago/Turabian StyleMaycotte-Cervantes, María Luisa, Adriana Aguilar-Galarza, Miriam Aracely Anaya-Loyola, Ma. de Lourdes Anzures-Cortes, Lorenza Haddad-Talancón, Akram Sharim Méndez-Rangel, Teresa García-Gasca, Víctor Manuel Rodríguez-García, and Ulisses Moreno-Celis. 2020. "Influence of Single Nucleotide Polymorphisms of ELOVL on Biomarkers of Metabolic Alterations in the Mexican Population" Nutrients 12, no. 11: 3389. https://doi.org/10.3390/nu12113389
APA StyleMaycotte-Cervantes, M. L., Aguilar-Galarza, A., Anaya-Loyola, M. A., Anzures-Cortes, M. d. L., Haddad-Talancón, L., Méndez-Rangel, A. S., García-Gasca, T., Rodríguez-García, V. M., & Moreno-Celis, U. (2020). Influence of Single Nucleotide Polymorphisms of ELOVL on Biomarkers of Metabolic Alterations in the Mexican Population. Nutrients, 12(11), 3389. https://doi.org/10.3390/nu12113389